Engineering Protein–Peptide Interfaces via Combinatorial Mutagenesis and Mass Photometric Screening
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Library Design
3.2. Library Synthesis
3.3. Analysis of the Native SpyCatcher–SpyTag Interaction by Mass Photometry
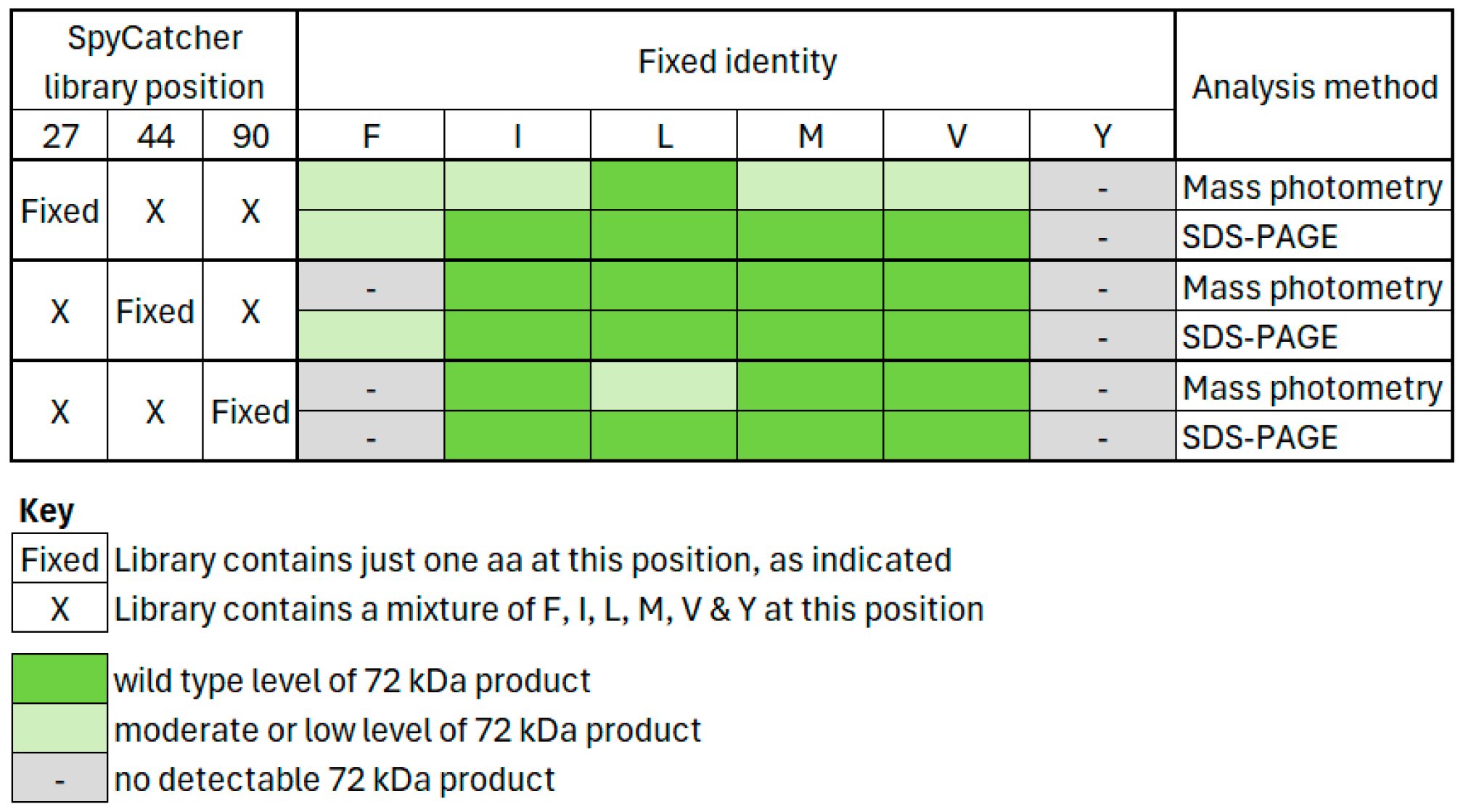
3.4. Many Hydrophobic Variants of SpyCatcher Can React with Native SpyTag
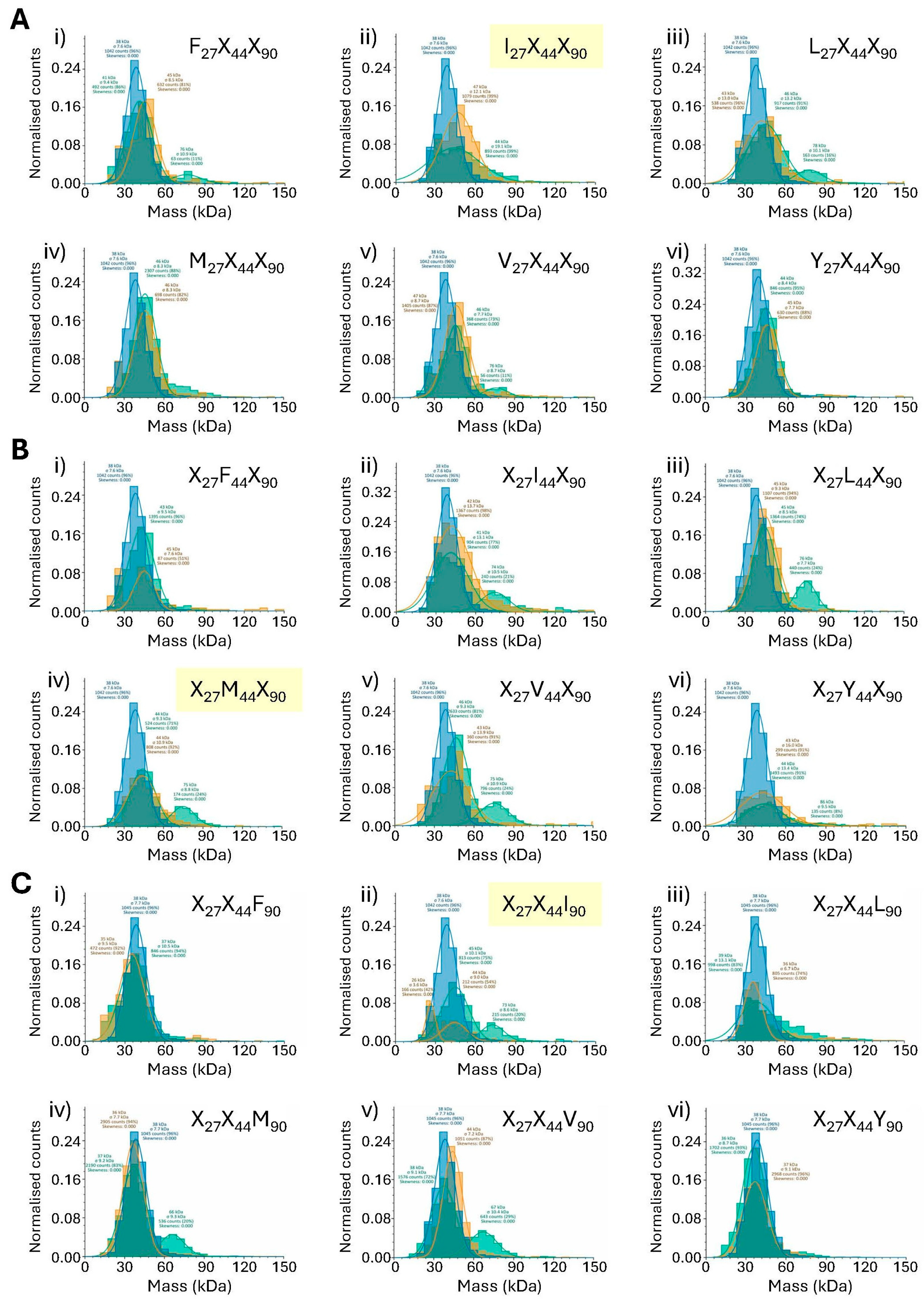
3.4.1. Analysis of SpyCatcher Position 27 (Native SpyCatcher = Isoleucine)
3.4.2. Analysis of SpyCatcher Position 44 (Native SpyCatcher = Methionine)
3.4.3. Analysis of SpyCatcher Position 90 (Native SpyCatcher = Isoleucine)
3.4.4. SDS-PAGE Validates Mass Photometry Data
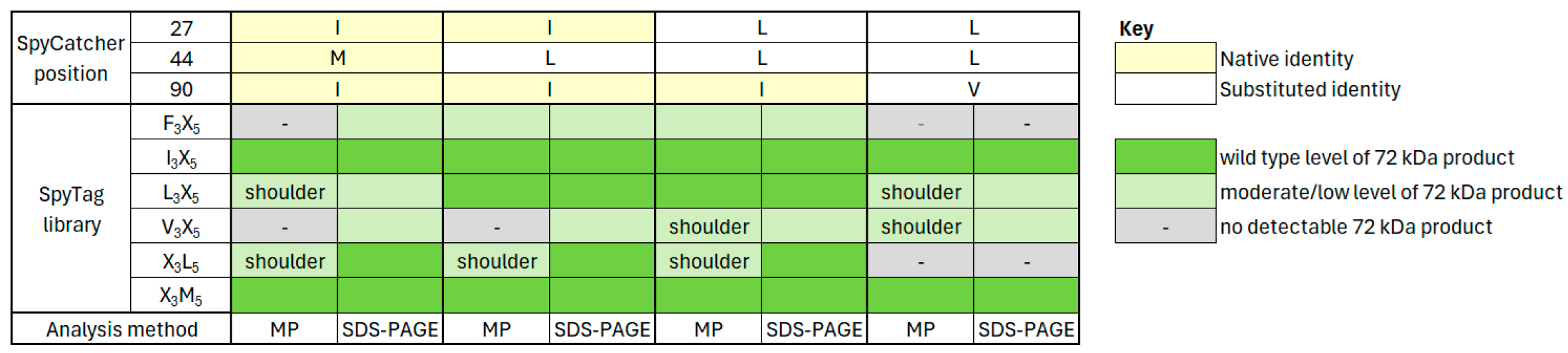
3.5. Application of SpyCatcher Library Data
3.6. Specificity of Native and Substituted SpyCatcher Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IMAC | Immobilised Metal Affinity Chromatography |
References
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, E690–E697. [Google Scholar] [CrossRef]
- Li, L.; Fierer, J.O.; Rapoport, T.A.; Howarth, M. Structural analysis and optimization of the covalent association between SpyTag and SpyCatcher. J. Mol. Biol. 2014, 426, 309–317. [Google Scholar] [CrossRef]
- Reddington, S.C.; Howarth, M. Secrets of a covalent interaction for biomolecular engineering: SpyTag and SpyCatcher. Curr. Opin. Chem. Biol. 2015, 29, 94–99. [Google Scholar] [CrossRef]
- Schoene, C.; Fierer, J.O.; Bennett, S.P.; Howarth, M. SpyTag/SpyCatcher cyclization confers resilience to boiling on a mesophilic enzyme. Angew. Chem. Int. Ed. 2014, 53, 6101–6104. [Google Scholar] [CrossRef]
- Williams, D.M.; Kaufman, G.; Izadi, H.; Gahm, A.E.; Prophet, S.M.; Vanderlick, K.T.; Osuji, C.O.; Regan, L. Facile Protein Immobilization Using Engineered Surface-Active Biofilm Proteins. ACS Appl. Nano Mater. 2018, 1, 2483–2488. [Google Scholar] [CrossRef]
- Liu, Z.; Zhou, H.; Wang, W.; Tan, W.; Fu, Y.-Z.; Zhu, M. A novel method for synthetic vaccine construction based on protein assembly. Sci. Rep. 2014, 4, 7266. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zhang, W.-B.; Mahdavi, A.; Arnold, F.H.; Tirrell, D.A. Synthesis of bioactive protein hydrogels by geneticallyencoded SpyTag-SpyCatcher chemistry. Proc. Natl. Acad. Sci. USA 2014, 111, 11269–11274. [Google Scholar] [CrossRef] [PubMed]
- Keeble, A.H.; Banerjee, A.; Ferla, M.P.; Reddington, S.C.; Anuar, I.N.; Howarth, M. Evolving Accelerated Amidation by SpyTag/SpyCatcher to Analyze Membrane Dynamics. Angew. Chem. Int. Ed. Engl. 2017, 56, 16521–16525. [Google Scholar] [CrossRef]
- Keeble, A.H.; Turkki, P.; Stokes, S.; Khairil Anuar, I.N.A.; Rahikainen, R.; Hytönen, V.P.; Howarth, M. Approaching infinite affinity through engineering of peptide–protein interaction pairs. Proc. Natl. Acad. Sci. USA 2019, 116, 26523–26533. [Google Scholar] [CrossRef]
- Houghten, R.A.; Pinilla, C.; Blondelle, S.E.; Appel, J.R.; Dooley, C.T.; Cuervo, J.H. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature 1991, 354, 84–86. [Google Scholar] [CrossRef]
- Hughes, M.D.; Zhang, Z.-R.; Sutherland, A.J.; Santos, A.F.; Hine, A.V. Discovery of active proteins directly from combinatorial randomized protein libraries without display, purification or sequencing: Identification of novel zinc finger proteins. Nucleic Acids Res. 2005, 33, e32. [Google Scholar] [CrossRef] [PubMed]
- Kille, S.; Acevedo-Rocha, C.G.; Parra, L.P.; Zhang, Z.G.; Opperman, D.J.; Reetz, M.T.; Acevedo, J.P. Reducing codon redundancy and screening effort in combinatorial protein libraries created by saturation mutagenesis. ACS Synth. Biol. 2013, 2, 83–92. [Google Scholar] [CrossRef]
- Soltermann, F.; Foley, E.D.B.; Pagnoni, V.; Galpin, M.; Benesch, J.L.P.; Kukura, P.; Struwe, W.B. Quantifying protein–protein interactions by mass photometry. Angew. Chem. Int. Ed. 2020, 59, 10774–10779. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Piszczek, G. Standard protocol for mass photometry experiments. Eur. Biophys. J. 2021, 50, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Young, G.; Hundt, N.; Cole, D.; Fineberg, A.; Andrecka, J.; Tyler, A.; Olerinyova, A.; Ansari, A.; Marklund, E.G.; Collier, M.P.; et al. Quantitative mass imaging of single biological macromolecules. Science 2018, 360, 423–427. [Google Scholar] [CrossRef]
- Wu, D.; Piszczek, G. Measuring the affinity of protein-protein interactions on a single-molecule level by mass photometry. Anal. Biochem. 2020, 592, 113575. [Google Scholar] [CrossRef]
- Refeyn Ltd. How Does Mass Photometry Work? 2025. Available online: https://refeyn.com/post/how-does-mass-photometry-work (accessed on 8 August 2025).
- Hundt, N. Label-free, mass-sensitive single-molecule imaging using interferometric scattering microscopy. Essays Biochem. 2020, 65, 81–91. [Google Scholar] [CrossRef]
- Asor, R.; Kukura, P. Characterising biomolecular interactions and dynamics with mass photometry. Curr. Opin. Chem. Biol. 2022, 68, 102132. [Google Scholar] [CrossRef]
- Graciani, G.; Yoon, T.Y. The new kid on the block: Mass photometry. Mol. Cells 2023, 46, 187–189. [Google Scholar] [CrossRef]
- Veggiani, G.; Nakamuraa, T.; Brenner, M.D.; Gayet, R.V.; Yand, J.; Robinson, C.V.; Howarth, M. Programmable polyproteams built using twinpeptide superglues. Proc. Natl. Acad. Sci. USA 2016, 113, 1202–1207. [Google Scholar] [CrossRef]
- Hughes, M.D.; Nagel, D.A.; Santos, A.F.; Sutherland, A.J.; Hine, A.V. Removing the redundancy from randomised gene libraries. J. Mol. Biol. 2003, 331, 973–979. [Google Scholar] [CrossRef]
- Chembath, A.; Wagstaffe, B.P.G.; Ashraf, A.; Ferreira Amaral, M.M.; Frigotto, L.; Hine, A.V. Nondegenerate Saturation Mutagenesis: Library Construction and Analysis via MAX and ProxiMAX Randomization. In Directed Evolution Methods and Protocols; Currin, A., Swainston, N., Eds.; Humana Press: Totowa, NJ, USA. Meth. Mol. Biol. 2022, 2461, 19–41. [Google Scholar] [CrossRef]
- Hagan, R.M.; Björnsson, R.; McMahon, S.A.; Schomburg, B.; Braithwaite, V.; Bühl, M.; Naismith, J.H.; Schwarz-Linek, U. NMR Spectroscopic and Theoretical Analysis of a Spontaneously Formed Lys–Asp Isopeptide Bond. Angewante Chemie Int. Ed. 2010, 49, 8421–8425. [Google Scholar] [CrossRef]
- Hosseini, B. Creation of Novel Proteins for Peptide Recognition and Interaction. Doctoral Thesis, Aston University, Birmingham, UK, 2024. [Google Scholar] [CrossRef]
- Van Zyl, W.F.; Van Staden, A.D.; Dicks, L.M.T.; Trindade, M. Use of the mCherry fluorescent protein to optimize the expression of class I lanthipeptides in Escherichia coli. Microb. Cell Fact. 2023, 22, 149. [Google Scholar] [CrossRef]
- Claasen, M.; Kofinova, Z.; Contino, M.; Struwe, W.B. Analysis of protein complex formation at micromolar concentrations by coupling microfluidics with mass photometry. J. Vis. Exp. 2024, 203, e65772. [Google Scholar] [CrossRef] [PubMed]
- Kratochvíl, J.; Asor, R.; Helmi, S.; Struwe, W.B.; Kukura, P. Lifting the Concentration Limit of Mass Photometry by PEG Nanopatterning. Nano Lett. 2024, 24, 10032–10039. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Struwe, W.B.; Kukura, P. Single molecule mass photometry of nucleic acids. Nucleic Acids Res. 2020, 48, e97. [Google Scholar] [CrossRef] [PubMed]
- Young, J.W.; Pfitzner, E.; van Wee, R.; Kirschbaum, C.; Kukura, P.; Robinson, C.V. Characterization of membrane protein interactions by peptidisc-mediated mass photometry. iScience 2024, 27, 108785. [Google Scholar] [CrossRef]
- Olerinyova, A.; Sonn-Segev, A.; Gault, J.; Eichmann, C.; Schimpf, J.; Kopf, A.H.; Rudden, L.S.P.; Ashkinadze, D.; Bomba, R.; Frey, L.; et al. Mass Photometry of Membrane Proteins. Chem 2021, 7, 224–236. [Google Scholar] [CrossRef]
- Kofinova, Z.; Karunanithy, G.; Ferreira, A.S.; Struwe, W.B. Measuring protein-protein interactions and quantifying their dissociation constants with mass photometry. Curr. Protocols 2024, 4, e962. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, D.; Yang, W.; Wu, X.-L.; Lai, L.; Zhang, W.-B. Tuning SpyTag–SpyCatcher mutant pairs toward orthogonal reactivity encryption. Chem. Sci. 2017, 8, 6577–6582. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hosseini, B.; Ashraf, M.; Kitchen, P.; Chembath, A.; Collighan, R.; Spickett, C.M.; Regan, L.; Hine, A.V. Engineering Protein–Peptide Interfaces via Combinatorial Mutagenesis and Mass Photometric Screening. Biomolecules 2025, 15, 1183. https://doi.org/10.3390/biom15081183
Hosseini B, Ashraf M, Kitchen P, Chembath A, Collighan R, Spickett CM, Regan L, Hine AV. Engineering Protein–Peptide Interfaces via Combinatorial Mutagenesis and Mass Photometric Screening. Biomolecules. 2025; 15(8):1183. https://doi.org/10.3390/biom15081183
Chicago/Turabian StyleHosseini, Bitasadat, Mohammed Ashraf, Philip Kitchen, Anupama Chembath, Russell Collighan, Corinne M. Spickett, Lynne Regan, and Anna V. Hine. 2025. "Engineering Protein–Peptide Interfaces via Combinatorial Mutagenesis and Mass Photometric Screening" Biomolecules 15, no. 8: 1183. https://doi.org/10.3390/biom15081183
APA StyleHosseini, B., Ashraf, M., Kitchen, P., Chembath, A., Collighan, R., Spickett, C. M., Regan, L., & Hine, A. V. (2025). Engineering Protein–Peptide Interfaces via Combinatorial Mutagenesis and Mass Photometric Screening. Biomolecules, 15(8), 1183. https://doi.org/10.3390/biom15081183







