Structural and Functional Effects of the Interaction Between an Antimicrobial Peptide and Its Analogs with Model Bacterial and Erythrocyte Membranes
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Peptide Synthesis
2.2.2. Peptide Solutions
2.2.3. Model Membrane Composition and Preparation of Large Unilamellar Vesicles (LUVs)
2.2.4. Fluorescence Studies of Peptide–Membrane Binding
2.2.5. Circular Dichroism (CD) of Peptide–Membrane Interaction
2.2.6. Evaluation of Vesicle Size, Polydispersity, and Zeta Potential: Dynamic Light Scattering (DLS)
2.2.7. 5(6)-Carboxyfluoresceine (CF) Incorporation into LUVs
2.2.8. LUV Permeabilization by Peptides: Leakage of 5(6)-Carboxyfluorescein (CF)
2.2.9. Optical Microscopy of Giant Unilamellar Vesicles (GUVs)
2.2.10. Minimum Inhibitory Concentration (MIC) Determination
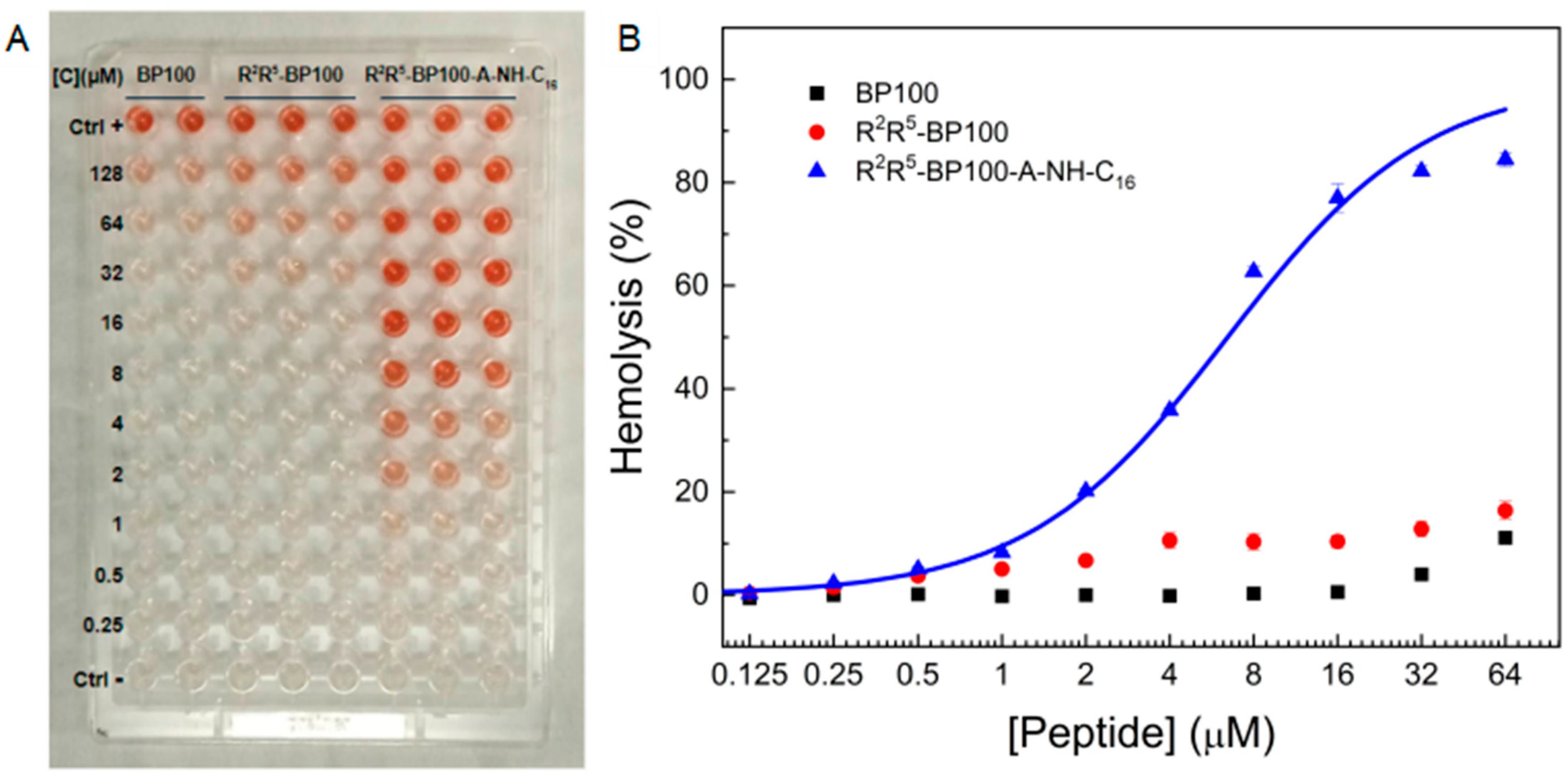
2.2.11. Hemolytic Activity (HA)
3. Results
3.1. Monitoring Peptide Binding via Tyr Fluorescence
3.2. Effect of LUVs on Peptide Conformations
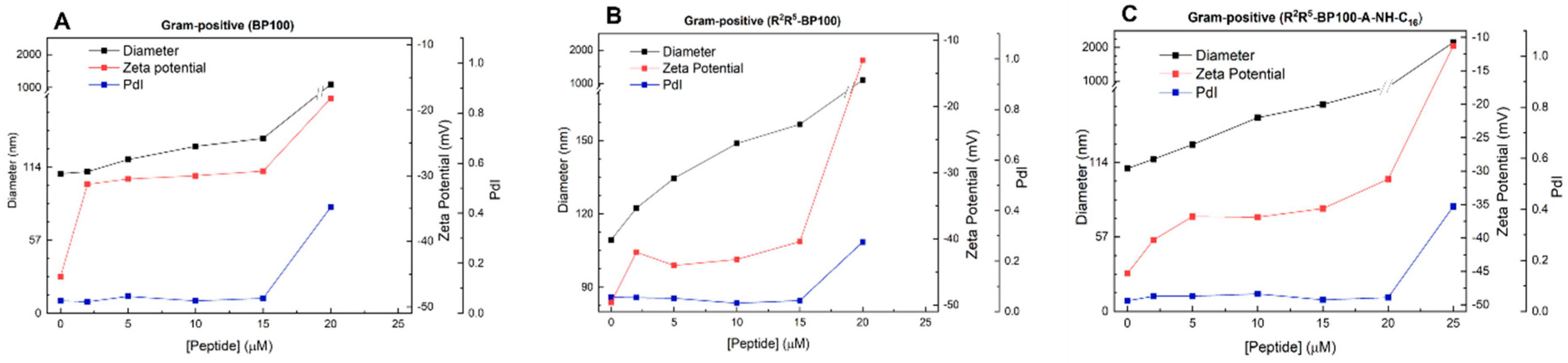
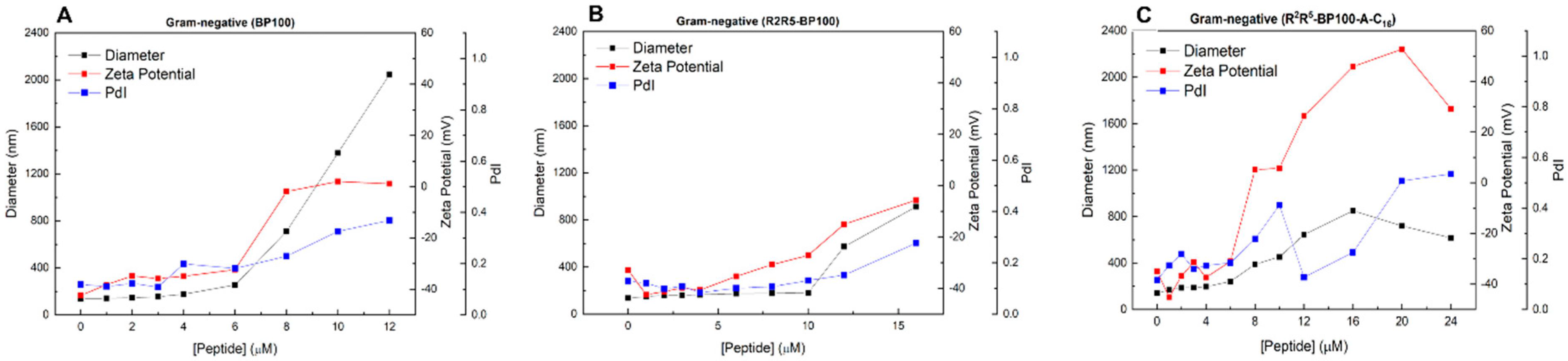
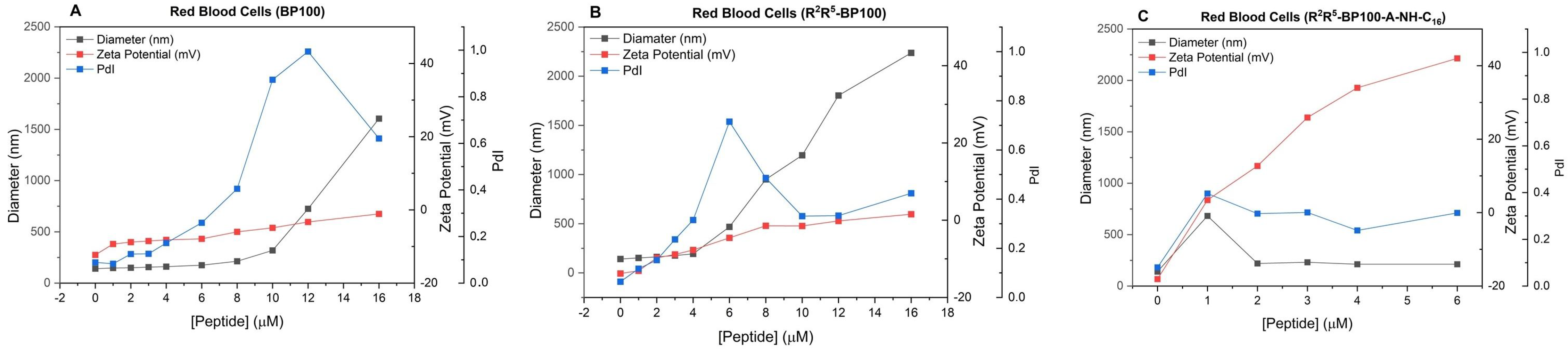
3.3. Effect of Peptide Binding on Vesicle Diameter (Dh), Polydispersity (PDI), and Zeta Potential (ζ): Dynamic Light Scattering (DLS) Studies
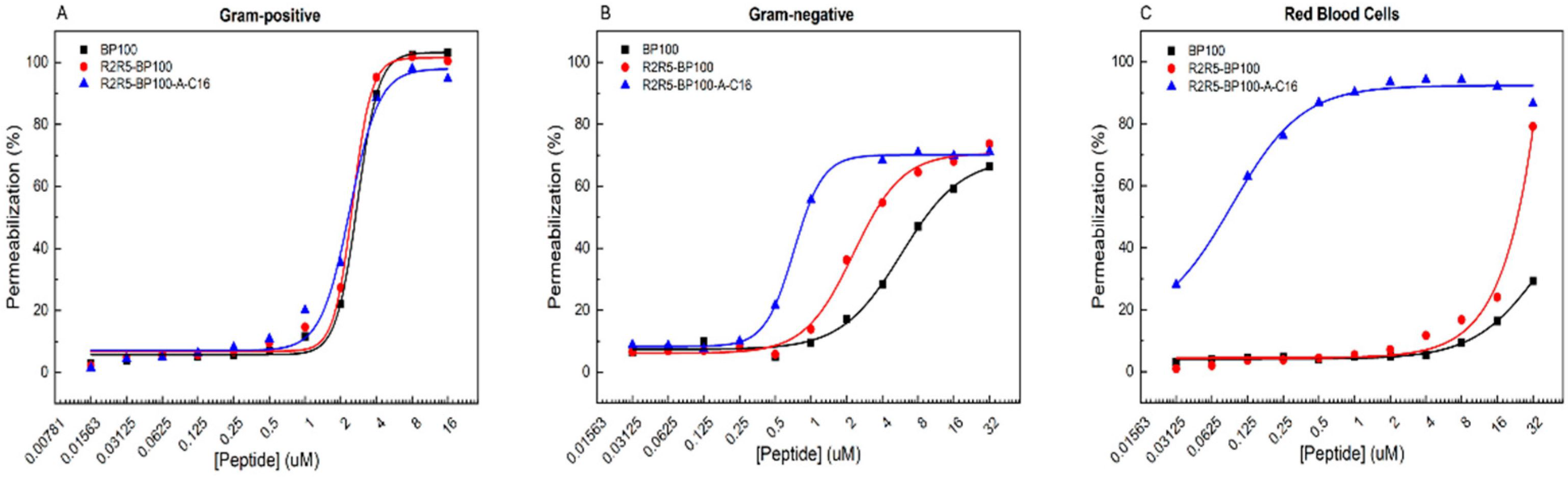
3.4. Peptide-Promoted LUV Permeabilization
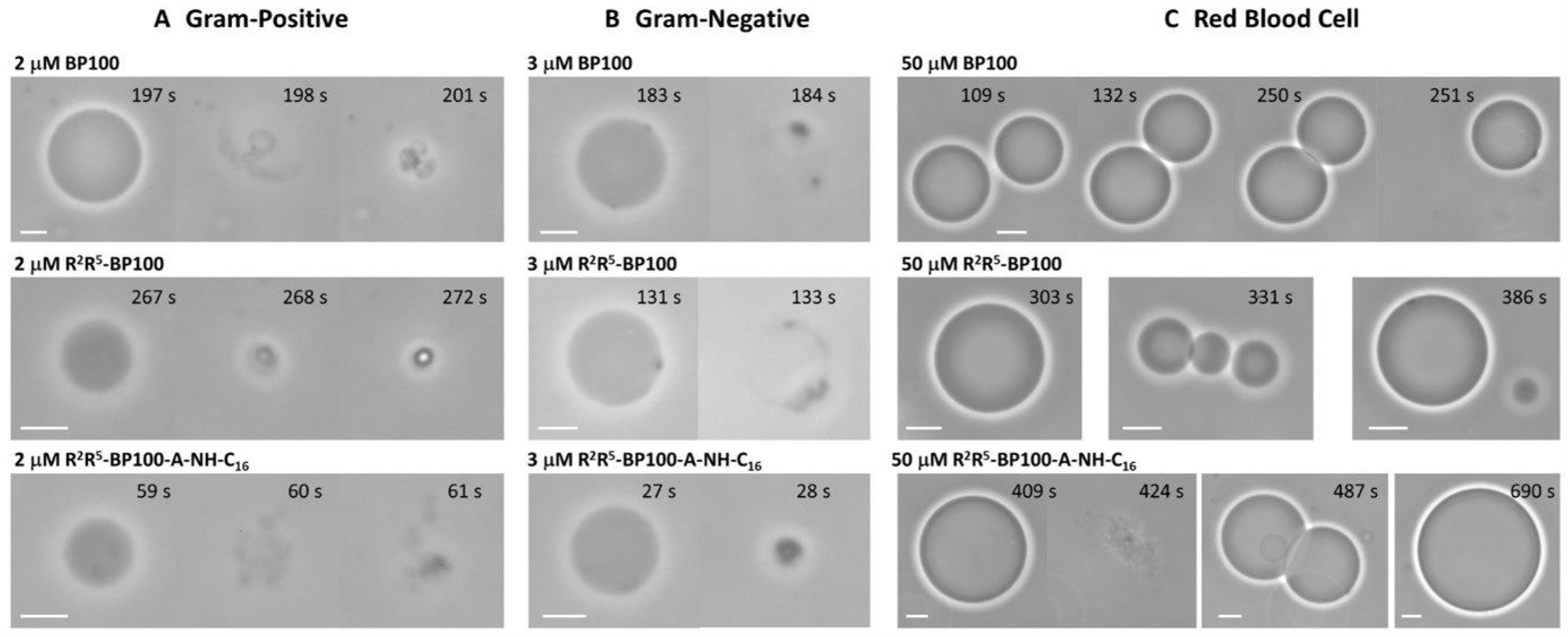
3.5. Peptide Effects on Giant Unilamellar Vesicles
3.6. Determination of Minimum Inhibitory Concentration, MIC, in Bacteria
3.7. Peptide Effect on Mammalian Cells: Red Blood Cell Hemolysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ukuhor, H.O. The interrelationships between antimicrobial resistance, COVID-19, past, and future pandemics. J. Infect. Public. Health 2021, 14, 53–60. [Google Scholar] [CrossRef]
- Muteeb, G.; Rehman, T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef]
- García-Olmedo, F.; Molina, A.; Alamillo, J.M.; Rodríguez-Palenzuéla, P. Plant defense peptides. Biopolymers 1998, 47, 479–491. [Google Scholar] [CrossRef]
- Padovan, L.; Scocchi, M.; Tossi, A. Structural Aspects of Plant Antimicrobial Peptides. Curr. Protein Pept. Sci. 2010, 11, 210–219. [Google Scholar] [CrossRef]
- Bulet, P.; Stöcklin, R.; Menin, L. Anti-microbial peptides: From invertebrates to vertebrates. Immunol. Rev. 2004, 198, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Hancock, Peptide Antimicrobial Agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef]
- Fontoura, R.; Spada, J.C.; Silveira, S.T.; Tsai, S.M.; Brandelli, A. Purification and characterization of an antimicrobial peptide produced by Pseudomonas sp. strain 4B. World J. Microbiol. Biotechnol. 2009, 25, 205–213. [Google Scholar] [CrossRef]
- Chileveru, H.R.; Lim, S.A.; Chairatana, P.; Wommack, A.J.; Chiang, I.-L.; Nolan, E.M. Visualizing Attack of Escherichia coli by the Antimicrobial Peptide Human Defensin 5. Biochemistry 2015, 54, 1767–1777. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Mojsoska, B.; Carretero, G.; Larsen, S.; Mateiu, R.V.; Jenssen, H. Peptoids successfully inhibit the growth of gram negative E. coli causing substantial membrane damage. Sci. Rep. 2017, 7, 42332. [Google Scholar] [CrossRef] [PubMed]
- Mader, J.S.; Hoskin, D.W. Cationic antimicrobial peptides as novel cytotoxic agents for cancer treatment. Expert Opin. Investig. Drugs 2006, 15, 933–946. [Google Scholar] [CrossRef]
- Steinbuch, K.B.; Fridman, M. Mechanisms of resistance to membrane-disrupting antibiotics in Gram-positive and Gram-negative bacteria. Medchemcomm 2016, 7, 86–102. [Google Scholar] [CrossRef]
- El Shazely, B.; Yu, G.; Johnston, P.R.; Rolff, J. Resistance Evolution Against Antimicrobial Peptides in Staphylococcus aureus Alters Pharmacodynamics Beyond the MIC. Front. Microbiol. 2020, 11, 103. [Google Scholar] [CrossRef] [PubMed]
- Wang GuangShun, W.G. Discovery, classification and functional diversity of antimicrobial peptides. In Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies; CABI: Ceremonial, UK, 2017; pp. 1–19. [Google Scholar] [CrossRef]
- Hancock, R.E. Peptide antibiotics. Lancet 1997, 349, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, A.; Martinez, M.; Maturana, P.; Semorile, L.C.; Maffia, P.C. Antimicrobial Peptides: Interaction with Model and Biological Membranes and Synergism With Chemical Antibiotics. Front. Chem. 2018, 6, 204. [Google Scholar] [CrossRef]
- Oren, Z.; Shai, Y. Mode of action of linear amphipathic α-helical antimicrobial peptides. Biopolymers 1998, 47, 451–463. [Google Scholar] [CrossRef]
- Bechinger, B.; Lohner, K. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2006, 1758, 1529–1539. [Google Scholar] [CrossRef]
- Mihajlovic, M.; Lazaridis, T. Antimicrobial peptides bind more strongly to membrane pores. Biochim. Biophys. Acta Biomembr. 2010, 1798, 1494–1502. [Google Scholar] [CrossRef]
- Bardajı, E.; Montesinos, E.; Badosa, E.; Feliu, L.; Planas, M.; Ferre, R. Antimicrobial Linear Peptides. Patent No. P200601098 7 February 2017. [Google Scholar]
- Badosa, E.; Ferre, R.; Planas, M.; Feliu, L.; Besalú, E.; Cabrefiga, J.; Bardají, E.; Montesinos, E. A library of linear undecapeptides with bactericidal activity against phytopathogenic bacteria. Peptides 2007, 28, 2276–2285. [Google Scholar] [CrossRef]
- Ferre, R.; Melo, M.N.; Correia, A.D.; Feliu, L.; Bardají, E.; Planas, M.; Castanho, M. Synergistic Effects of the Membrane Actions of Cecropin-Melittin Antimicrobial Hybrid Peptide BP100. Biophys. J. 2009, 96, 1815–1827. [Google Scholar] [CrossRef] [PubMed]
- Torcato, I.M.; Huang, Y.-H.; Franquelim, H.G.; Gaspar, D.; Craik, D.J.; Castanho, M.A.; Henriques, S.T. Design and characterization of novel antimicrobial peptides, R-BP100 and RW-BP100, with activity against Gram-negative and Gram-positive bacteria. Biochim. Biophys. Acta Biomembr. 2013, 1828, 944–955. [Google Scholar] [CrossRef]
- Manzini, M.C.; Perez, K.R.; Riske, K.A.; Bozelli, J.C.; Santos, T.L.; da Silva, M.A.; Saraiva, G.K.; Politi, M.J.; Valente, A.P.; Almeida, F.C.; et al. Peptide:Lipid ratio and membrane surface charge determine the mechanism of action of the antimicrobial peptide BP100. Conformational and functional studies. Biochim. Biophys. Acta Biomembr. 2014, 1838, 1985–1999. [Google Scholar] [CrossRef]
- Carretero, G.P.B.; Vicente, E.F.; Cilli, E.M.; Alvarez, C.M.; Jenssen, H.; Schreier, S.; van der Wel, P. Dissecting the mechanism of action of actinoporins. Role of the N-terminal amphipathic α-helix in membrane binding and pore activity of sticholysins I and II. PLoS ONE 2018, 13, e0202981. [Google Scholar] [CrossRef] [PubMed]
- Park, P.; Matsubara, D.K.; Barzotto, D.R.; Lima, F.S.; Chaimovich, H.; Marrink, S.J.; Cuccovia, I.M. Vesicle protrusion induced by antimicrobial peptides suggests common carpet mechanism for short antimicrobial peptides. Sci. Rep. 2024, 14, 9701. [Google Scholar] [CrossRef]
- Klug, J.; Masone, D.; Del Pópolo, M.G. Molecular-level insight into the binding of arginine to a zwitterionic Langmuir monolayer. RSC Adv. 2017, 7, 30862–30869. [Google Scholar] [CrossRef]
- Riesco-Llach, G.; Llanet-Ferrer, S.; Planas, M.; Feliu, L. Deciphering the Mechanism of Action of the Antimicrobial Peptide BP100. Int. J. Mol. Sci. 2024, 25, 3456. [Google Scholar] [CrossRef]
- Chen, S.-P.; Chen, E.H.-L.; Yang, S.-Y.; Kuo, P.-S.; Jan, H.-M.; Yang, T.-C.; Hsieh, M.-Y.; Lee, K.-T.; Lin, C.-H.; Chen, R.P.-Y. A Systematic Study of the Stability, Safety, and Efficacy of the de novo Designed Antimicrobial Peptide PepD2 and Its Modified Derivatives Against Acinetobacter baumannii. Front. Microbiol. 2021, 12, 678330. [Google Scholar] [CrossRef]
- Kurtzhals, P.; Østergaard, S.; Nishimura, E.; Kjeldsen, T. Derivatization with fatty acids in peptide and protein drug discovery. Nat. Rev. Drug Discov. 2023, 22, 59–80. [Google Scholar] [CrossRef] [PubMed]
- Epand, R.M.; Epand, R.F. Bacterial membrane lipids in the action of antimicrobial agents. J. Pept. Sci. 2011, 17, 298–305. [Google Scholar] [CrossRef]
- Som, A.; Tew, G.N. Tew, Influence of Lipid Composition on Membrane Activity of Antimicrobial Phenylene Ethynylene Oligomers. J. Phys. Chem. B 2008, 112, 3495–3502. [Google Scholar] [CrossRef] [PubMed]
- Navas, B.P.; Lohner, K.; Deutsch, G.; Sevcsik, E.; Riske, K.; Dimova, R.; Garidel, P.; Pabst, G. Composition dependence of vesicle morphology and mixing properties in a bacterial model membrane system. Biochim. Biophys. Acta Biomembr. 2005, 1716, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, H.P.; Cesar, C.L.; Barjas-Castro, M.d.L. Barjas-Castro, Electrical properties of the red blood cell membrane and immunohematological investigation. Rev. Bras. Hematol. Hemoter. 2011, 33, 297–301. [Google Scholar] [CrossRef]
- Chou, H.-T.; Wen, H.-W.; Kuo, T.-Y.; Lin, C.-C.; Chen, W.-J. Interaction of cationic antimicrobial peptides with phospholipid vesicles and their antibacterial activity. Peptides 2010, 31, 1811–1820. [Google Scholar] [CrossRef]
- Van Deenen, L.L.M.; De Gier, J. Lipids of the Red Cell Membrane. In The Red Blood Cell, 2nd ed.; Surgenor, D.M.N., Ed.; Avademic Press: New York, NY, USA, 1974; Volume I, Chapter 4; pp. 147–211. [Google Scholar]
- Carretero, G.P.; Saraiva, G.K.; Cauz, A.C.; Rodrigues, M.A.; Kiyota, S.; Riske, K.A.; dos Santos, A.A.; Pinatto-Botelho, M.F.; Bemquerer, M.P.; Gueiros-Filho, F.J.; et al. Synthesis, biophysical and functional studies of two BP100 analogues modified by a hydrophobic chain and a cyclic peptide. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1502–1516. [Google Scholar] [CrossRef]
- Rouser, G.; Fleischer, S.; Yamamoto, A. Two dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 1970, 5, 494–496. [Google Scholar] [CrossRef]
- Angelova, M.I.; Dimitrov, D.S. Liposome electroformation. Faraday Discuss. Chem. Soc. 1986, 81, 303. [Google Scholar] [CrossRef]
- Weinberger, A.; Tsai, F.-C.; Koenderink, G.H.; Schmidt, T.F.; Itri, R.; Meier, W.; Schmatko, T.; Schröder, A.; Marques, C. Gel-Assisted Formation of Giant Unilamellar Vesicles. Biophys. J. 2013, 105, 154–164. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Oddo, A.; Hansen, P.R. Hemolytic Activity of Antimicrobial Peptides. In Antimicrobial Peptides: Methods and Protocols; Springer: New York, NY, USA, 2017; pp. 427–435. [Google Scholar] [CrossRef]
- Poveda, J.A.; Prieto, M.; Encinar, J.A.; González-Ros, J.M.; Mateo, C.R. Intrinsic Tyrosine Fluorescence as a Tool to Study the Interaction of the Shaker B ‘Ball’ Peptide with Anionic Membranes. Biochemistry 2003, 42, 7124–7132. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schreier, S.; Barbosa, S.R.; Vieira, R.d.F.F.; Cilli, E.M.; Paiva, A.C.M.; Casallanovo, F.; Nakaie, C.R. Conformational basis for the biological activity of TOAC-labeled angiotensin II and bradykinin: Electron paramagnetic resonance, circular dichroism, and fluorescence studies. Biopolymers 2004, 74, 389–402. [Google Scholar] [CrossRef]
- Vermeer, L.S.; Marquette, A.; Schoup, M.; Fenard, D.; Galy, A.; Bechinger, B. Simultaneous Analysis of Secondary Structure and Light Scattering from Circular Dichroism Titrations: Application to Vectofusin-1. Sci. Rep. 2016, 6, 39450. [Google Scholar] [CrossRef]
- Mink, C.; Strandberg, E.; Wadhwani, P.; Melo, M.N.; Reichert, J.; Wacker, I.; Castanho, M.A.R.B.; Ulrich, A.S. Overlapping Properties of the Short Membrane-Active Peptide BP100 with (i) Polycationic TAT and (ii) α-helical Magainin Family Peptides. Front. Cell. Infect. Microbiol. 2021, 11, 609542. [Google Scholar] [CrossRef]
- Miles, A.J.; Wallace, B.A. Circular dichroism spectroscopy of membrane proteins. Chem. Soc. Rev. 2016, 45, 4859–4872. [Google Scholar] [CrossRef] [PubMed]
- Islam, Z.; Alam, J.M.; Tamba, Y.; Karal, M.A.S.; Yamazaki, M. The single GUV method for revealing the functions of antimicrobial, pore-forming toxin, and cell-penetrating peptides or proteins. Phys. Chem. Chem. Phys. 2014, 16, 15752–15767. [Google Scholar] [CrossRef]
- Riske, K.A. Optical Microscopy of Giant Vesicles as a Tool to Reveal the Mechanism of Action of Antimicrobial Peptides and the Specific Case of Gomesin; Academic Press: Cambridge, MA, USA, 2015; pp. 99–129. [Google Scholar] [CrossRef]
- Tamba, Y.; Yamazaki, M. Single Giant Unilamellar Vesicle Method Reveals Effect of Antimicrobial Peptide Magainin 2 on Membrane Permeability. Biochemistry 2005, 44, 15823–15833. [Google Scholar] [CrossRef]
- Billah, M.; Ahmed, M.; Islam, Z.; Yamazaki, M. Processes and mechanisms underlying burst of giant unilamellar vesicles induced by antimicrobial peptides and compounds. Biochim. Biophys. Acta Biomembr. 2024, 1866, 184330. [Google Scholar] [CrossRef]
- Carretero, G.P.B.; Saraiva, G.K.V.; Rodrigues, M.A.; Kiyota, S.; Bemquerer, M.P.; Chaimovich, H.; Cuccovia, I.M. Naphthalimide-Containing BP100 Leads to Higher Model Membranes Interactions and Antimicrobial Activity. Biomolecules 2021, 11, 542. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.R.; Ferreira, M.; Nunes, C.; Reis, S.; Teixeira, C.; Gomes, P.; Gameiro, P. The Unusual Aggregation and Fusion Activity of the Antimicrobial Peptide W-BP100 in Anionic Vesicles. Membranes 2023, 13, 138. [Google Scholar] [CrossRef] [PubMed]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Pept. Sci. 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Ojah, E.O.; Gneid, H.; Herschede, S.R.; Busschaert, N. Structure-Activity Relationships in Supramolecular Hosts Targeting Bacterial Phosphatidylethanolamine (PE) Lipids. Chem. A Eur. J. 2024, 30, e202402698. [Google Scholar] [CrossRef] [PubMed]
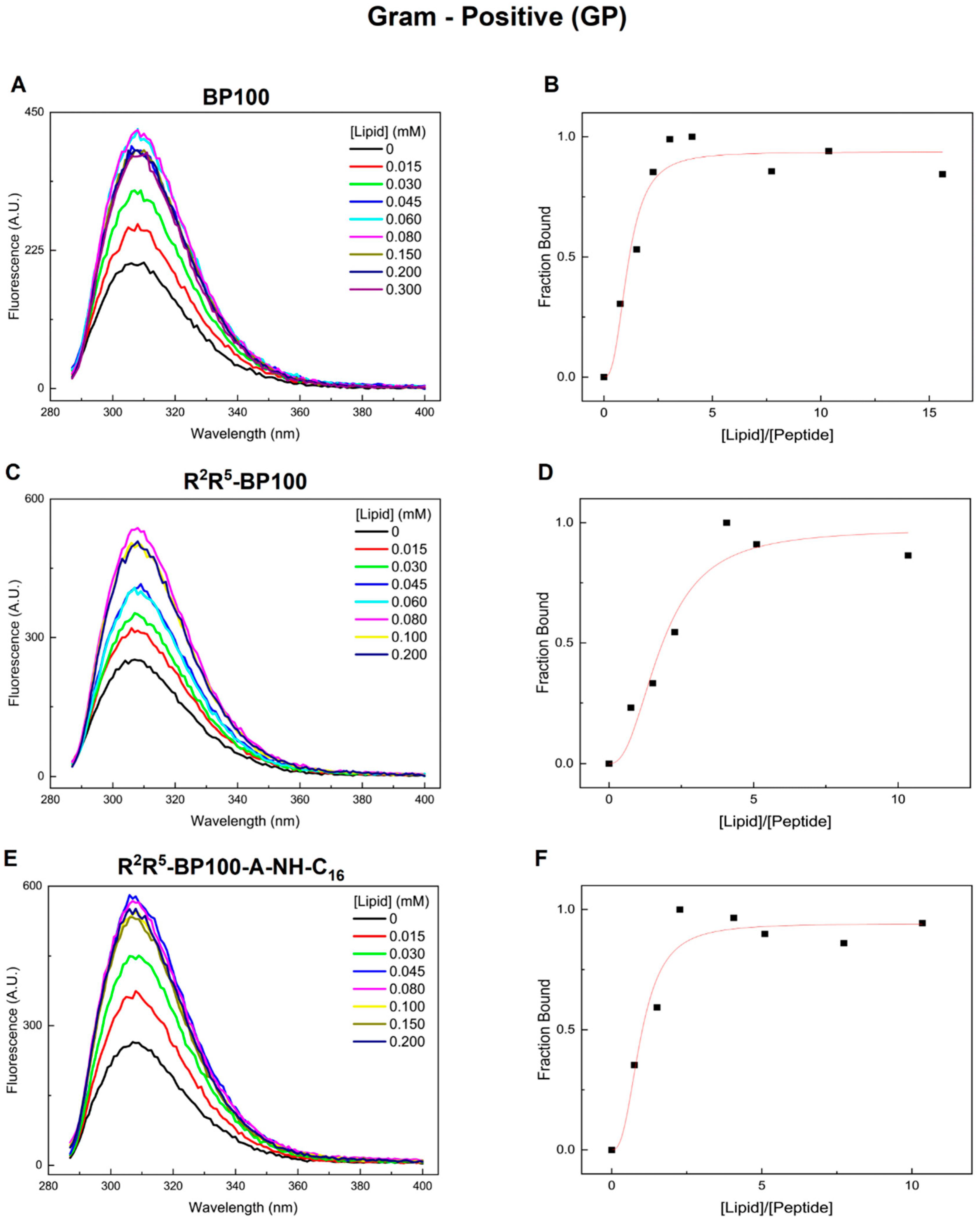
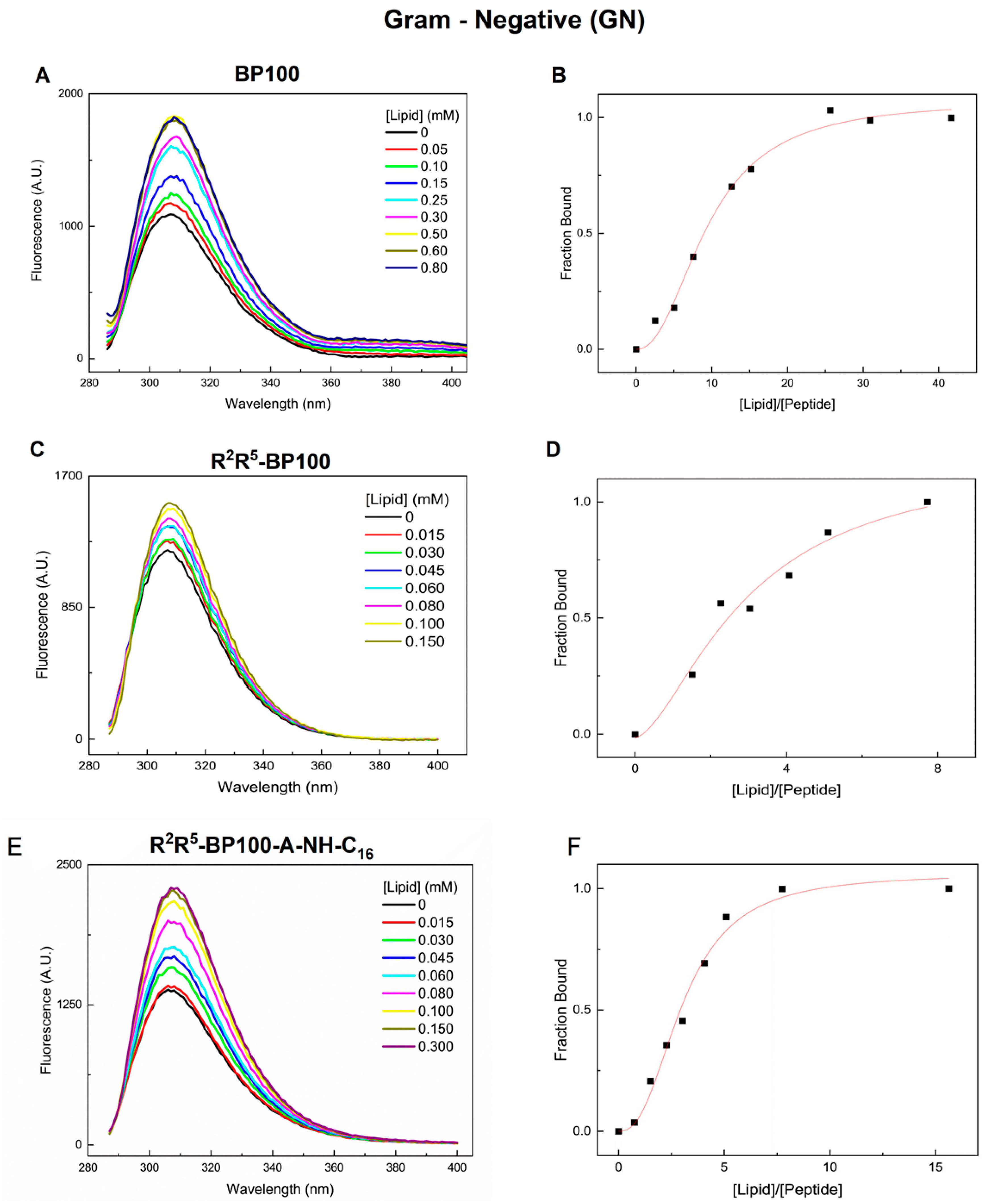
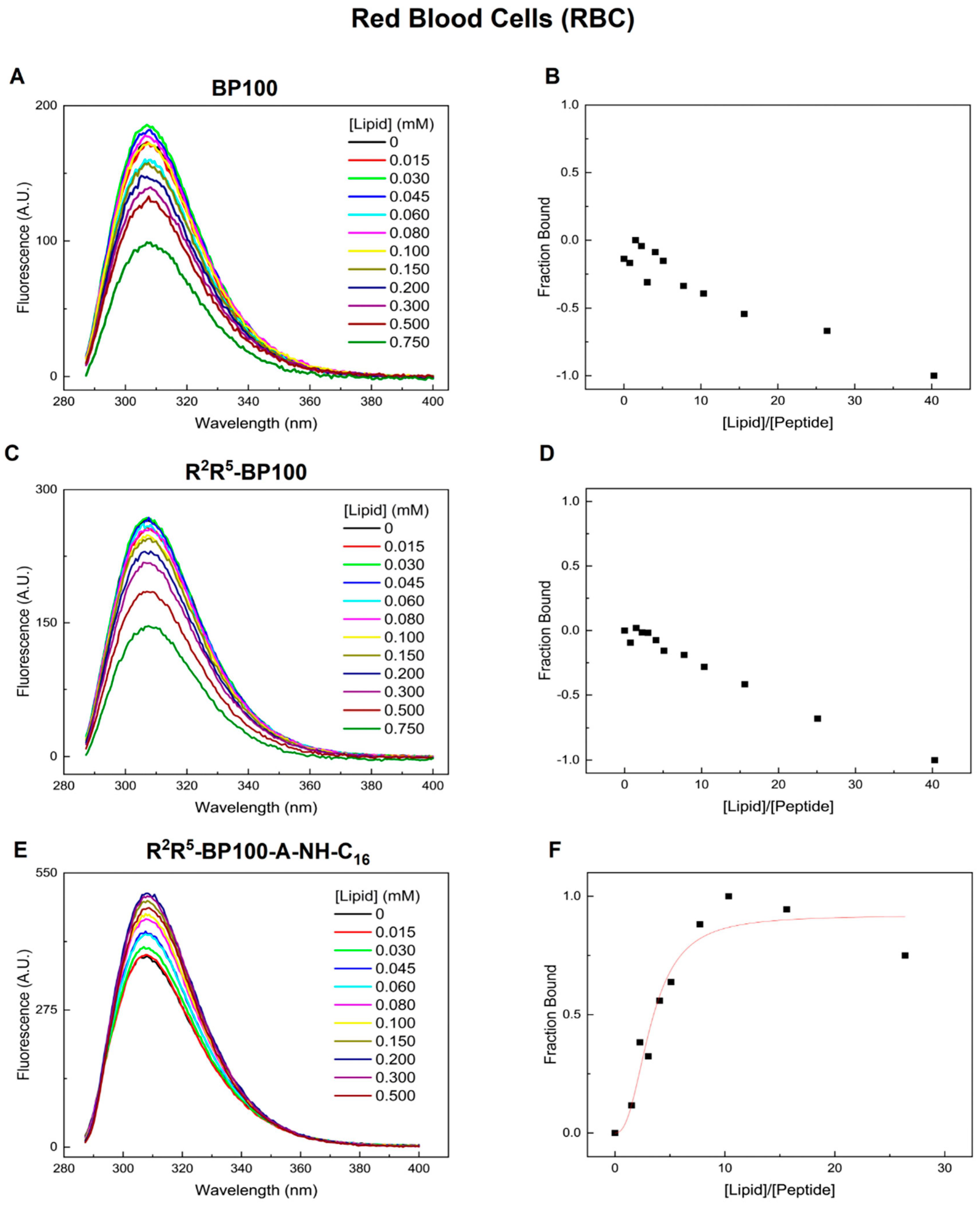
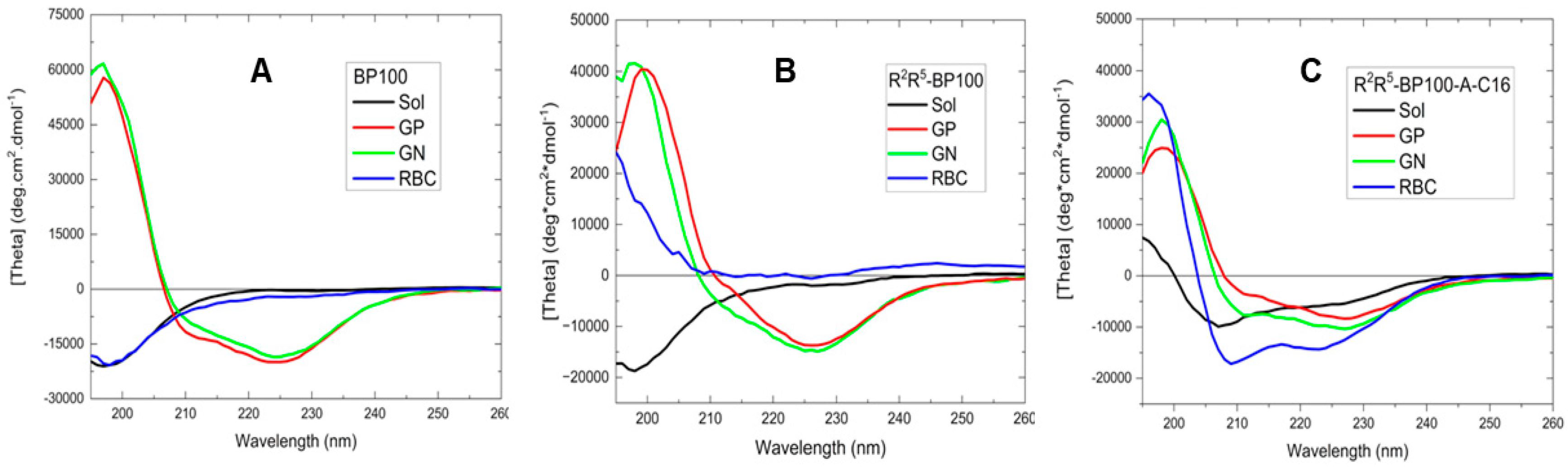
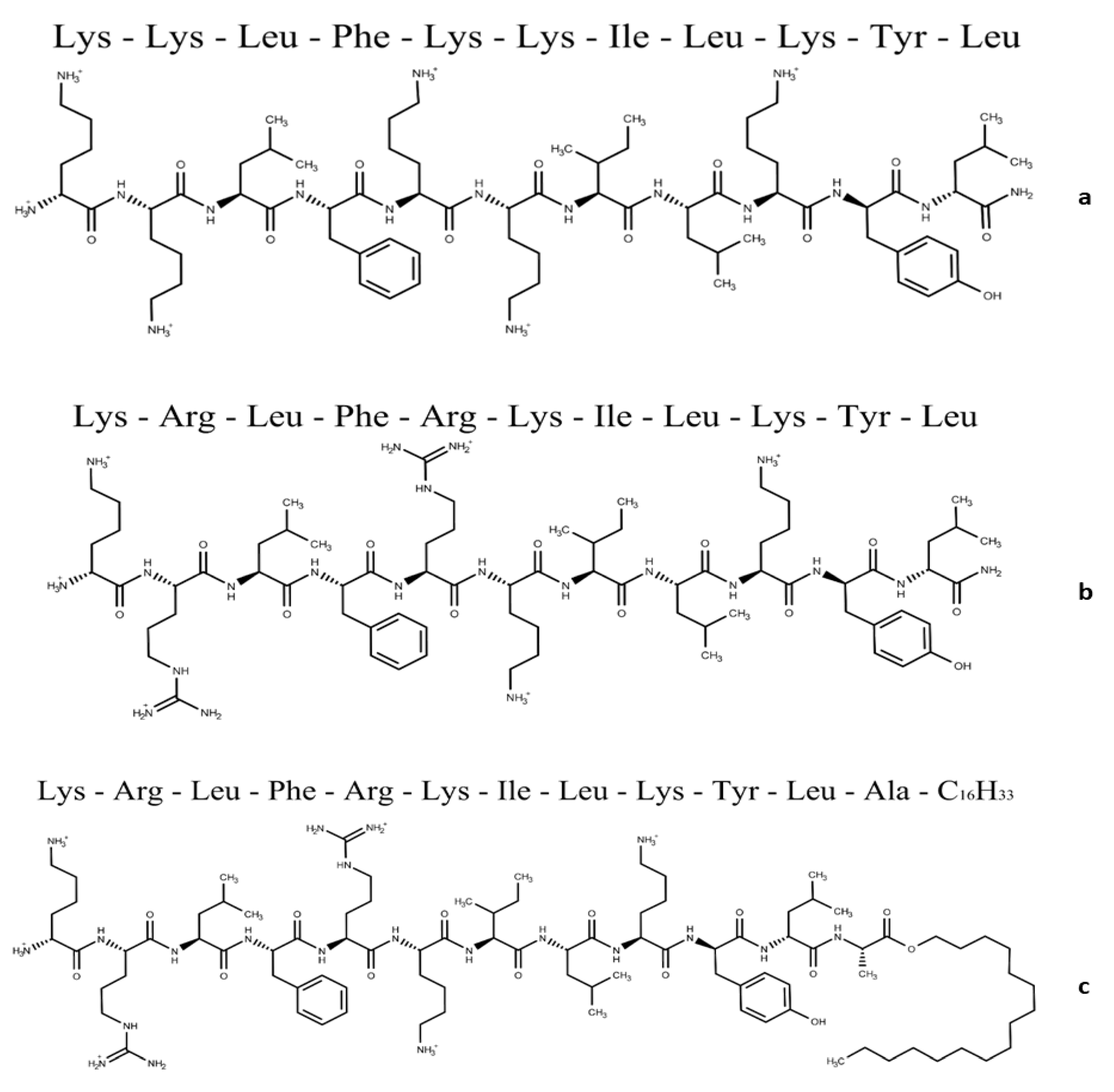
| Peptide | Charge at pH 7 | Monoisotopic Mass (m/z) |
|---|---|---|
| BP100 | +6 | 1421.0 |
| R2R5-BP100 | +6 | 1477.0 |
| R2R5-BP100-A-NH-C16 | +6 | 1772.1 |
| Model Membrane | POPE | POPG | CL | POPC | CHOL | SPH |
|---|---|---|---|---|---|---|
| Gram-negative, GN | 75 | 15 | 10 | - | - | - |
| Gram-positive, GP | - | 58 | 42 | - | - | - |
| Red Blood Cells, RBCs | 20 | - | 25 | 30 | 25 |
| Peptide | P50/L | ||
|---|---|---|---|
| Gram-Positive | Gram-Negative | Red Blood Cell | |
| BP100 | 0.90 | 0.11 | -------- |
| R2R5-BP100 | 0.55 | 0.34 | --------- |
| R2R5-BP100-A-NH-C16 | 1.02 | 0.33 | 0.30 |
| C50 (μM) | |||
|---|---|---|---|
| Peptide | GP | GN | RBC |
| BP100 | 2.8 | 5.8 | 40.5 * |
| R2R5-BP100 | 2.5 | 2.3 | 23.6 * |
| R2R5–BP100-A-NH-C16 | 2.4 | 0.7 | 0.09 |
| Bacteria | MIC (μM) | |||
|---|---|---|---|---|
| BP100 | R2R5-BP100 | R2R5-BP100-A-NH-C16 | Gram | |
| Escherichia coli ATCC 35218 | 2 (1) | 2 (2) | 32 (>32) | - |
| Staphylococcus aureus N315 | 16 | 8 | 16 | + |
| Staphylococcus aureus ATCC 25923 | 2 | 2 | 32 | + |
| Bacillus subtilis PY79 | 2 (1) | 1 (1) | 16 (16) | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furuya, M.L.; Carretero, G.P.; Bemquerer, M.P.; Kiyota, S.; Rodrigues, M.A.; Gaziri, T.J.d.N.; Zuluaga, N.L.B.; Matsubara, D.K.; Wandermuren, M.N.; Riske, K.A.; et al. Structural and Functional Effects of the Interaction Between an Antimicrobial Peptide and Its Analogs with Model Bacterial and Erythrocyte Membranes. Biomolecules 2025, 15, 1143. https://doi.org/10.3390/biom15081143
Furuya ML, Carretero GP, Bemquerer MP, Kiyota S, Rodrigues MA, Gaziri TJdN, Zuluaga NLB, Matsubara DK, Wandermuren MN, Riske KA, et al. Structural and Functional Effects of the Interaction Between an Antimicrobial Peptide and Its Analogs with Model Bacterial and Erythrocyte Membranes. Biomolecules. 2025; 15(8):1143. https://doi.org/10.3390/biom15081143
Chicago/Turabian StyleFuruya, Michele Lika, Gustavo Penteado Carretero, Marcelo Porto Bemquerer, Sumika Kiyota, Magali Aparecida Rodrigues, Tarcillo José de Nardi Gaziri, Norma Lucia Buritica Zuluaga, Danilo Kiyoshi Matsubara, Marcio Nardelli Wandermuren, Karin A. Riske, and et al. 2025. "Structural and Functional Effects of the Interaction Between an Antimicrobial Peptide and Its Analogs with Model Bacterial and Erythrocyte Membranes" Biomolecules 15, no. 8: 1143. https://doi.org/10.3390/biom15081143
APA StyleFuruya, M. L., Carretero, G. P., Bemquerer, M. P., Kiyota, S., Rodrigues, M. A., Gaziri, T. J. d. N., Zuluaga, N. L. B., Matsubara, D. K., Wandermuren, M. N., Riske, K. A., Chaimovich, H., Schreier, S., & Cuccovia, I. M. (2025). Structural and Functional Effects of the Interaction Between an Antimicrobial Peptide and Its Analogs with Model Bacterial and Erythrocyte Membranes. Biomolecules, 15(8), 1143. https://doi.org/10.3390/biom15081143






