Transcriptome Analysis Unravels CD4+ T-Cell and Treg-Cell Differentiation in Ovarian Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Handling
2.2. Clustering of scRNA-Seq Data
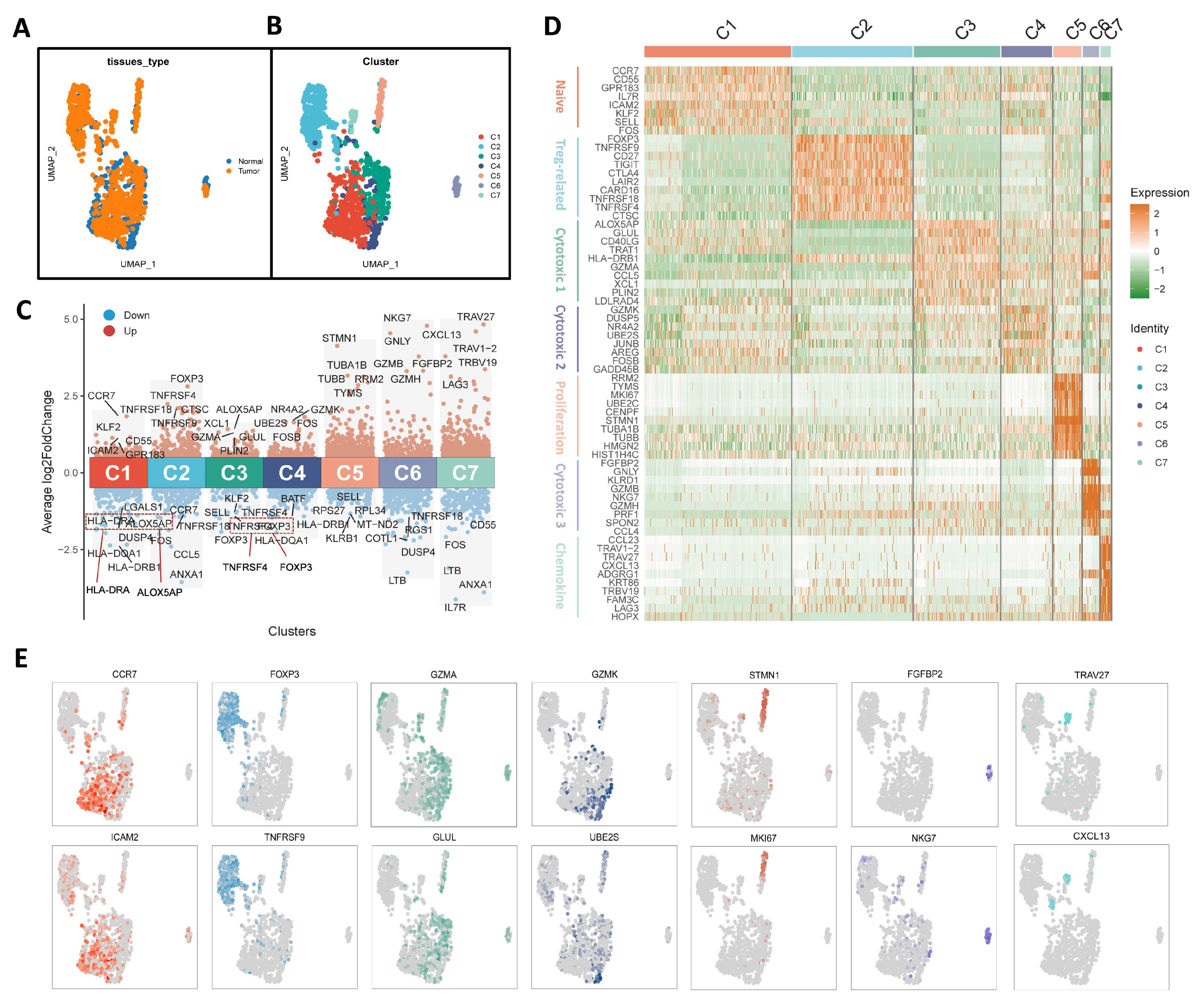
2.3. Cell-Type Identification Analysis of Tumor-Infiltrating CD4+ T Cells
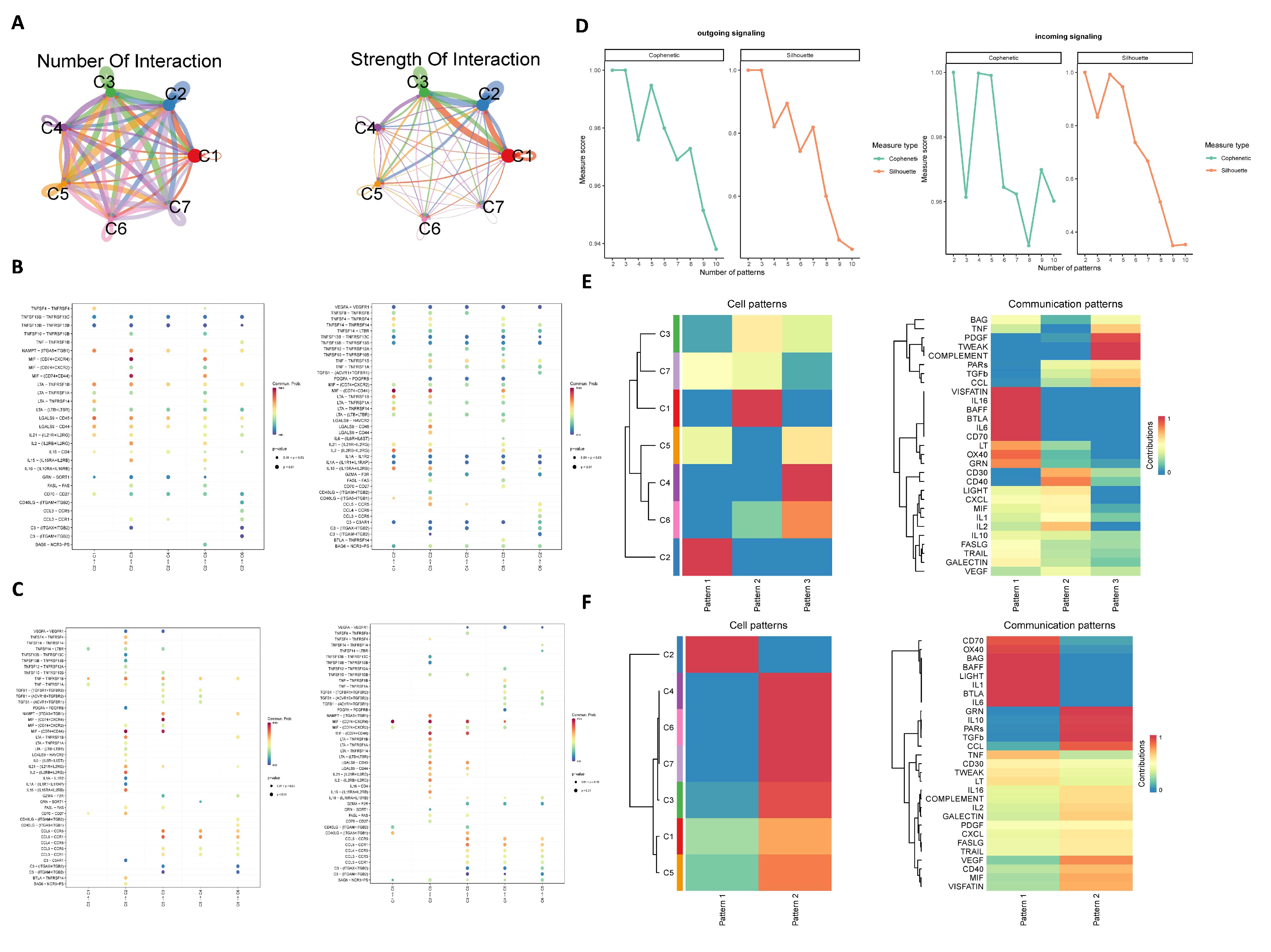
2.4. Analysis of Cell Communication
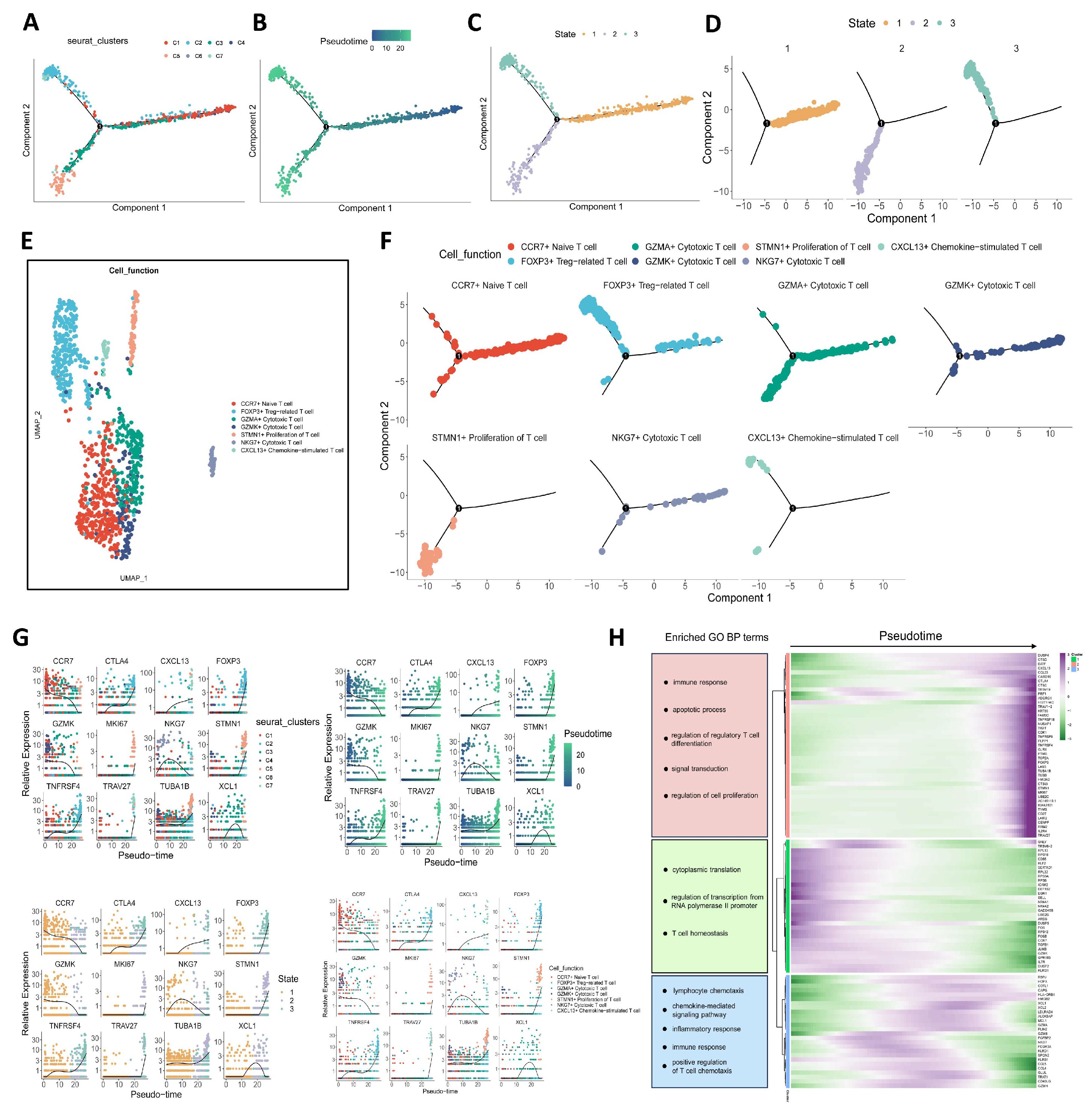
2.5. Trajectory Analysis
2.6. Gene Set Enrichment in Treg Cluster of OCSCDs
2.7. Assessment of Treg Cells in OCSCDs
2.8. Differential Expression and Functional Enrichment Analysis in Low Treg Group and High Treg Group
2.9. Mutated Gene and Immune Subtype Analysis in Low Treg Group and High Treg Group
2.10. Immune Signature Analysis and Prediction of Treatment Sensitivity
2.11. Survival Analysis Related to Low Treg Group and High Treg Group
2.12. Cell Culture and Sample Extraction
2.13. RNA Extraction and RT-qPCR
2.14. CCK-8 and Transwell Assay
3. Results
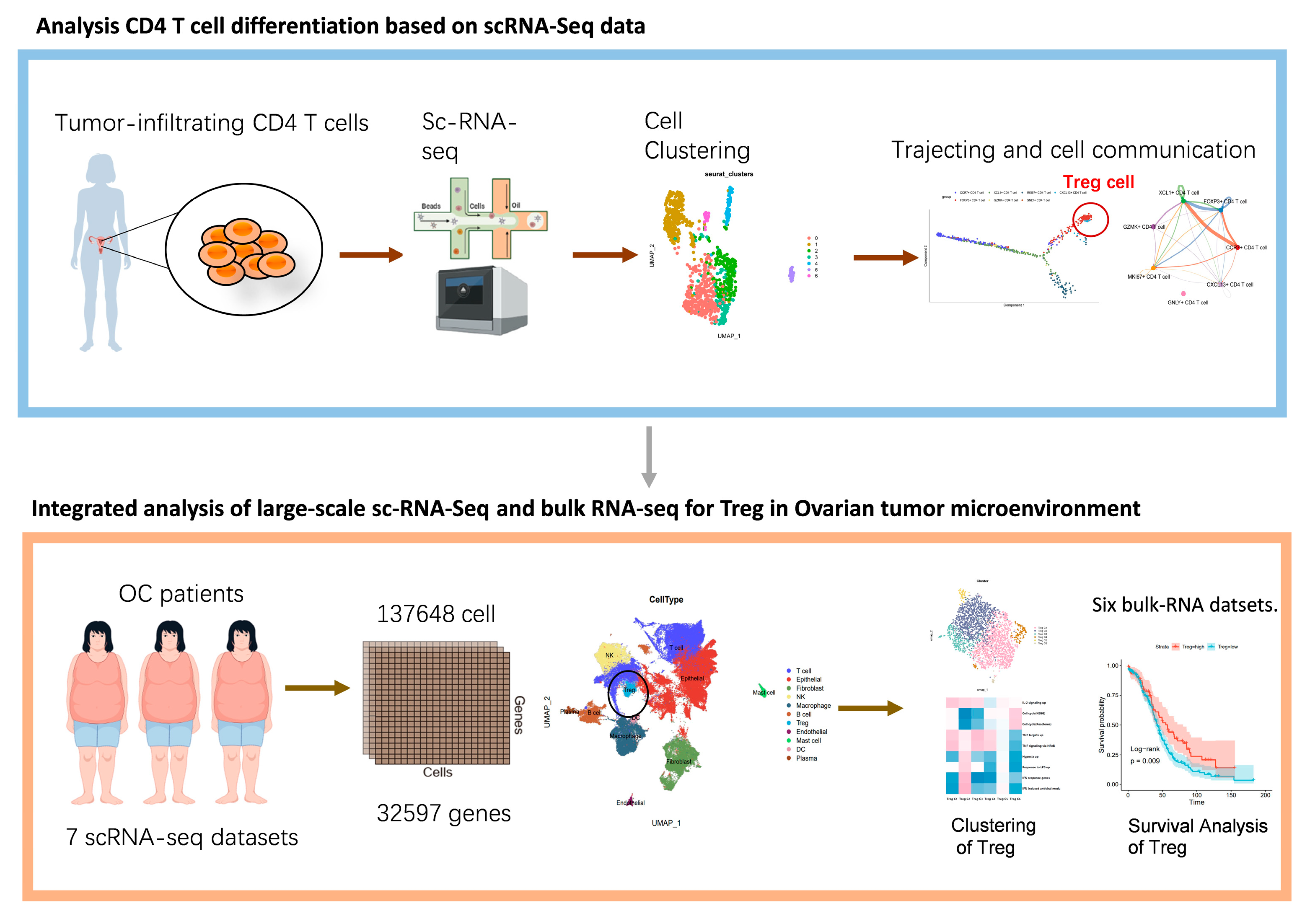
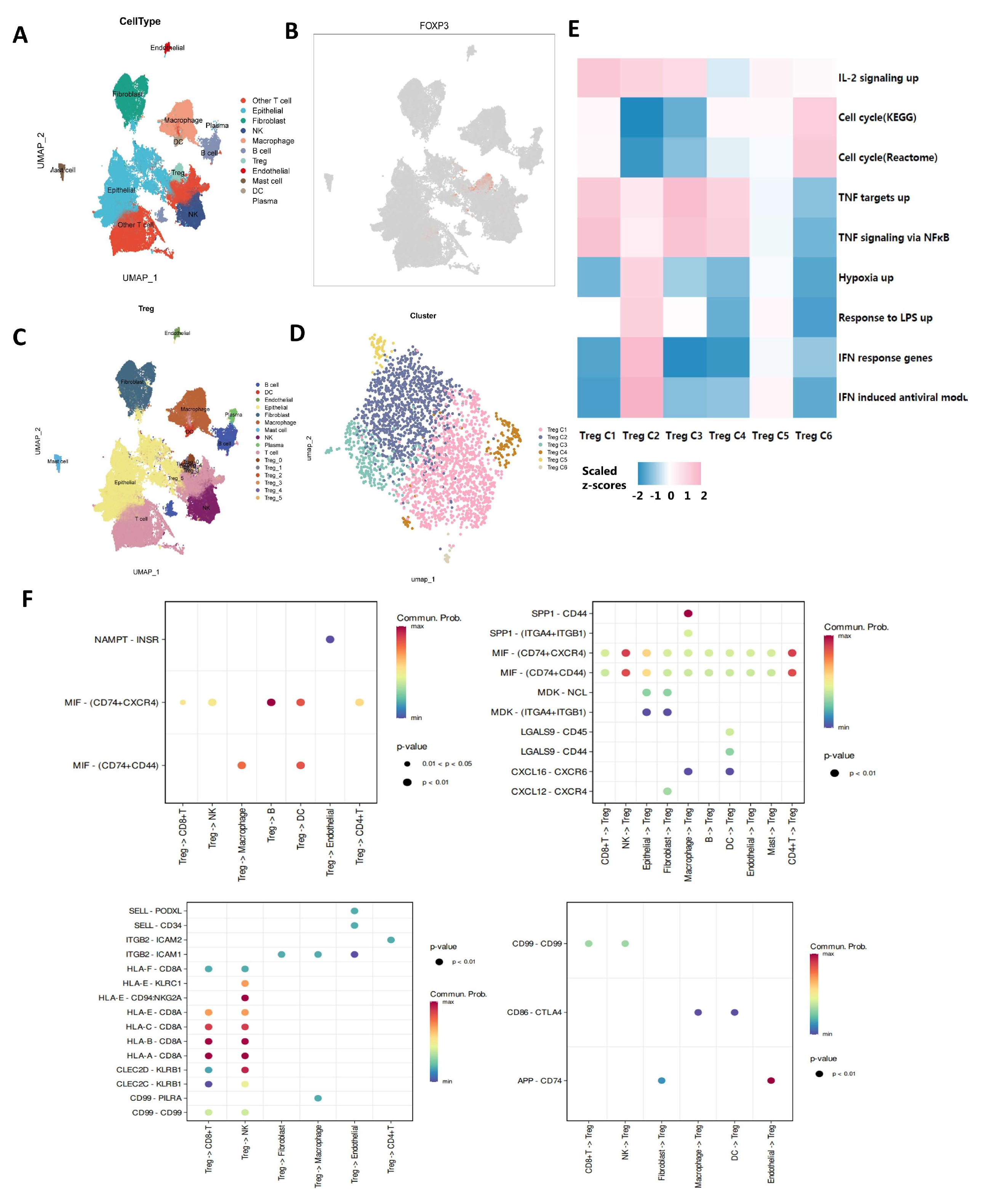
3.1. Characterization of Tumor-Infiltrating CD4+ T Cells in OC
3.2. Trajectory of Tumor-Infiltrating CD4+ T Cells
3.3. Cell–Cell Communication Analysis
3.4. Characterization and Cell–Cell Communication of Treg Cells in OCSCDs
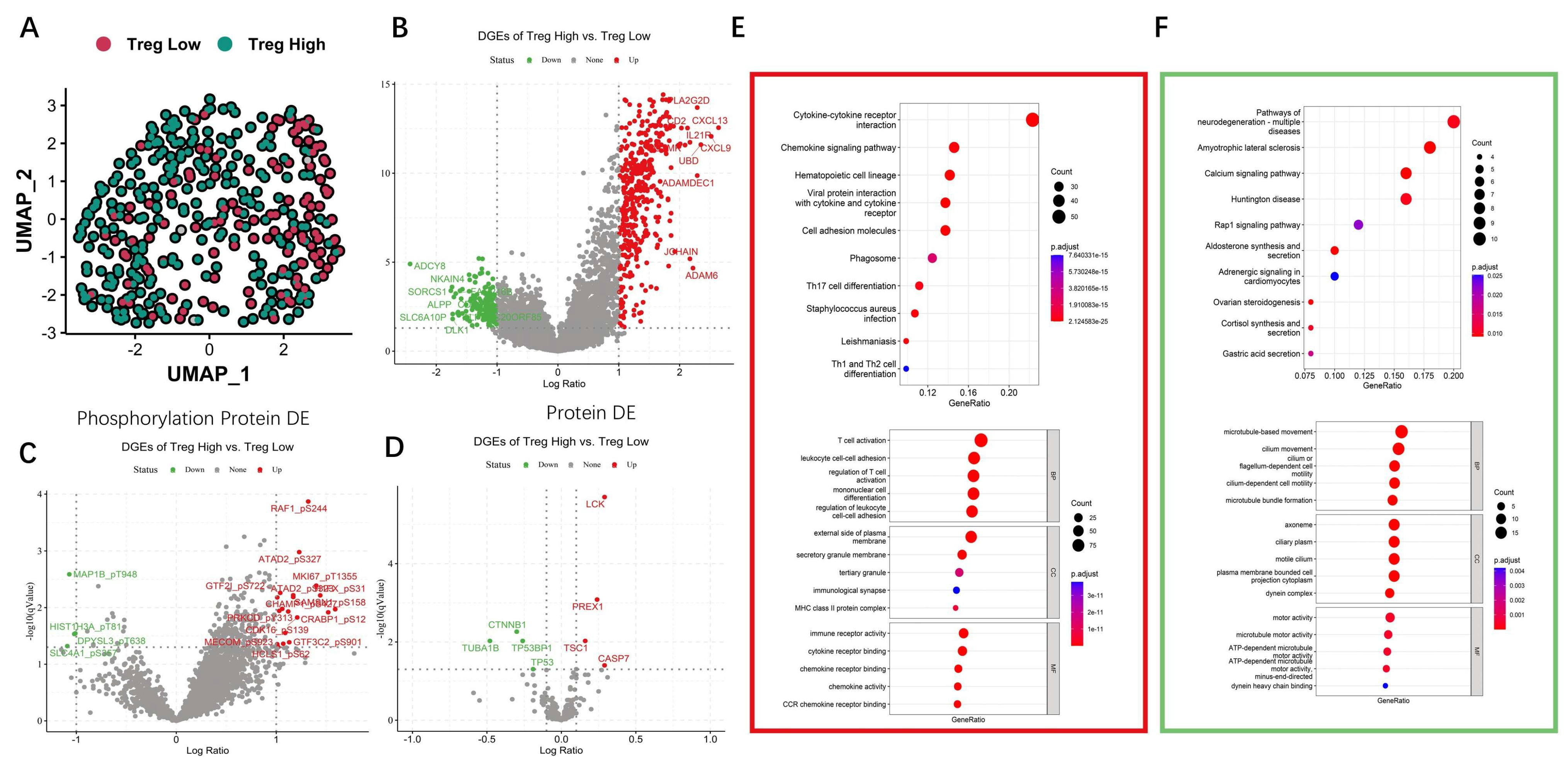
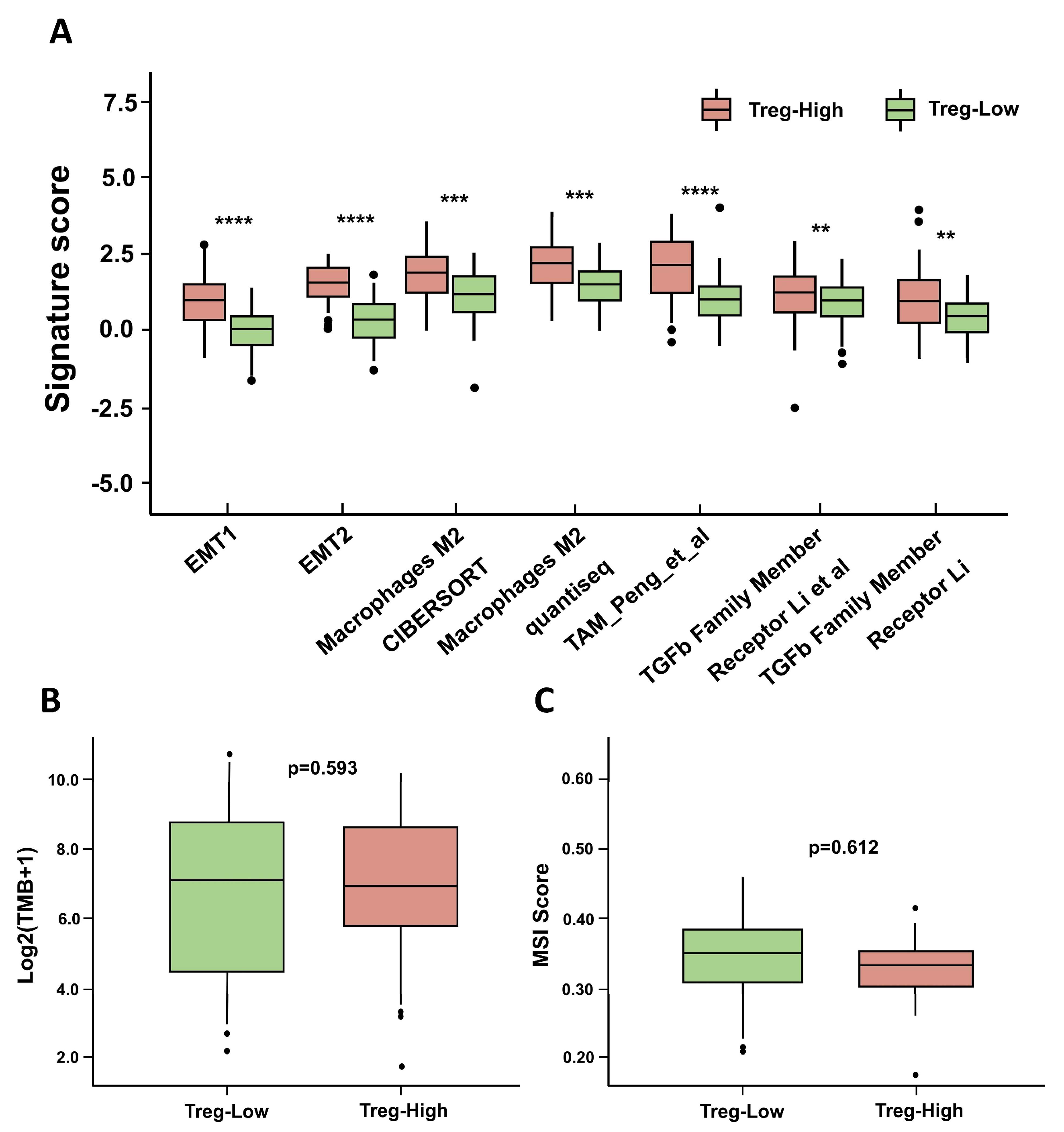
3.5. Differential and Enrichment Analysis Related to Treg Cells
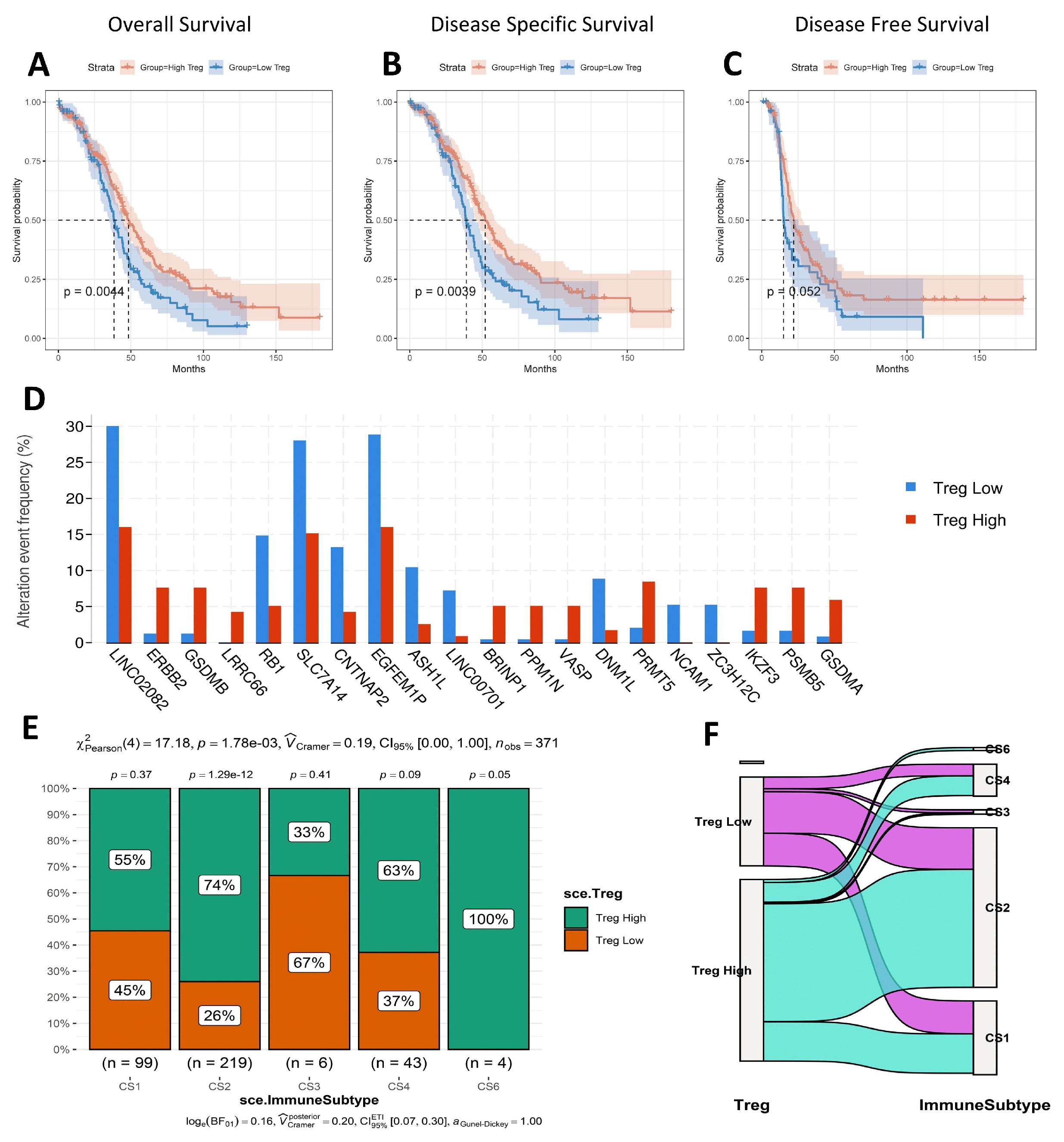
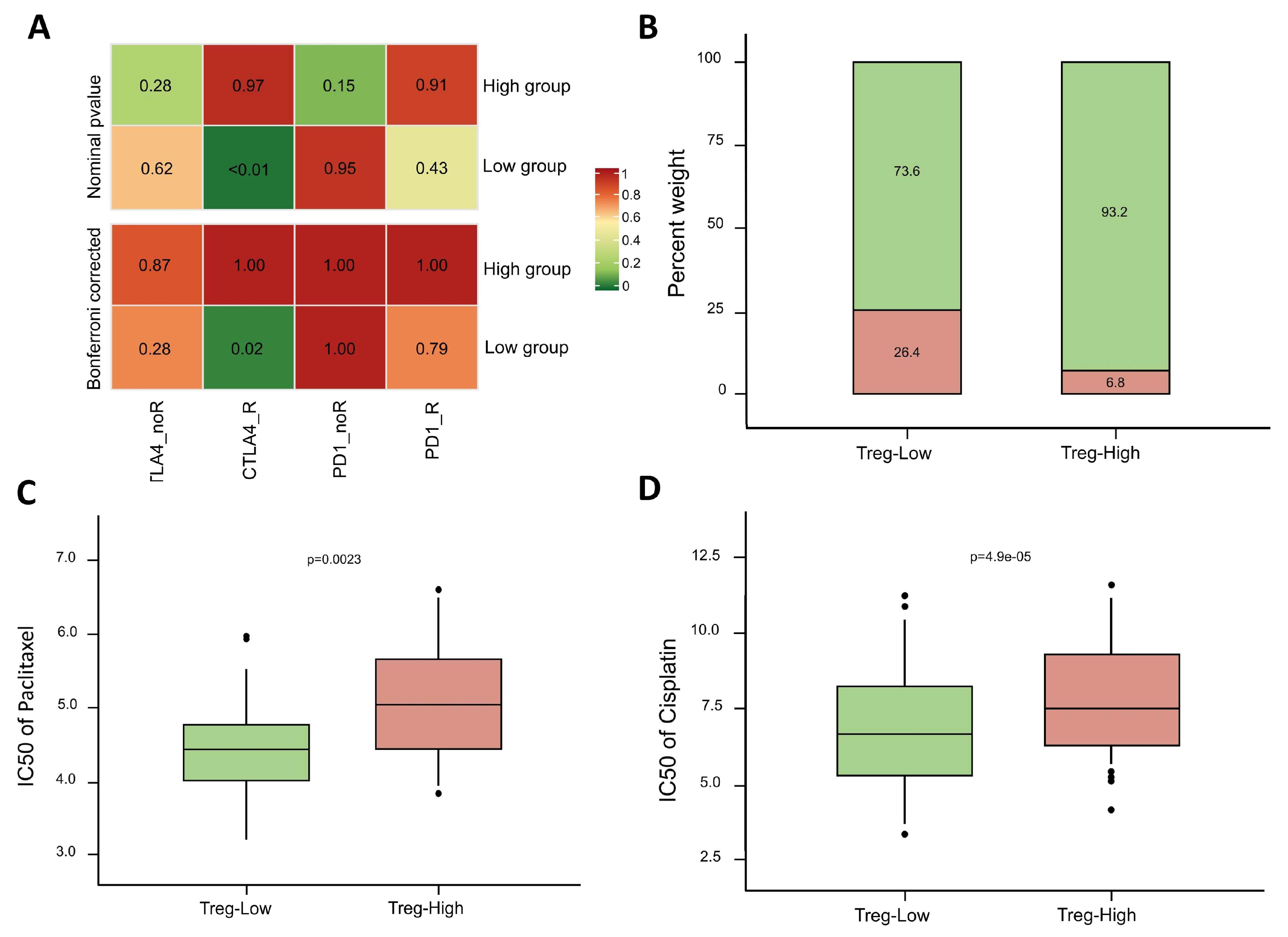
3.6. Survival Analysis, Mutated Gene, and Immune Subtype Analysis
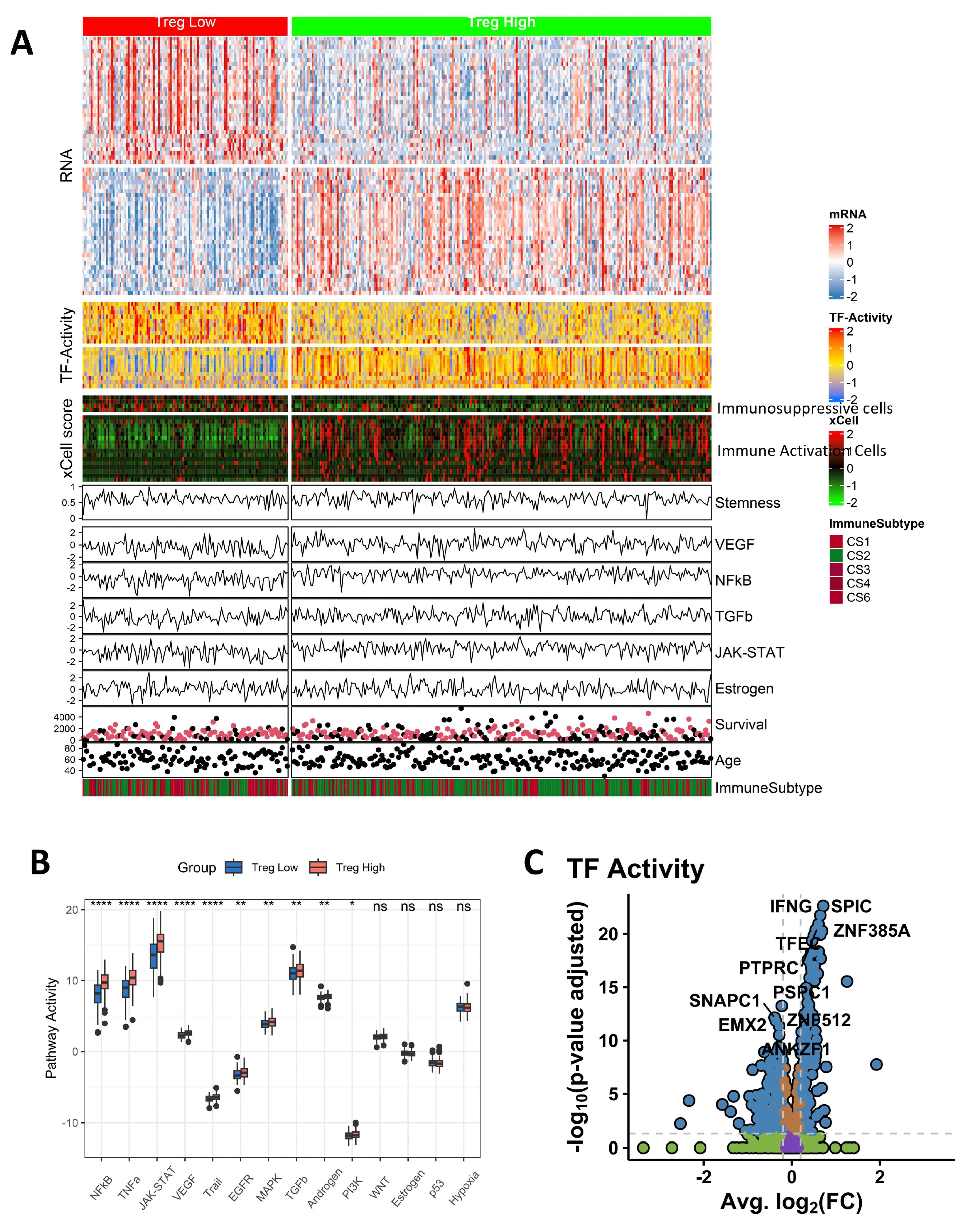
3.7. Pathway Activity and TF Activity Analysis
3.8. The Immune Landscape of Different Groups
3.9. Immunotherapy and Drug Sensitivity Prediction
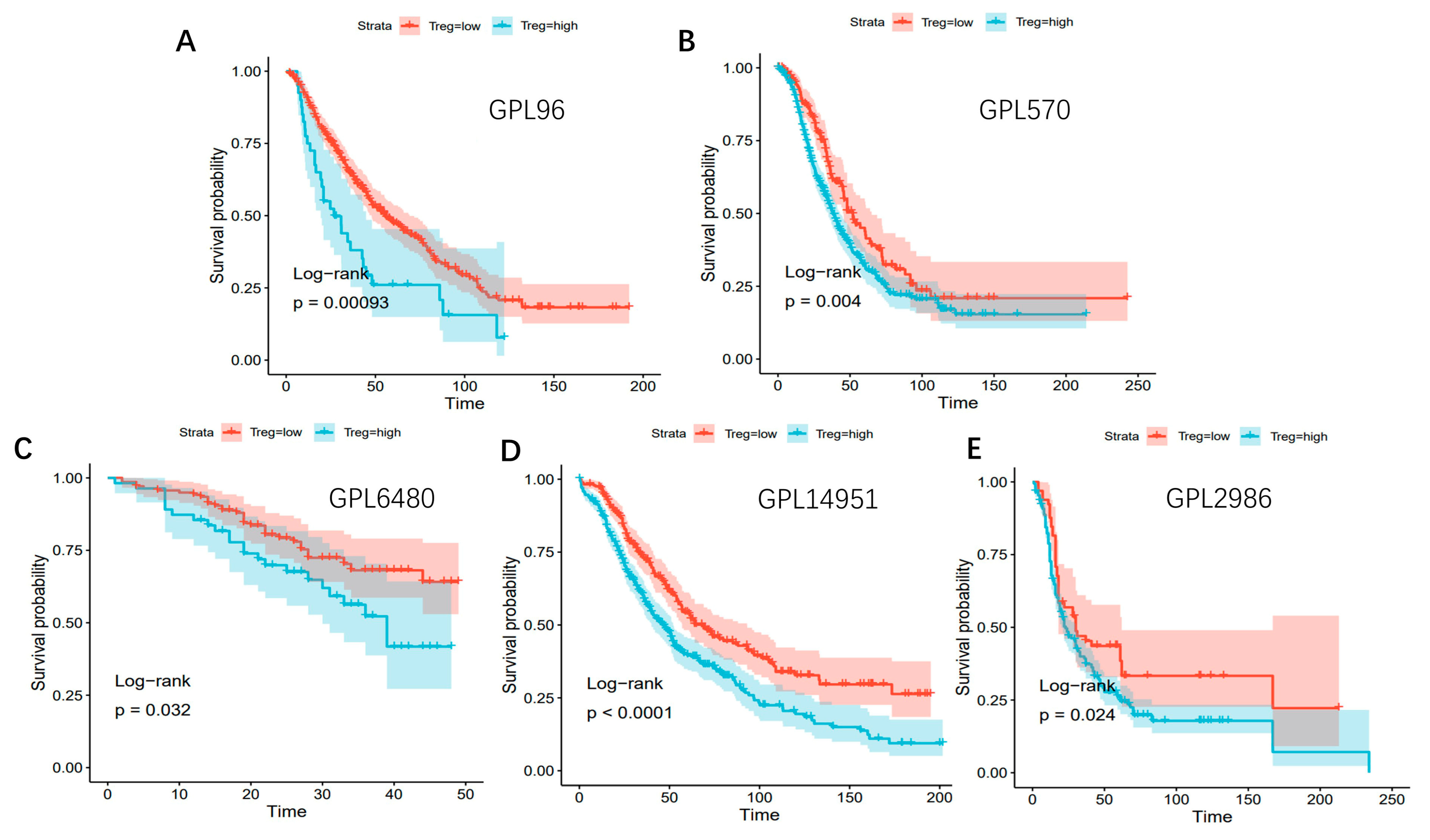
3.10. Survival Analysis Related to Treg Cells Based on Different Bulk RNA-Seq Datasets
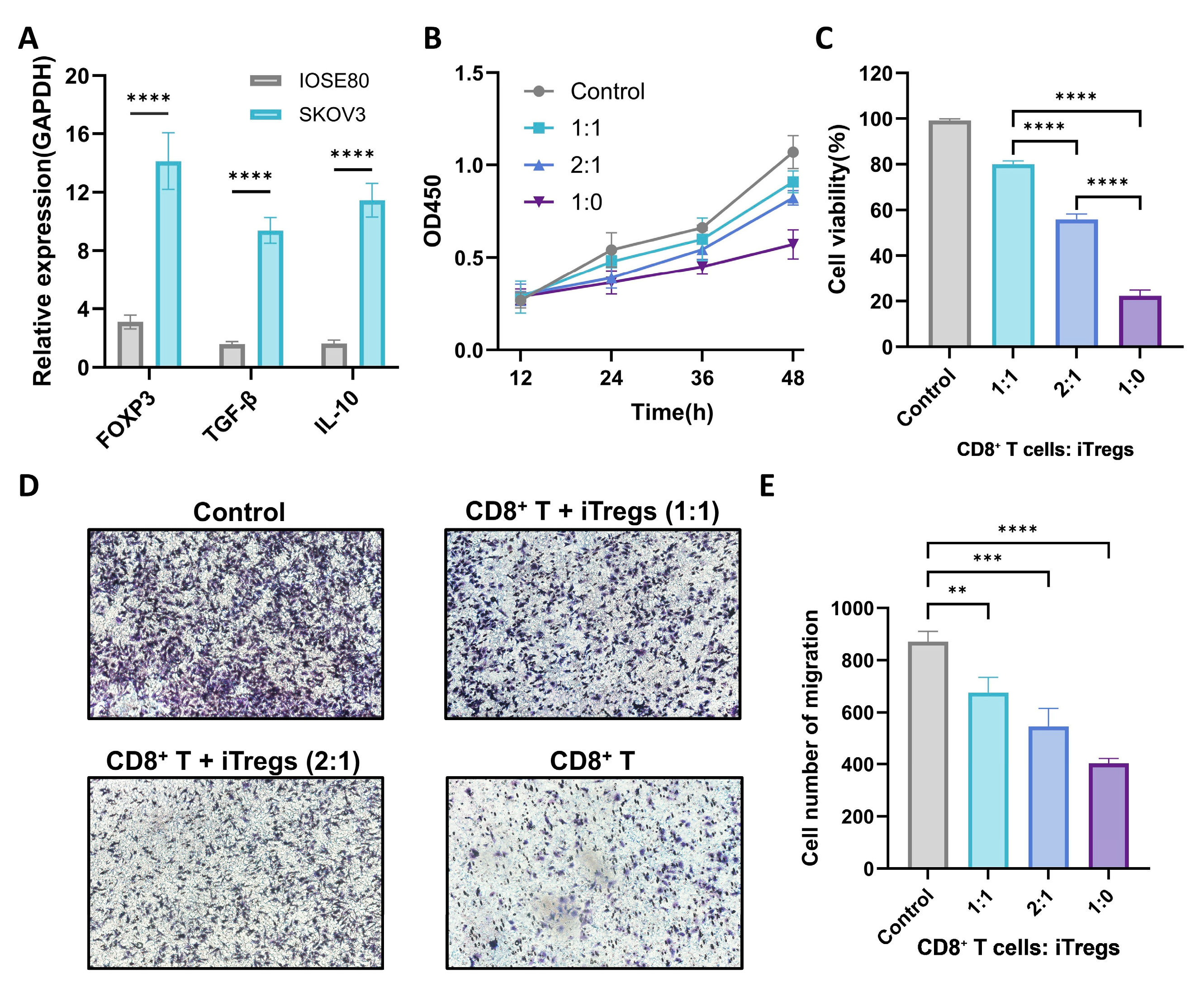
3.11. Experimental Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sideris, M.; Menon, U.; Manchanda, R. Screening and prevention of ovarian cancer. Med. J. Aust. 2024, 220, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Webb, P.M.; Jordan, S.J. Epidemiology of epithelial ovarian cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 41, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Cortez, A.J.; Tudrej, P.; Kujawa, K.A.; Lisowska, K.M. Advances in ovarian cancer therapy. Cancer Chemother. Pharmacol. 2017, 81, 17–38. [Google Scholar] [CrossRef] [PubMed]
- Jessmon, P.; Boulanger, T.; Zhou, W.; Patwardhan, P. Epidemiology and treatment patterns of epithelial ovarian cancer. Expert Rev. Anticancer Ther. 2017, 17, 427–437. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, C.; Zhou, S. Targeting tumor microenvironment in ovarian cancer: Premise and promise. Biochim. Biophys. Acta (BBA) Rev. Cancer 2020, 1873, 188361. [Google Scholar] [CrossRef]
- Hu, X.; Bian, C.; Zhao, X.; Yi, T. Efficacy evaluation of multi-immunotherapy in ovarian cancer: From bench to bed. Front. Immunol. 2022, 13, 1034903. [Google Scholar] [CrossRef]
- Rodriguez, G.; Galpin, K.; McCloskey, C.; Vanderhyden, B. The Tumor Microenvironment of Epithelial Ovarian Cancer and Its Influence on Response to Immunotherapy. Cancers 2018, 10, 242. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, Y.; Yang, J.; Zhao, X.; Wei, X. Tumor Microenvironment in Ovarian Cancer: Function and Therapeutic Strategy. Front. Cell Dev. Biol. 2020, 8, 758. [Google Scholar] [CrossRef]
- Liu, Y.-T.; Sun, Z.-J. Turning cold tumors into hot tumors by improving T-cell infiltration. Theranostics 2021, 11, 5365–5386. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef]
- Mirza, M.R.; Coleman, R.L.; González-Martín, A.; Moore, K.N.; Colombo, N.; Ray-Coquard, I.; Pignata, S. The forefront of ovarian cancer therapy: Update on PARP inhibitors. Ann. Oncol. 2020, 31, 1148–1159. [Google Scholar] [CrossRef]
- Liu, D.; Schilling, B.; Liu, D.; Sucker, A.; Livingstone, E.; Jerby-Arnon, L.; Zimmer, L.; Gutzmer, R.; Satzger, I.; Loquai, C.; et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 2019, 25, 1916–1927. [Google Scholar] [CrossRef] [PubMed]
- Alspach, E.; Lussier, D.M.; Miceli, A.P.; Kizhvatov, I.; DuPage, M.; Luoma, A.M.; Meng, W.; Lichti, C.F.; Esaulova, E.; Vomund, A.N.; et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 2019, 574, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Marty Pyke, R.; Thompson, W.K.; Salem, R.M.; Font-Burgada, J.; Zanetti, M.; Carter, H. Evolutionary Pressure against MHC Class II Binding Cancer Mutations. Cell 2018, 175, 416–428.e413. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Yu, Q.; Zhou, Y.; Zheng, S.; Tao, J.; Jiang, Q.; Yuan, G. Functionally impaired follicular helper T cells induce regulatory B cells and CD14+ human leukocyte antigen-DR− cell differentiation in non-small cell lung cancer. Cancer Sci. 2018, 109, 3751–3761. [Google Scholar] [CrossRef]
- Kumar, P.; Bhattacharya, P.; Prabhakar, B.S. A comprehensive review on the role of co-signaling receptors and Treg homeostasis in autoimmunity and tumor immunity. J. Autoimmun. 2018, 95, 77–99. [Google Scholar] [CrossRef]
- Dal Molin, A.; Di Camillo, B. How to design a single-cell RNA-sequencing experiment: Pitfalls, challenges and perspectives. Brief. Bioinform. 2019, 20, 1384–1394. [Google Scholar] [CrossRef]
- Wen, L.; Tang, F. Recent advances in single-cell sequencing technologies. Precis. Clin. Med. 2022, 5, pbac002. [Google Scholar] [CrossRef]
- Suvà, M.L.; Tirosh, I. Single-Cell RNA Sequencing in Cancer: Lessons Learned and Emerging Challenges. Mol. Cell 2019, 75, 7–12. [Google Scholar] [CrossRef]
- Hao, Y.H.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.W.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef]
- Jin, S.Q.; Guerrero-Juarez, C.F.; Zhang, L.H.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.Q.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef]
- Wu, D.; Smyth, G.K. Camera: A competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012, 40, e133. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.Y.; Wang, Z.; Pe’er, D.; Danko, C.G. Cell type and gene expression deconvolution with BayesPrism enables Bayesian integrative analysis across bulk and single-cell RNA sequencing in oncology. Nat. Cancer 2022, 3, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.C.; Wang, L.G.; Han, Y.Y.; He, Q.Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics-A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Roux, M. A Comparative Study of Divisive and Agglomerative Hierarchical Clustering Algorithms. J. Classif. 2018, 35, 345–366. [Google Scholar] [CrossRef]
- Dong, Q.Z.; Ye, Z.L.; Shen, R.F.; Yu, G.C.; Wu, J.N.; Xiong, Y.; Zhou, R.; Qiu, W.J.; Huang, N.; Li, S.; et al. IOBR: Multi-Omics Immuno-Oncology Biological Research to Decode Tumor Microenvironment and Signatures. Front. Immunol. 2021, 12, 687975. [Google Scholar] [CrossRef]
- Hoshida, Y.; Brunet, J.-P.; Tamayo, P.; Golub, T.R.; Mesirov, J.P. Subclass mapping: Identifying common subtypes in independent disease data sets. PLoS ONE 2007, 2, e1195. [Google Scholar] [CrossRef]
- Sawant, D.V.; Yano, H.; Chikina, M.; Zhang, Q.X.; Liao, M.T.; Liu, C.; Callahan, D.J.; Sun, Z.; Sun, T.; Tabib, T.; et al. Adaptive plasticity of IL-10+ and IL-35+ Treg cells cooperatively promotes tumor T cell exhaustion. Nat. Immunol. 2019, 20, 724–735. [Google Scholar] [CrossRef]
- Huang, J.J.; Chan, W.C.; Ngai, C.H.; Lok, V.; Zhang, L.; Lucero-Prisno, D.E.; Xu, W.H.; Zheng, Z.J.; Elcarte, E.; Withers, M.; et al. Worldwide Burden, Risk Factors, and Temporal Trends of Ovarian Cancer: A Global Study. Cancers 2022, 14, 2230. [Google Scholar] [CrossRef]
- Guo, X.Y.; Zhang, Y.Y.; Zheng, L.T.; Zheng, C.H.; Song, J.T.; Zhang, Q.M.; Kang, B.X.; Liu, Z.Z.R.; Jin, L.; Xing, R.; et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 2018, 24, 978–985. [Google Scholar] [CrossRef]
- Vasanthakumar, A.; Liao, Y.; Teh, P.; Pascutti, M.F.; Oja, A.E.; Garnham, A.L.; Gloury, R.; Tempany, J.C.; Sidwell, T.; Cuadrado, E.; et al. The TNF Receptor Superfamily-NF-κB Axis Is Critical to Maintain Effector Regulatory T Cells in Lymphoid and Non-lymphoid Tissues. Cell Rep. 2017, 20, 2906–2920. [Google Scholar] [CrossRef]
- Overacre-Delgoffe, A.E.; Chikina, M.; Dadey, R.E.; Yano, H.; Brunazzi, E.A.; Shayan, G.; Horne, W.; Moskovitz, J.M.; Kolls, J.K.; Sander, C.; et al. Interferon-γ Drives Treg Fragility to Promote Anti-tumor Immunity. Cell 2017, 169, 1130–1141.e11. [Google Scholar] [CrossRef] [PubMed]
- Gaud, G.; Lesourne, R.; Love, P.E. Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 2018, 18, 485–497. [Google Scholar] [CrossRef]
- Lo, W.L.; Shah, N.H.; Ahsan, N.; Horkova, V.; Stepanek, O.; Salomon, A.R.; Kuriyan, J.; Weiss, A. Lck promotes Zap70-dependent LAT phosphorylation by bridging Zap70 to LAT. Nat. Immunol. 2018, 19, 733–741. [Google Scholar] [CrossRef]
- Qin, Z.; Hou, P.; Lin, H.Z.; Chen, M.H.; Wang, R.N.; Xu, T. Inhibition of Lck/Fyn kinase activity promotes the differentiation of induced Treg cells through AKT/mTOR pathway. Int. Immunopharmacol. 2024, 134, 112237. [Google Scholar] [CrossRef] [PubMed]
- Shevach, E.M.; Thornton, A.M. tTregs, pTregs, and iTregs: Similarities and differences. Immunol. Rev. 2014, 259, 88–102. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, B.; Sun, B.; Xiao, Z. Transcriptome Analysis Unravels CD4+ T-Cell and Treg-Cell Differentiation in Ovarian Cancer. Biomolecules 2025, 15, 1241. https://doi.org/10.3390/biom15091241
Shao B, Sun B, Xiao Z. Transcriptome Analysis Unravels CD4+ T-Cell and Treg-Cell Differentiation in Ovarian Cancer. Biomolecules. 2025; 15(9):1241. https://doi.org/10.3390/biom15091241
Chicago/Turabian StyleShao, Baoyi, Bo Sun, and Zhongdang Xiao. 2025. "Transcriptome Analysis Unravels CD4+ T-Cell and Treg-Cell Differentiation in Ovarian Cancer" Biomolecules 15, no. 9: 1241. https://doi.org/10.3390/biom15091241
APA StyleShao, B., Sun, B., & Xiao, Z. (2025). Transcriptome Analysis Unravels CD4+ T-Cell and Treg-Cell Differentiation in Ovarian Cancer. Biomolecules, 15(9), 1241. https://doi.org/10.3390/biom15091241




