Piezo1 Channel Activators Yoda1 and Yoda2 in the Context of Red Blood Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Blood Collection and Preparation
2.2. Automated Patch Clamp Recordings
2.3. Calcium Imaging and Flow Cytometry
2.4. Membrane Potential Measurements
2.5. Data Analysis and Statistics
3. Results
3.1. Patch Clamp Recordings
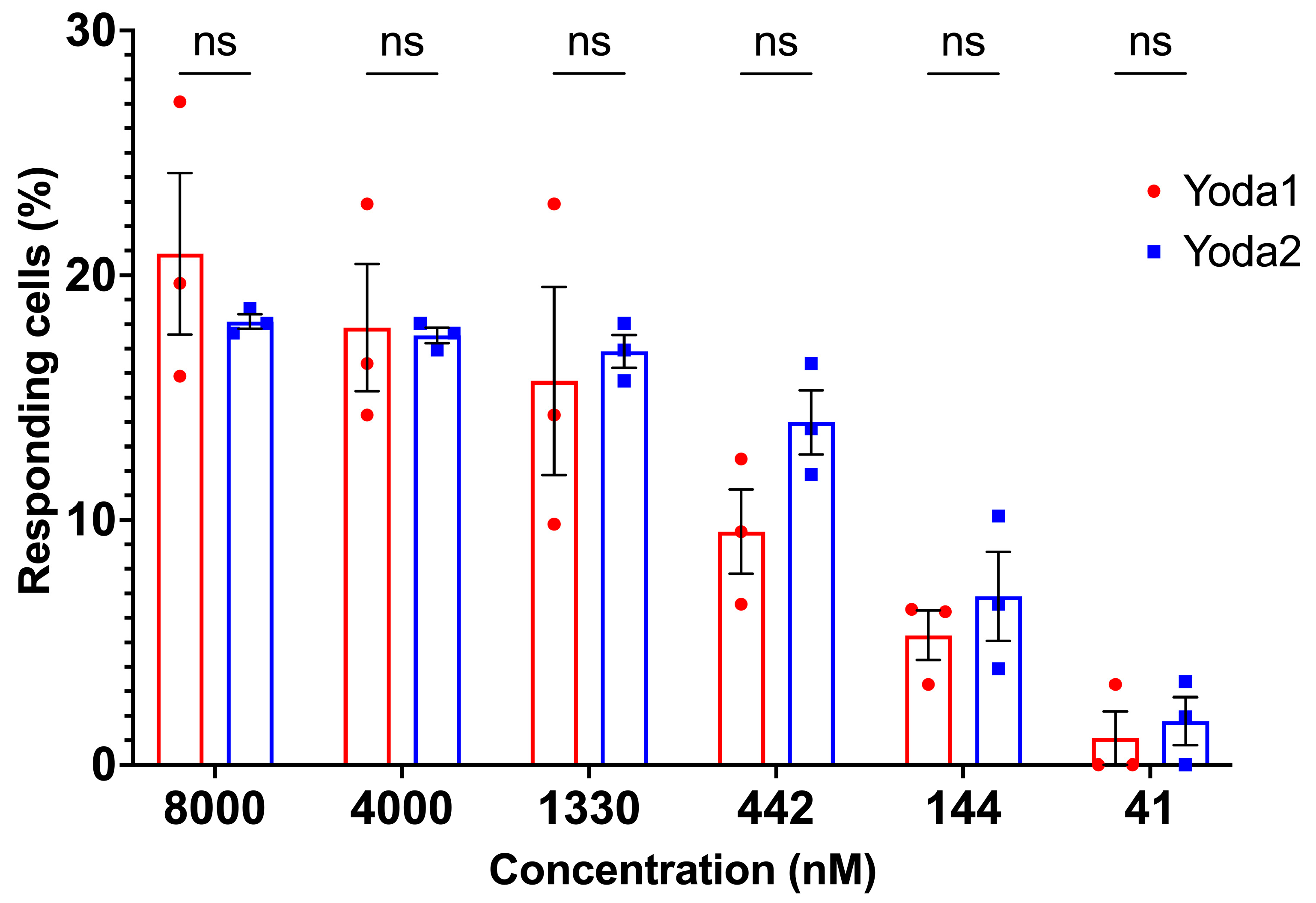
3.2. Calcium Measurements
3.3. Membrane Potential Measurements
4. Discussion
- (i)
- The Ca2+ entry, and hence the fluorescence of the dye, accumulates over time, i.e., the kinetic properties of Piezo1 or, more precisely, the kinetic modulation of Piezo1 by the different Yoda variants come into play.
- (ii)
- There are further players inside the RBC that influence the free Ca2+ concentration (in a possibly Ca2+ concentration-dependent manner), most notably the Ca2+ pumps (mainly PMCA4) [34].
- (i)
- Although all measurements relate to the activity of Piezo1, they are differently linked to the channel activity, as already discussed above. Patch clamp is the direct measurement, the Ca2+ increase is the response of the channel opening but modulated by other factors, and the activity of the Gárdos channel is even further downstream with the potential for further modulations.
- (ii)
- The nature of the measurements is vastly different. While patch clamp and Ca2+ measurements are single-cell techniques, the MBE method is a cell population measurement presenting an average value of all cells, whereas in the patch clamp recordings, non-responding cells are not considered. In the whole-cell configuration, the intracellular compartment is connected to a reservoir containing the internal solution, resulting in a wash-out of the cytosol. In contrast, Ca2+ and MBE measurements are performed on intact cells, just modulated by the abundance of Fluo-4 in the cytosol or CCCP in the cell membrane, respectively.
- (iii)
- The concrete experimental conditions can be different. This is less about different personnel or laboratory locations, but rather about the fact that different methods require different conditions. This can be the composition of the ionic solutions, e.g., patch clamp recordings require a high fluoride concentration (110 mM) for an efficient seal formation, or the nature of the MBE method requires a pH-unbuffered solution. Also, patch clamp recordings and Ca2+ measurements are performed at room temperature (due to historical reasons and technical limitations, respectively), whereas the MBE method needs to be performed at 37 °C. Furthermore, for all three types of measurements, the effect of the Yoda compounds is convoluted with mechanical stress on the RBCs. All three major methods applied (patch-clamp technique, flow cytometry, and MBE method) induce some but different mechanical stress to the RBCs (cell suction-induced membrane curvature change, flow in tubes and chambers, stirring of cell suspension, respectively). Although this stress was present in the control condition (without Yoda stimulation), we cannot exclude a different effect of the mechanical stress on the Yoda-induced activity of Piezo1.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| a.u. | arbitrary units |
| DMSO | Di-Methyl-Sulf-Oxide |
| MBE | Macey–Bennekou–Egée |
| ns | not significant |
| RBCs | red blood cells |
| RCF | relative centrifugal force |
Appendix A
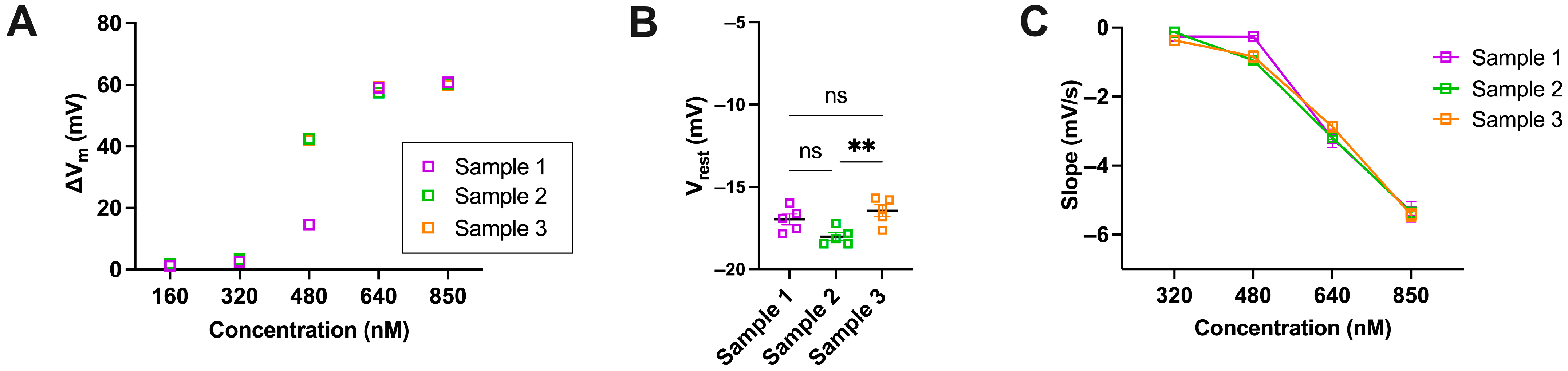
Additional Data Referring to the Patch Clamp Measurements
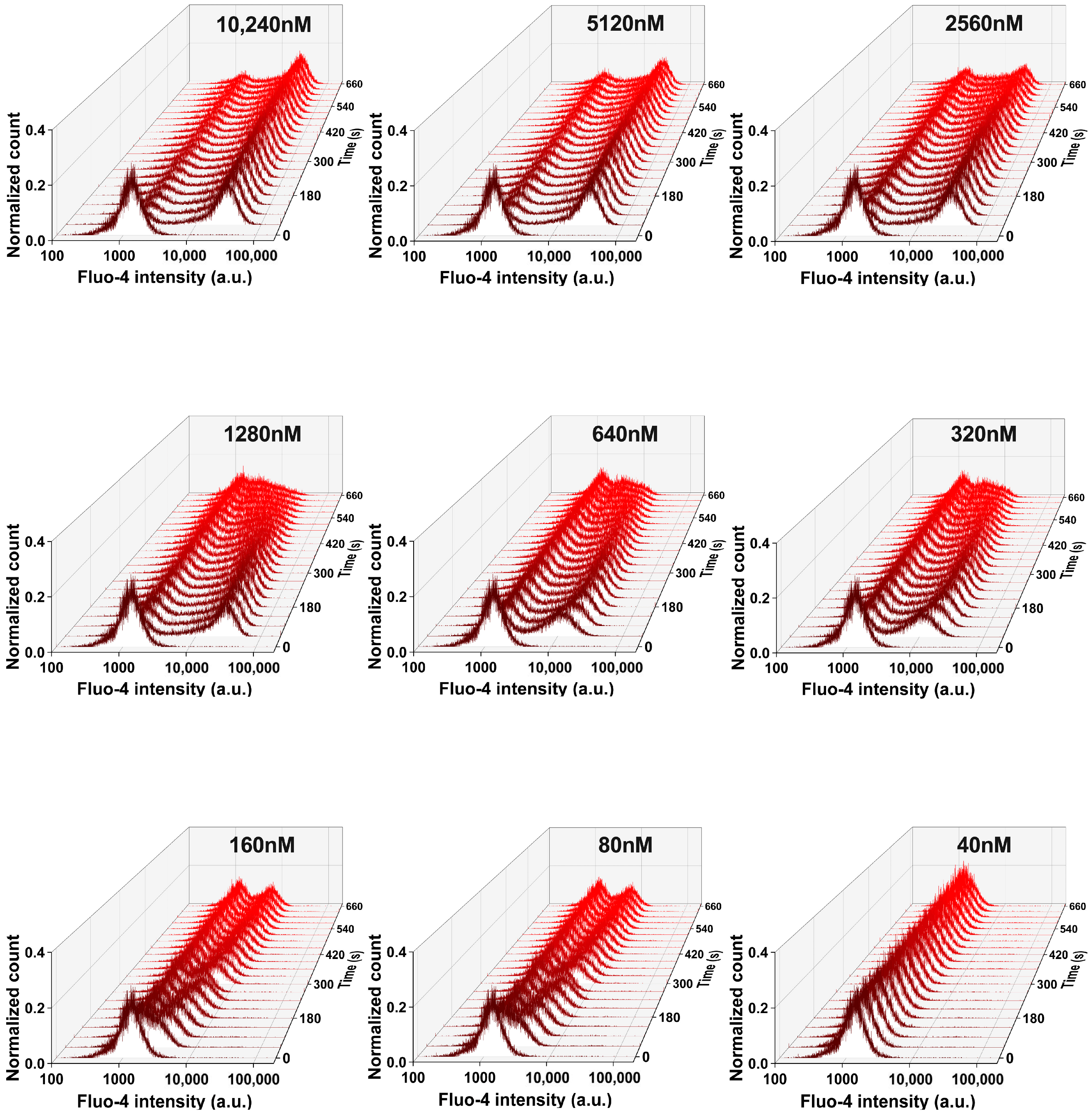
Appendix B
Additional Data Referring to Ca2+ Measurements
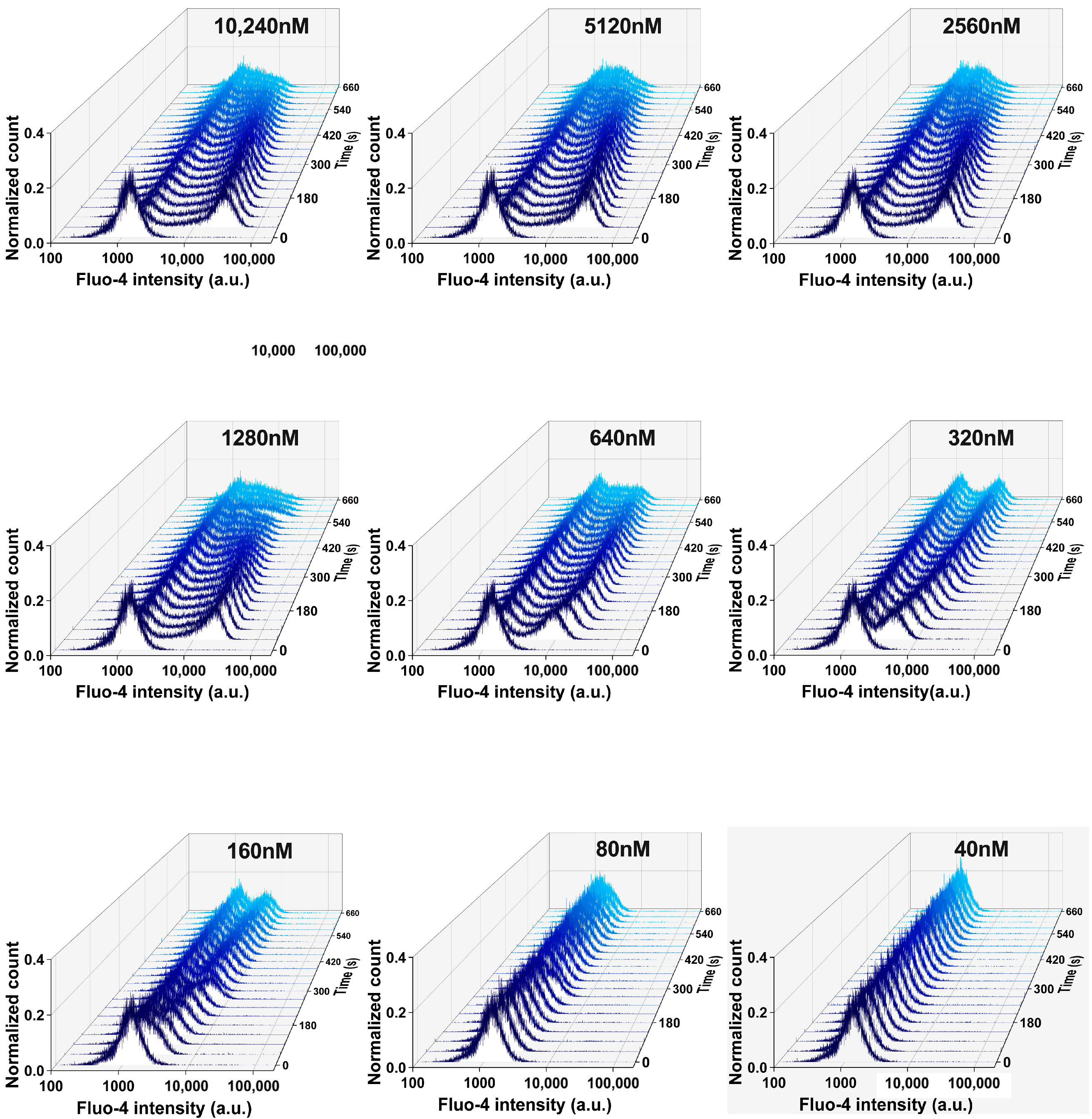
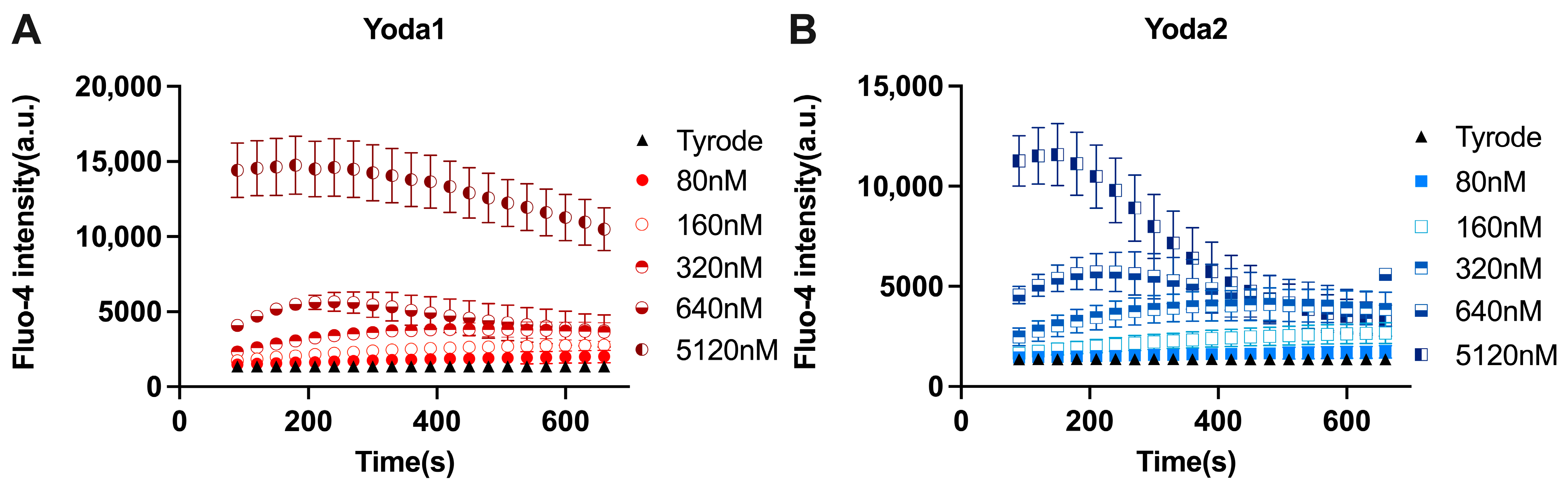
References
- Coste, B.; Mathur, J.; Schmidt, M.; Earley, T.J.; Ranade, S.; Petrus, M.J.; Dubin, A.E.; Patapoutian, A. Piezo1 and Piezo2 Are Essential Components of Distinct Mechanically Activated Cation Channels. Science 2010, 330, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Coste, B.; Xiao, B.; Santos, J.S.; Syeda, R.; Grandl, J.; Spencer, K.S.; Kim, S.E.; Schmidt, M.; Mathur, J.; Dubin, A.E.; et al. Piezo Proteins Are Pore-Forming Subunits of Mechanically Activated Channels. Nature 2012, 483, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Ranade, S.S.; Qiu, Z.; Woo, S.-H.; Hur, S.S.; Murthy, S.E.; Cahalan, S.M.; Xu, J.; Mathur, J.; Bandell, M.; Coste, B.; et al. Piezo1, a Mechanically Activated Ion Channel, Is Required for Vascular Development in Mice. Proc. Natl. Acad. Sci. USA 2014, 111, 10347–10352. [Google Scholar] [CrossRef]
- Gudipaty, S.A.; Lindblom, J.; Loftus, P.D.; Redd, M.J.; Edes, K.; Davey, C.F.; Krishnegowda, V.; Rosenblatt, J. Mechanical Stretch Triggers Rapid Epithelial Cell Division through Piezo1. Nature 2017, 543, 118–121. [Google Scholar] [CrossRef]
- He, L.; Si, G.; Huang, J.; Samuel, A.D.T.; Perrimon, N. Mechanical Regulation of Stem-Cell Differentiation by the Stretch-Activated Piezo Channel. Nature 2018, 555, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Martinac, B. Mechanisms of PIEZO Channel Inactivation. Int. J. Mol. Sci. 2023, 24, 14113. [Google Scholar] [CrossRef]
- Cahalan, S.M.; Lukacs, V.; Ranade, S.S.; Chien, S.; Bandell, M.; Patapoutian, A. Piezo1 Links Mechanical Forces to Red Blood Cell Volume. eLife 2015, 4, e07370. [Google Scholar] [CrossRef]
- Kuck, L.; Peart, J.N.; Simmonds, M.J. Piezo1 Regulates Shear-Dependent Nitric Oxide Production in Human Erythrocytes. Am. J. Physiol.-Heart Circ. Physiol. 2022, 323, H24–H37. [Google Scholar] [CrossRef]
- Lew, V.L. The Calcium Homeostasis of Human Red Blood Cells in Health and Disease: Interactions of PIEZO1, the Plasma Membrane Calcium Pump, and Gardos Channels. Annu. Rev. Physiol. 2025, 87, 257–277. [Google Scholar] [CrossRef]
- Zarychanski, R.; Schulz, V.P.; Houston, B.L.; Maksimova, Y.; Houston, D.S.; Smith, B.; Rinehart, J.; Gallagher, P.G. Mutations in the Mechanotransduction Protein PIEZO1 Are Associated with Hereditary Xerocytosis. Blood 2012, 120, 1908–1915. [Google Scholar] [CrossRef]
- Albuisson, J.; Murthy, S.E.; Bandell, M.; Coste, B.; Louis-dit-Picard, H.; Mathur, J.; Fénéant-Thibault, M.; Tertian, G.; De Jaureguiberry, J.-P.; Syfuss, P.-Y.; et al. Dehydrated Hereditary Stomatocytosis Linked to Gain-of-Function Mutations in Mechanically Activated PIEZO1 Ion Channels. Nat. Commun. 2013, 4, 1884. [Google Scholar] [CrossRef]
- Rotordam, M.G.; Fermo, E.; Becker, N.; Barcellini, W.; Brüggemann, A.; Fertig, N.; Egée, S.; Rapedius, M.; Bianchi, P.; Kaestner, L. A Novel Gain-of-Function Mutation of Piezo1 Is Functionally Affirmed in Red Blood Cells by High-Throughput Patch Clamp. Haematologica 2019, 104, e179–e183. [Google Scholar] [CrossRef]
- Jankovsky, N.; Caulier, A.; Demagny, J.; Guitton, C.; Djordjevic, S.; Lebon, D.; Ouled-Haddou, H.; Picard, V.; Garçon, L. Recent Advances in the Pathophysiology of PIEZO1-related Hereditary Xerocytosis. Am. J. Hematol. 2021, 96, 1017–1026. [Google Scholar] [CrossRef]
- Dryllis, G.; Russo, R.; Andolfo, I.; Iolascon, A.; Rosato, B.E.; Konstantopoulos, K. Hereditary Hemolytic Anemia Due to PIEZO1 Red Blood Cell Membrane Defect. Hemoglobin 2024, 48, 357–359. [Google Scholar] [CrossRef]
- Cox, C.D.; Bae, C.; Ziegler, L.; Hartley, S.; Nikolova-Krstevski, V.; Rohde, P.R.; Ng, C.-A.; Sachs, F.; Gottlieb, P.A.; Martinac, B. Removal of the Mechanoprotective Influence of the Cytoskeleton Reveals PIEZO1 Is Gated by Bilayer Tension. Nat. Commun. 2016, 7, 10366. [Google Scholar] [CrossRef]
- Lewis, A.H.; Grandl, J. Mechanical Sensitivity of Piezo1 Ion Channels Can Be Tuned by Cellular Membrane Tension. eLife 2015, 4, e12088. [Google Scholar] [CrossRef]
- Ridone, P.; Pandzic, E.; Vassalli, M.; Cox, C.D.; Macmillan, A.; Gottlieb, P.A.; Martinac, B. Disruption of Membrane Cholesterol Organization Impairs the Activity of PIEZO1 Channel Clusters. J. Gen. Physiol. 2020, 152, e201912515. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Allegrini, B.; Rapetti-Mauss, R.; Picard, V.; Garçon, L.; Kohl, P.; Soriani, O.; Peyronnet, R.; Guizouarn, H. Hereditary Xerocytosis: Differential Behavior of PIEZO1 Mutations in the N-Terminal Extracellular Domain Between Red Blood Cells and HEK Cells. Front. Physiol. 2021, 12, 736585. [Google Scholar] [CrossRef] [PubMed]
- Caulier, A.; Garçon, L. PIEZO1, Sensing the Touch during Erythropoiesis. Curr. Opin. Hematol. 2022, 29, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Garcia, L.R. Regulation of PIEZO Channels by Membrane Lipids. Ph.D. Thesis, University of Tennessee Health Science Center, Memphis, TN, USA, 2024. [Google Scholar] [CrossRef]
- Syeda, R.; Xu, J.; Dubin, A.E.; Coste, B.; Mathur, J.; Huynh, T.; Matzen, J.; Lao, J.; Tully, D.C.; Engels, I.H.; et al. Chemical Activation of the Mechanotransduction Channel Piezo1. eLife 2015, 4, e07369. [Google Scholar] [CrossRef] [PubMed]
- Botello-Smith, W.M.; Zhang, H.; Ozkan, A.D.; Jiang, W.; Pham, C.N.; Luo, Y.; Lacroix, J.J. Structural Bases for Chemical and Mechanical Gating in the Piezo1 Channel. Biophys. J. 2019, 116, 478a–479a. [Google Scholar] [CrossRef]
- Yang, S.; Miao, X.; Arnold, S.; Li, B.; Ly, A.T.; Wang, H.; Wang, M.; Guo, X.; Pathak, M.M.; Zhao, W.; et al. Membrane Curvature Governs the Distribution of Piezo1 in Live Cells. Nat. Commun. 2022, 13, 7467. [Google Scholar] [CrossRef] [PubMed]
- Petkova-Kirova, P.; Murciano, N.; Iacono, G.; Jansen, J.; Simionato, G.; Qiao, M.; Van Der Zwaan, C.; Rotordam, M.G.; John, T.; Hertz, L.; et al. The Gárdos Channel and Piezo1 Revisited: Comparison between Reticulocytes and Mature Red Blood Cells. Int. J. Mol. Sci. 2024, 25, 1416. [Google Scholar] [CrossRef]
- Hatem, A.; Esperti, S.; Murciano, N.; Qiao, M.; Giustina Rotordam, M.; Becker, N.; Nader, E.; Maurer, F.; Pérès, L.; Bouyer, G.; et al. Adverse Effects of Delta-9-tetrahydrocannabinol on Sickle Red Blood Cells. Am. J. Hematol. 2023, 98, E383–E386. [Google Scholar] [CrossRef]
- Macey, R.I.; Adorante, J.S.; Orme, F.W. Erythrocyte Membrane Potentials Determined by Hydrogen Ion Distribution. Biochim. Et. Biophys. Acta BBA Biomembr. 1978, 512, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Baunbæk, M.; Bennekou, P. Evidence for a Random Entry of Ca2+ into Human Red Cells. Bioelectrochemistry 2008, 73, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Alonso, D.M.; Pérès, L.; Hatem, A.; Bouyer, G.; Egée, S. The Chloride Conductance Inhibitor NS3623 Enhances the Activity of a Non-Selective Cation Channel in Hyperpolarizing Conditions. Front. Physiol. 2021, 12, 743094. [Google Scholar] [CrossRef]
- Jansen, J.; Qiao, M.; Hertz, L.; Wang, X.; Fermo, E.; Zaninoni, A.; Colombatti, R.; Bernhardt, I.; Bianchi, P.; Kaestner, L. Mechanistic Ion Channel Interactions in Red Cells of Patients with Gárdos Channelopathy. Blood. Adv. 2021, 5, 3303–3308. [Google Scholar] [CrossRef]
- Danielczok, J.G.; Terriac, E.; Hertz, L.; Petkova-Kirova, P.; Lautenschläger, F.; Laschke, M.W.; Kaestner, L. Red Blood Cell Passage of Small Capillaries Is Associated with Transient Ca2+-Mediated Adaptations. Front. Physiol. 2017, 8, 979. [Google Scholar] [CrossRef]
- Wadud, R.; Hannemann, A.; Rees, D.C.; Brewin, J.N.; Gibson, J.S. Yoda1 and Phosphatidylserine Exposure in Red Cells from Patients with Sickle Cell Anaemia. Sci. Rep. 2020, 10, 20110. [Google Scholar] [CrossRef]
- Hatem, A.; Poussereau, G.; Gachenot, M.; Pérès, L.; Bouyer, G.; Egée, S. Dual Action of Dooku1 on PIEZO1 Channel in Human Red Blood Cells. Front. Physiol. 2023, 14, 1222983. [Google Scholar] [CrossRef]
- Bogdanova, A.; Makhro, A.; Wang, J.; Lipp, P.; Kaestner, L. Calcium in Red Blood Cells—A Perilous Balance. Int. J. Mol. Sci. 2013, 14, 9848–9872. [Google Scholar] [CrossRef] [PubMed]
- Kaestner, L.; Bogdanova, A.; Egee, S. Calcium Channels and Calcium-Regulated Channels in Human Red Blood Cells. Adv. Exp. Med. Biol. 2020, 1131, 625–648. [Google Scholar] [PubMed]
- Faucherre, A.; Kissa, K.; Nargeot, J.; Mangoni, M.E.; Jopling, C. Piezo1 Plays a Role in Erythrocyte Volume Homeostasis. Haematologica 2014, 99, 70–75. [Google Scholar] [CrossRef]
- Bae, C.; Sachs, F.; Gottlieb, P.A. Protonation of the Human PIEZO1 Ion Channel Stabilizes Inactivation. J. Biol. Chem. 2015, 290, 5167–5173. [Google Scholar] [CrossRef]
- Glogowska, E.; Schneider, E.R.; Maksimova, Y.; Schulz, V.P.; Lezon-Geyda, K.; Wu, J.; Radhakrishnan, K.; Keel, S.B.; Mahoney, D.; Freidmann, A.M.; et al. Novel Mechanisms of PIEZO1 Dysfunction in Hereditary Xerocytosis. Blood 2017, 130, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
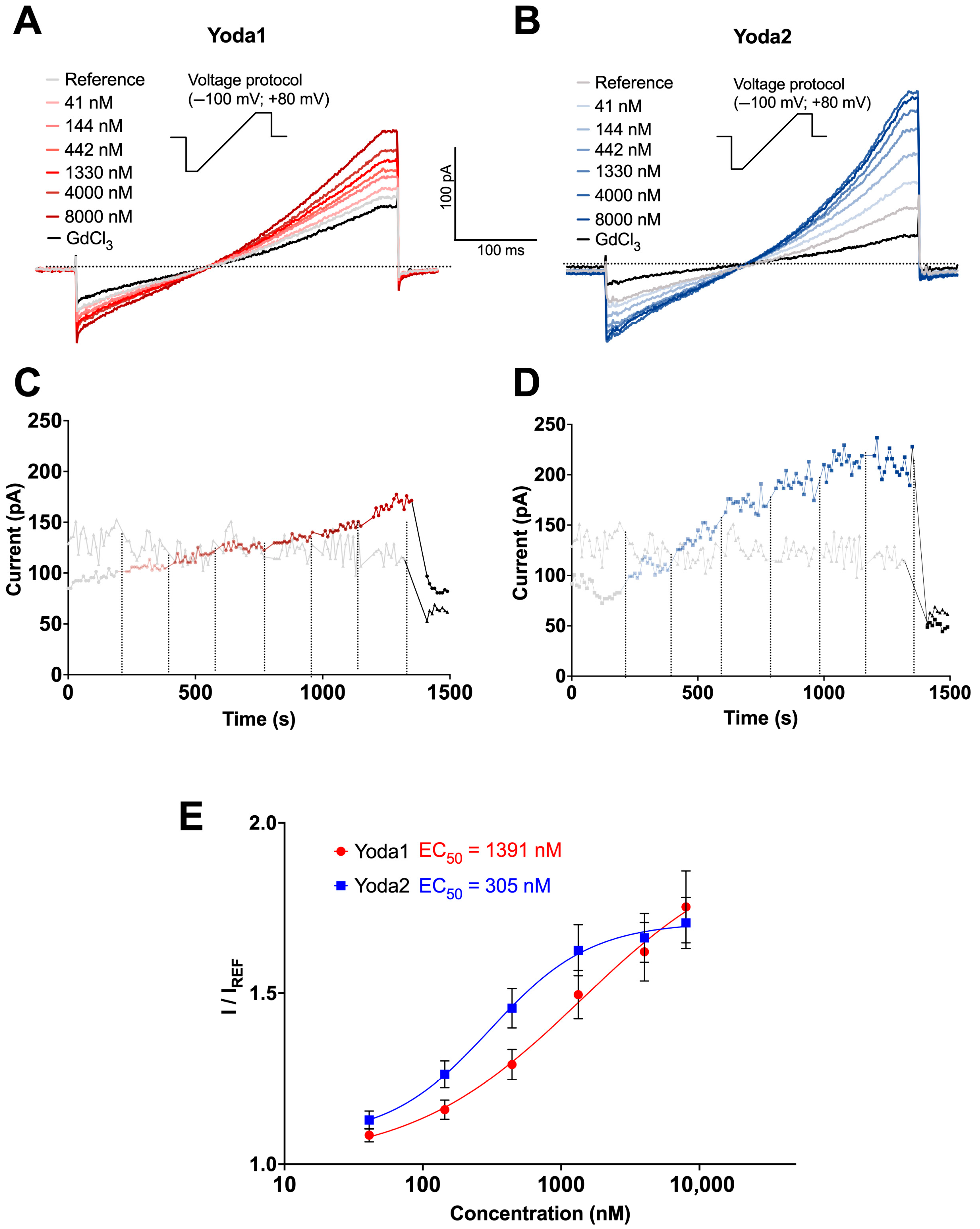
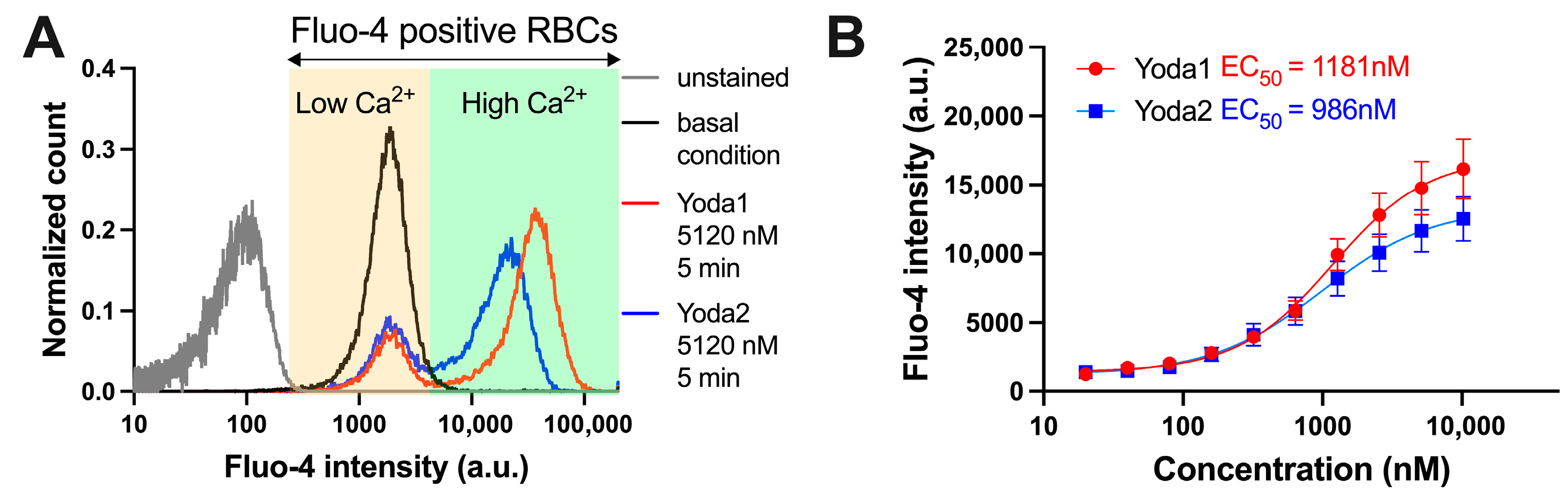
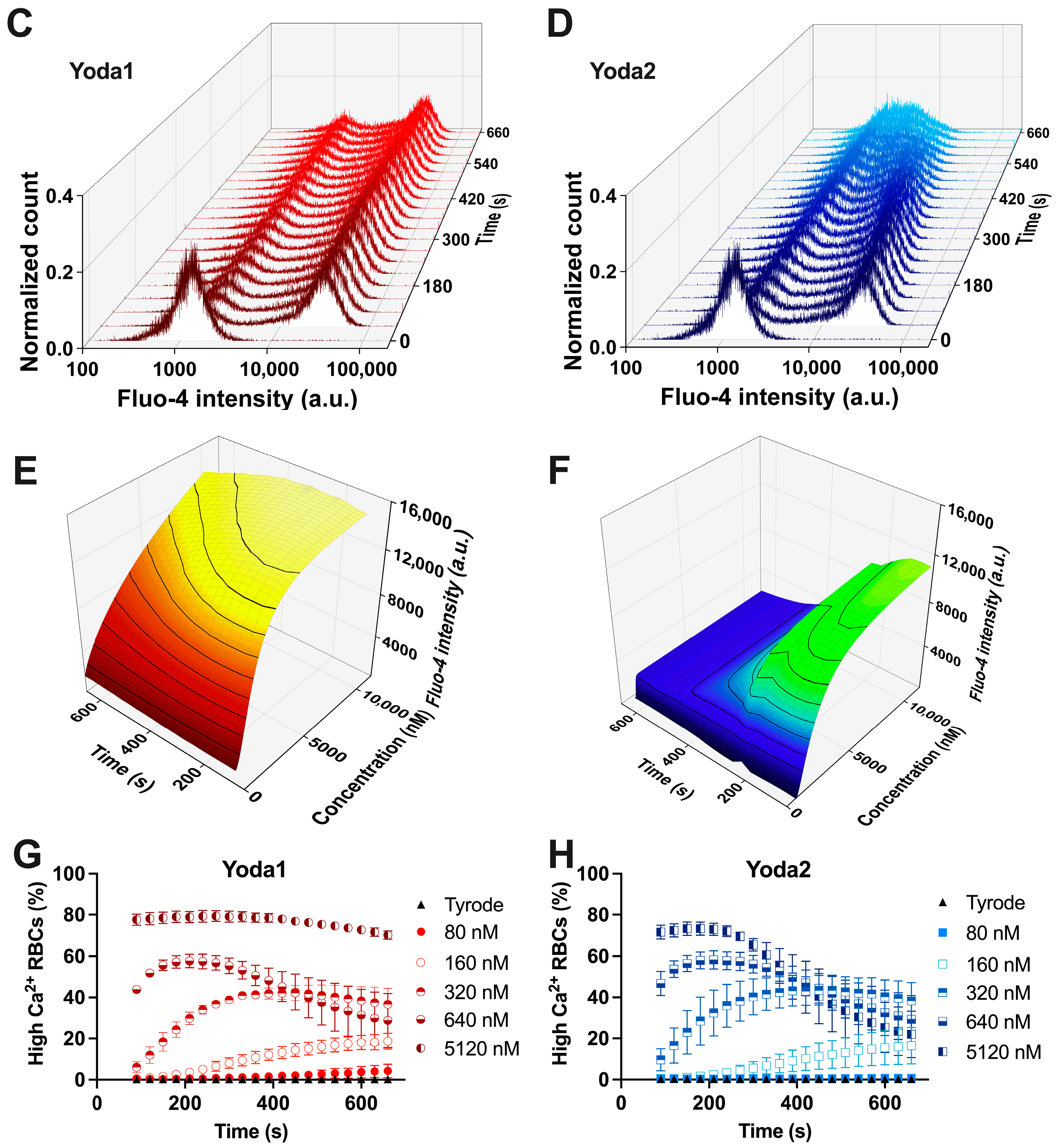
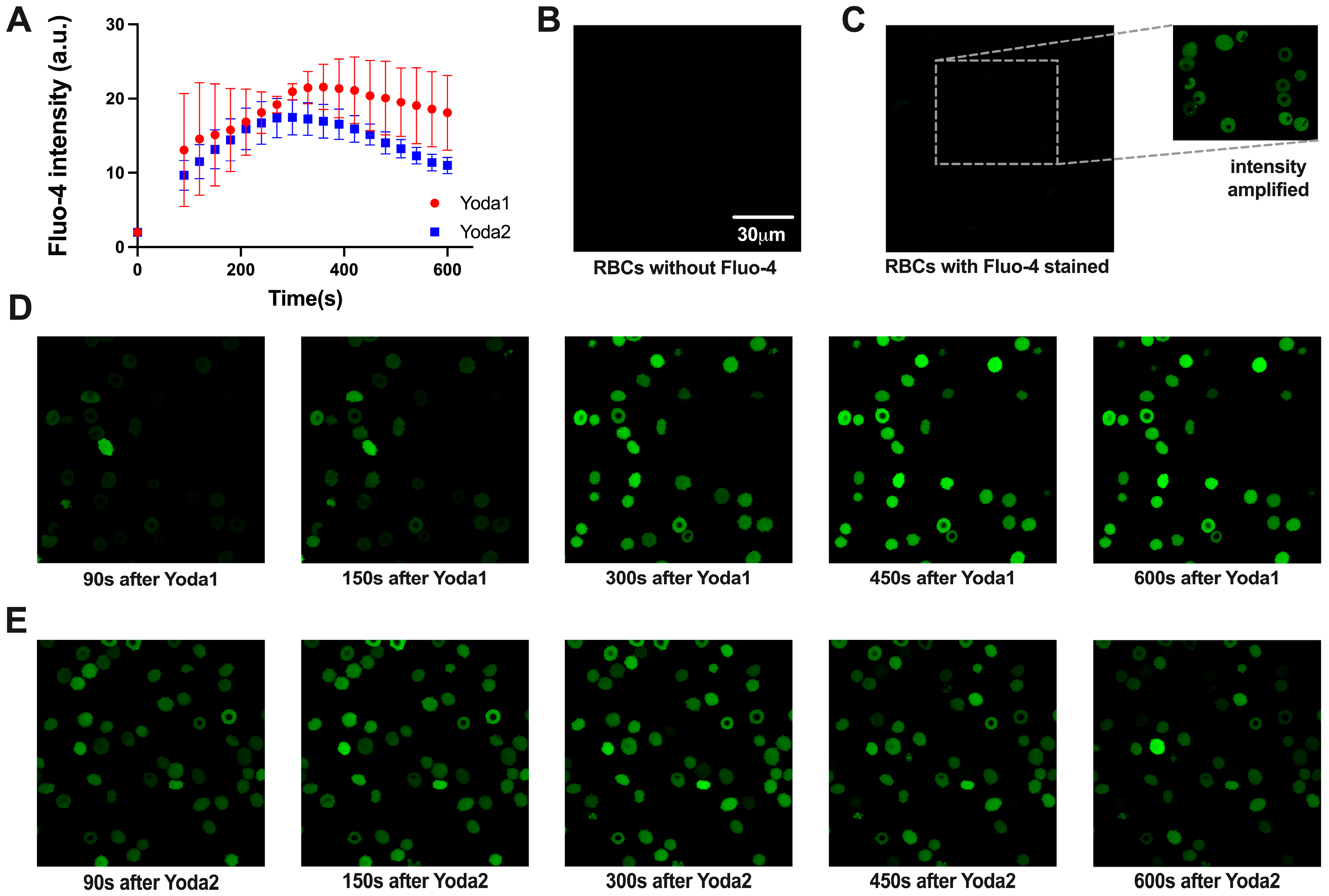
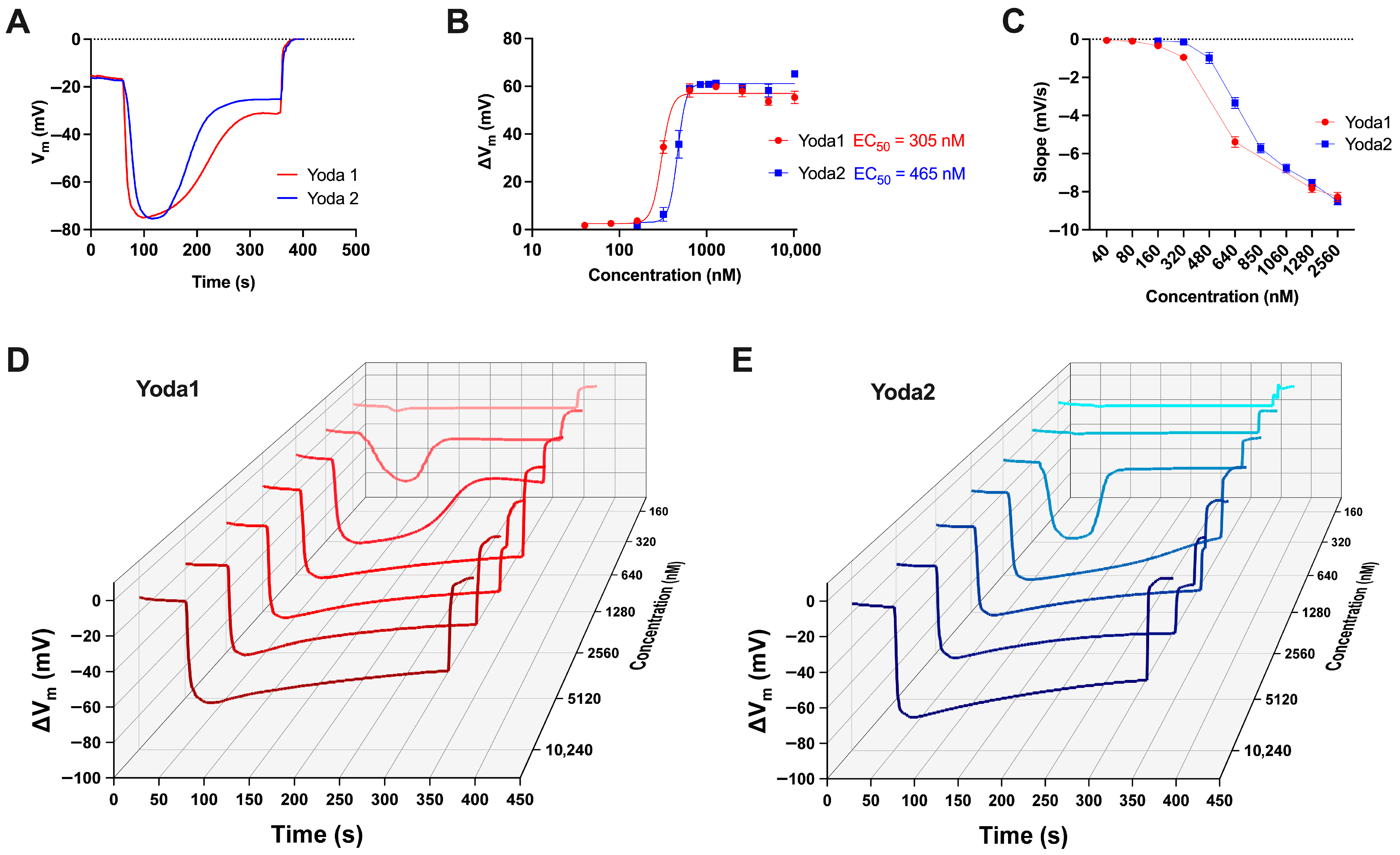
| Compound | EC50 (Patch Clamp) [nM] | EC50 (Ca2+ Measurements) [nM] | EC50 (Membrane Potential) [nM] |
|---|---|---|---|
| Yoda1 | 1391 | 1181 | 305 |
| Yoda2 | 305 | 986 | 465 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, M.; Penttinen, R.; Coli, A.; Murciano, N.; Maurer, F.M.; Wagner, C.; Rotordam, M.G.; Kaestner, L. Piezo1 Channel Activators Yoda1 and Yoda2 in the Context of Red Blood Cells. Biomolecules 2025, 15, 1110. https://doi.org/10.3390/biom15081110
Qiao M, Penttinen R, Coli A, Murciano N, Maurer FM, Wagner C, Rotordam MG, Kaestner L. Piezo1 Channel Activators Yoda1 and Yoda2 in the Context of Red Blood Cells. Biomolecules. 2025; 15(8):1110. https://doi.org/10.3390/biom15081110
Chicago/Turabian StyleQiao, Min, Reetta Penttinen, Ariel Coli, Nicoletta Murciano, Felix M. Maurer, Christian Wagner, Maria Giustina Rotordam, and Lars Kaestner. 2025. "Piezo1 Channel Activators Yoda1 and Yoda2 in the Context of Red Blood Cells" Biomolecules 15, no. 8: 1110. https://doi.org/10.3390/biom15081110
APA StyleQiao, M., Penttinen, R., Coli, A., Murciano, N., Maurer, F. M., Wagner, C., Rotordam, M. G., & Kaestner, L. (2025). Piezo1 Channel Activators Yoda1 and Yoda2 in the Context of Red Blood Cells. Biomolecules, 15(8), 1110. https://doi.org/10.3390/biom15081110







