The Multifaceted Role of miR-211 in Health and Disease
Abstract
1. Introduction
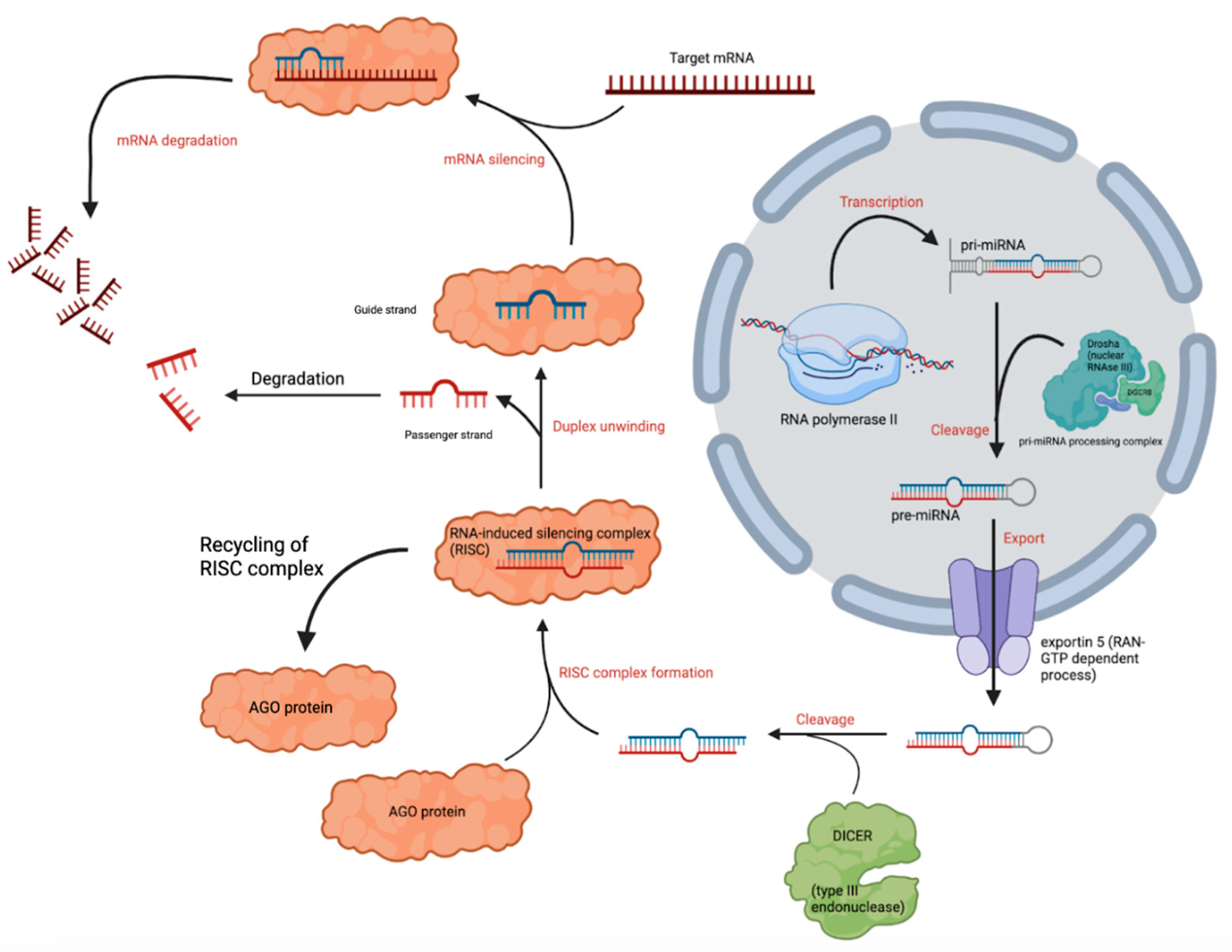
2. Biogenesis of miRNA Variants
3. Role of miR-211 in Normal Human Biology and Physiology
3.1. miR-211 Regulation of TGF-β Signaling and Cell Cycle Control
3.2. miR-211 Targets in Chromatin Regulation and PI3K/AKT Signaling
| Gene | Pathological Interaction with miR-211 | Normal Function | Tissue Expression | Primary Cell Types | Ref. |
|---|---|---|---|---|---|
| KCNMA1 | Upregulated; promotes cancer progression | Regulates membrane potential and Ca2+ signaling | Brain, smooth muscle, endocrine tissues | Neurons, smooth muscle cells, adrenal gland cells | [33] |
| IGF2R | Upregulated; enhances tumor growth | Mediates uptake of IGF-2 | Liver, kidney, muscle | Hepatocytes, renal tubules, myocytes | [34] |
| TGFBR2 | Increased expression contributes to metastasis | Receptor in TGF-β pathway; regulates growth and differentiation | Lung, liver, heart, immune cells | Alveolar cells, hepatocytes, cardiomyocytes, T cells | [35] |
| TCF12 | Upregulated; promotes cancer progression | Transcriptional regulation of development | Embryonic and adult tissues | Stem cells, differentiating cells | [53,54,55] |
| SOX11 | Elevated in tumors | Neurogenesis and differentiation | Developing nervous system | Neural progenitors, differentiating neurons | [50] |
| SOX4 | Upregulated; contributes to metastasis | Cell fate determination | Bone marrow, lymphoid tissue | HSCs, lymphocytes, mesenchymal cells | [50] |
| SPARC | Promotes cancer migration | Cell–matrix remodeling | Bone, skin, connective tissues | Osteoblasts, fibroblasts, stromal cells | [51] |
| SNAI1 | Induces EMT | EMT regulation | Embryonic and tumor tissues | Epithelial cells, cancer stem cells | [24] |
| ZEB2 | Upregulated; drives EMT and metastasis | EMT and neural crest development | Neural crest, epithelia | Neural crest cells, epithelial cells | [24] |
| ACSL4 | Elevated expression affects lipid metabolism in cancer | Long-chain fatty acid activation | Liver, brain, adipose tissue | Hepatocytes, neurons, adipocytes | [52] |
| SSRP1 | Promotes chromatin remodeling in tumor cells | Chromatin regulation via FACT complex | Proliferating tissues | Tumor cells, chromatin-regulating cells | [44,45,46,47] |
| Runx2 | Upregulated; contributes to osteosarcoma | Osteoblast differentiation and bone formation | Bone, cartilage | Osteoblasts, chondrocytes | [31,56,57,58] |
| Signaling Pathway | Validated or Predicted Target(s) | Biological Context | Functional Consequence | Reference(s) |
|---|---|---|---|---|
| TGF-β/SMAD | TGFBR2 | Melanocytes, renal epithelium, T cells | Represses TGF-β signaling, limits SMAD2/3 activation, prevents fibrosis and apoptosis | [35,38,40] |
| PI3K/AKT | PI3K-associated factors (e.g., via SSRP1) | Synoviocytes, chondrocytes, epithelial cells | Reduces AKT activation; controls inflammation and abnormal proliferation | [48,49] |
| Cell Cycle Regulation | Cyclin D1, CDK6, CDC25B | Cancer cells, synoviocytes | Induces G0/G1 arrest; inhibits proliferation and promotes cell cycle checkpoint activation | [24,25,26,27] |
| Chromatin Remodeling | SSRP1 | RA synoviocytes, epithelial progenitors | Inhibits FACT complex; restrains NF-κB and p53 pathway activation | [44,59] |
| EMT/Metastasis | SNAI1, ZEB2, SPARC | Cancer (e.g., cervical, renal, oral) | Inhibits EMT, reduces cell migration and metastatic potential | [24,51] |
| Metabolic Regulation | ACSL4, pyruvate metabolism enzymes | Melanoma, retina | Regulates oxidative metabolism; protects against metabolic stress and ferroptosis | [52,60,61] |
| BMP2/TGF-β Superfamily | BMP2 | Pancreatic cancer | Suppresses tumor growth and invasion | [50] |
| STAT3 (indirect) | Indirect via SSRP1 and PI3K/AKT | Synovial fibroblasts (RA) | Reduces inflammatory cytokines; restores apoptosis sensitivity | [59] |
3.3. miR-211 and Bone Physiology
3.4. miR-211 and Human Eye Development
4. Role of miR-211 in Disease Pathology
4.1. miR-211’s Role in Renal Hypoxia/Reoxygenation and Ischemia/Reperfusion Injury
4.2. miR-211’s Role in Atherosclerosis and Vascular Calcification
4.3. miR-211 Immunological Disease
4.4. miR-211 in Cancer
5. Future Directions and Limitations
5.1. miR-211: Therapeutic Potential and Translational Challenges
5.2. Dual Roles of miR-211 in Cancer: Context-Dependent Mechanisms
5.3. Extracellular Vesicle-Based Delivery of miR-211 and Other Therapeutic miRNAs
5.4. Gaps in Knowledge of miR-211 Biology
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Miska, E.A.; Alvarez-Saavedra, E.; Abbott, A.L.; Lau, N.C.; Hellman, A.B.; McGonagle, S.M.; Bartel, D.P.; Ambros, V.R.; Horvitz, H.R. Most Caenorhabditis elegans microRNAs Are Individually Not Essential for Development or Viability. PLoS Genet. 2007, 3, e215. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Gurtan, A.M.; Sharp, P.A. The role of miRNAs in regulating gene expression networks. J. Mol. Biol. 2013, 425, 3582–3600. [Google Scholar] [CrossRef]
- Jame-Chenarboo, F.; Ng, H.H.; Macdonald, D.; Mahal, L.K. High-Throughput Analysis Reveals miRNA Upregulating α-2,6-Sialic Acid through Direct miRNA–mRNA Interactions. ACS Cent. Sci. 2022, 8, 1527–1536. [Google Scholar] [CrossRef]
- Bai, J.; Xiao, R.; Jiang, D.; Luo, X.; Tang, Y.; Cui, M.; You, L.; Zhao, Y. Sialic Acids: Sweet modulators fueling cancer cells and domesticating the tumor microenvironment. Cancer Lett. 2025, 596, 217773. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear Export of MicroRNA Precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.-O.; Tomari, Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef]
- Zamudio, J.R.; Kelly, T.J.; Sharp, P.A. Argonaute-bound small RNAs from promoter-proximal RNA Polymerase II. Cell 2014, 156, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 Mediates RNA Cleavage Targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef]
- Hiers, N.M.; Li, T.; Traugot, C.M.; Xie, M. Target-directed microRNA degradation: Mechanisms, significance, and functional implications. Trends Biochem. Sci. 2024, 49, 433–447. [Google Scholar] [CrossRef]
- Pawlica, P.; Sheu-Gruttadauria, J.; MacRae, I.J.; Steitz, J.A. How complementary targets expose the microRNA 3′ end for tailing and trimming during target-directed microRNA degradation. Mol. Cell 2019, 75, 1247–1261.e5. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Nam, J.-W.; Heo, I.; Rhee, J.-K.; Sohn, S.Y.; Cho, Y.; Zhang, B.-T.; Kim, V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef]
- Tomasello, L.; Distefano, R.; Nigita, G.; Croce, C.M. The MicroRNA Family Gets Wider: The IsomiRs Classification and Role. Front. Cell Dev. Biol. 2021, 9, 668648. [Google Scholar] [CrossRef] [PubMed]
- Bofill-De Ros, X.; Kasprzak, W.K.; Bhandari, Y.; Fan, L.; Cavanaugh, Q.; Jiang, M.; Dai, L.; Yang, A.; Shao, T.J.; Shapiro, B.A.; et al. Structural Differences between Pri-miRNA Paralogs Promote Alternative Drosha Cleavage and Expand Target Repertoires. Cell Rep. 2019, 26, 447–459.e4. [Google Scholar] [CrossRef]
- Wang, J.-W.; Zhang, W.; Zhang, Y.; Zhou, J.; Li, J.; Zhang, M.; Wen, S.; Gao, X.; Zhou, N.; Li, H.; et al. Reproducible and high sample throughput isomiR next-generation sequencing for cancer diagnosis. J. Clin. Oncol. 2024, 42, e15013. [Google Scholar] [CrossRef]
- Ye, L.; Wang, F.; Wang, J.; Wu, H.; Yang, H.; Yang, Z.; Huang, H. Role and mechanism of miR-211 in human cancer. J Cancer 2022, 13, 2933–2944. [Google Scholar] [CrossRef]
- Xia, B.; Yang, S.; Liu, T.; Lou, G. miR-211 suppresses epithelial ovarian cancer proliferation and cell-cycle progression by targeting Cyclin D1 and CDK6. Mol. Cancer 2015, 14, 57. [Google Scholar] [CrossRef]
- Duan, X.; Wen, J.; Wang, Y.; Wang, L.; Zhang, Y.; Wu, Y.; Yu, X. microRNA-211 regulates cell proliferation, apoptosis and migration/invasion in human osteosarcoma cells via targeting EZRIN. Cell. Mol. Biol. Lett. 2019, 24, 48. [Google Scholar]
- NCBI Gene. MIR211 microRNA 211 [Homo sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/gene/406993 (accessed on 25 June 2025).
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Barbato, S.; Marrocco, E.; Intartaglia, D.; Pizzo, M.; Asteriti, S.; Naso, F.; Falanga, D.; Bhat, R.S.; Meola, N.; Carissimo, A.; et al. MiR-211 is essential for adult cone photoreceptor maintenance and visual function. Sci. Rep. 2017, 7, 17004. [Google Scholar] [CrossRef]
- Matsukawa, T.; Sakai, T.; Hiraiwa, H.; Higashiyama, R.; Fujita, K.; Tsubosaka, M.; Sato, T.; Ozaki, T. miR-211-5p contributes to chondrocyte differentiation by suppressing Fibulin-4 expression to play a role in osteoarthritis. J. Biochem. 2019, 166, 425–432. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, L.; Fan, Y.; Liao, L.; Ma, P.X.; Xiao, G.; Chen, D. The microRNAs miR-204 and miR-211 maintain joint homeostasis and protect against osteoarthritis progression. Nat. Commun. 2019, 10, 2876. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Cui, Y.; Yu, X.; Wu, Z.; Ding, G.; Cao, L. miR-211 suppresses hepatocellular carcinoma by downregulating SATB2. Oncotarget 2015, 6, 9457–9466. [Google Scholar] [CrossRef]
- Sun, T.; Yang, D.; Wu, Y.; Sheng, Q. The function of microRNA-211 expression in post-fracture bone cell apoptosis involving the transforming growth factor-β/phosphoinositide 3-kinase signaling pathway. J. Int. Med. Res. 2020, 48, 300060520926353. [Google Scholar] [CrossRef]
- Miller, J.P.; Melamed, N.; Deisseroth, K.; Salkoff, L.; Patel, M.K.; Kaczmarek, L.K.; Knaus, H.G.; Lacinová, L.; Lee, M.; Yeh, H.H.; et al. KCNMA1-linked channelopathy. J. Gen. Physiol. 2019, 151, 1173–1189. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Liu, J.; Kang, Y.; He, Y.; Liang, B.; Yang, P.; Yu, Z.; Zhang, Z.; Sun, L. miR-491-5p suppresses gastric cancer cell growth and migration through dual-targeting of EGFR and the IGF2/IGF1R signaling axis. Cell Death Dis. 2020, 11, 276. [Google Scholar] [CrossRef]
- Mazar, J.; DeYoung, K.; Khaitan, D.; Meister, E.; Almodovar, A.; Goydos, J.; Ray, A.; Perera, R.J. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. Mol. Cell 2010, 40, 841–849. [Google Scholar] [CrossRef]
- Levy, C.; Khaled, M.; Iliopoulos, D.; Janas, M.M.; Schubert, S.; Pinner, S.; Chen, P.-H.; Li, S.; Fletcher, A.L.; Yokoyama, S.; et al. Intronic miR-211 Assumes the Tumor Suppressive Function of Its Host Gene in Melanoma. Mol. Cell 2010, 40, 841–849. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, Y.; He, M.; Li, X.; Wang, R. Role of Circular RNAs in Pulmonary Fibrosis. Int. J. Mol. Sci. 2022, 23, 10493. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Lee, J.T.; Vergara, L.A.; Pienta, K.J.; Zhang, Y.E.; Sun, Y. Molecular imaging of TGFβ-induced Smad2/3 phosphorylation reveals a role for receptor tyrosine kinases in modulating TGFβ signaling. Clin. Cancer Res. 2011, 17, 7424–7432. [Google Scholar] [CrossRef]
- Bekenstein, U.; Mishra, N.; Milikovsky, D.Z.; Hanin, G.; Zelig, D.; Moran, A.; Barshack, I.; Shemer, A.; Yitzhaky, A.; Soreq, H. Dynamic changes in murine forebrain miR-211 expression associate with cholinergic imbalances and epileptiform activity. Proc. Natl. Acad. Sci. USA 2017, 114, E4996–E5005. [Google Scholar] [CrossRef]
- Chen, B.; Mu, C.; Zhang, Z.; He, X.; Liu, X. The love-hate relationship between TGF-β signaling and the immune system during development and tumorigenesis. Front. Immunol. 2022, 13, 891268. [Google Scholar] [CrossRef]
- Seoane, J.; Gomis, R.R. TGF-β Family Signaling in Tumor Suppression and Cancer Progression. Cold Spring Harb. Perspect. Biol. 2017, 9, a022277. [Google Scholar] [CrossRef]
- Güngör, M.Z.; Uysal, M.; Sentürk, Ş. The Bright and the Dark Side of TGF-β Signaling in Hepatocellular Carcinoma: Mechanisms, Dysregulation, and Therapeutic Implications. Cancers 2022, 14, 940. [Google Scholar] [CrossRef]
- Winkler, D.D.; Luger, K. The histone chaperone FACT: Structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 2011, 286, 18369–18374. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.X.; Dai, M.S.; Keller, D.M.; Lu, H. SSRP1 functions as a co-activator of the transcriptional activator p63. EMBO J. 2002, 21, 5487–5497. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Fan, L.; Wang, L.; Liu, Y.; Zhang, J.; Wang, B.; Chen, R.; Chen, X.; Zhuang, L.; Zhang, Y.; et al. SSRP1/SLC3A2 axis in arginine transport: A new target for overcoming immune evasion and tumor progression in peripheral T-cell lymphoma. Adv. Sci. 2025, 12, 202415698. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; He, K.; Luo, T.; Deng, Y.; Wang, H.; Liu, H.; Zhang, J.; Chen, K.; Xiao, J.; Duan, X.; et al. SSRP1 contributes to the malignancy of hepatocellular carcinoma and is negatively regulated by miR-497. Mol. Ther. 2016, 24, 903–914. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, S.; Li, Z.; Zhou, J. MicroRNA-211/BDNF axis regulates LPS-induced proliferation of normal human astrocyte through PI3K/AKT pathway. Biosci. Rep. 2017, 37, BSR20170755. [Google Scholar] [CrossRef]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef]
- Halloran, D.; Durbano, H.W.; Nohe, A. Bone Morphogenetic Protein-2 in Development and Bone Homeostasis. J. Dev. Biol. 2020, 8, 19. [Google Scholar] [CrossRef]
- Li, D.; Luo, C.; Deng, J.; Xu, Y.; Fu, S.; Liu, K.; Wu, J. MicroRNA 211-5p inhibits cancer cell proliferation and migration in pancreatic cancer by targeting BMP2. Aging 2023, 15, 14411–14421. [Google Scholar] [CrossRef]
- Yuan, M.; Mahmud, I.; Katsushima, K.; Joshi, K.; Saulnier, O.; Pokhrel, R.; Lee, B.; Liyanage, W.; Kunhiraman, H.; Stapleton, S.; et al. miRNA-211 maintains metabolic homeostasis in medulloblastoma through its target gene long-chain acyl-CoA synthetase 4. Acta Neuropathol. Commun. 2023, 11, 203. [Google Scholar] [CrossRef]
- An, L.; Li, X.; Yang, J. MicroRNA-211 attenuates cell proliferation in T-cell lymphoblastic lymphoma through targeting TCF12. Leuk Res. 2021, 110, 106653. [Google Scholar] [CrossRef]
- Chen, Y.-F.; Yang, C.C.; Kao, S.Y.; Li, C.J.; Lin, S.C.; Chang, K.W. MicroRNA-211 Enhances the Oncogenicity of Carcinogen-Induced Oral Carcinoma by Repressing TCF12 and Increasing Antioxidant Activity. Cancer Res. 2016, 76, 4872–4886. [Google Scholar] [CrossRef]
- Lee, C.-C.; Chen, W.S.; Chen, C.C.; Chen, L.L.; Lin, Y.S.; Fan, C.S.; Huang, T.S. TCF12 Protein Functions as Transcriptional Repressor of E-cadherin, and Its Overexpression Is Correlated with Metastasis of Colorectal Cancer. J. Biol. Chem. 2012, 287, 2798–2809. [Google Scholar] [CrossRef]
- Komori, T. Regulation of skeletal development and maintenance by Runx2 and Sp7. Int. J. Mol. Sci. 2024, 25, 10102. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, S.; McDonald, F. Runx2 and dental development. Eur. J. Oral Sci. 2006, 114, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Hallett, S.A.; Dixon, A.; Marrale, I.; Batoon, L.; Brenes, J.; Zhou, A.; Arbiv, A.; Kaartinen, V.; Allen, B.; Ono, W.; et al. RUNX2 is essential for maintaining synchondrosis chondrocytes and cranial base growth. Bone Res. 2025, 13, 57. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.-S.; Fan, K.-J.; Teng, H.; Chen, S.; Xu, B.-X.; Chen, D.; Wang, T.-Y. Mir204 and Mir211 suppress synovial inflammation and proliferation in rheumatoid arthritis by targeting Ssrp1. eLife 2022, 11, e78085. [Google Scholar] [CrossRef]
- Aït-Ali, N.; Fridlich, R.; Millet-Puel, G.; Clérin, E.; Delalande, F.; Jaillard, C.; Blond, F.; Perrocheau, L.; Reichman, S.; Byrne, L.C.; et al. Rod-Derived Cone Viability Factor Promotes Cone Survival by Stimulating Aerobic Glycolysis. Cell 2015, 161, 817–832. [Google Scholar] [CrossRef]
- Sahoo, A.; Sahoo, S.K.; Joshi, P.; Lee, B.; Perera, R.J. MicroRNA-211 loss promotes metabolic vulnerability and BRAF inhibitor sensitivity in melanoma. J. Investig. Dermatol. 2019, 139, 167–176. [Google Scholar] [CrossRef]
- Hu, Q.; Ecker, M. Overview of MMP-13 as a Promising Target for the Treatment of Osteoarthritis. Int. J. Mol. Sci. 2021, 22, 1742. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lai, Y.; Li, J.; Zhao, L. Loss of miR-204 and miR-211 shifts osteochondral balance and causes temporomandibular joint osteoarthritis. J. Cell Physiol. 2023, 238, 2668–2678. [Google Scholar] [CrossRef] [PubMed]
- Logan, M.; Martin, J.F.; Nagy, A.; Lobe, C.; Olson, E.N.; Tabin, C.J. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 2002, 33, 77–80. [Google Scholar] [CrossRef]
- Du, S.W.; Palczewski, K. MicroRNA regulation of critical retinal pigment epithelial functions. Trends Neurosci. 2022, 45, 78–90. [Google Scholar] [CrossRef]
- Catalani, E.; Brunetti, K.; Del Quondam, S.; Cervia, D. Targeting mitochondrial dysfunction and oxidative stress to prevent the neurodegeneration of retinal ganglion cells. Antioxidants 2023, 12, 2011. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, C.; Hong, Z.; Chen, H.; Chen, W.; Chen, G. C/EBP homologous protein (CHOP) mediates neuronal apoptosis in rats with spinal cord injury. Exp. Ther. Med. 2013, 5, 107–111. [Google Scholar] [CrossRef]
- Chitnis, N.; Pytel, D.; Bobrovnikova-Marjon, E.; Pant, D.; Zheng, H.; Maas, N.L.; Frederick, B.; Kushner, J.A.; Chodosh, L.A.; Koumenis, C.; et al. miR-211 is a pro-survival micro-RNA that regulates chop expression in a PERK-dependent manner. Mol. Cell 2012, 48, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Sun, S.; Zhang, L.; Hao, F.; Zhang, D. miR-211 alleviates ischaemia/reperfusion-induced kidney injury by targeting TGFβR2/TGF-β/SMAD3 pathway. Bioengineered 2020, 11, 547–557. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, H.; Xia, Y. The expression of miR-211-5p in atherosclerosis and its influence on diagnosis and prognosis. BMC Cardiovasc. Disord. 2021, 21, 371. [Google Scholar] [CrossRef]
- Panizo, S.; Naves-Díaz, M.; Carrillo-López, N.; Martínez-Arias, L.; Fernández-Martín, J.L.; Ruiz-Torres, M.P.; Cannata-Andía, J.B.; Rodríguez, I. MicroRNAs 29b, 133b, and 211 Regulate Vascular Smooth Muscle Calcification Mediated by High Phosphorus. J. Am. Soc. Nephrol. 2016, 27, 824–834. [Google Scholar] [CrossRef]
- Shahroudi, M.J.; Rezaei, M.; Mirzaeipour, M.; Saravani, M.; Shahraki-Ghadimi, H.; Arab, S. Association between miR-202, miR-211, and miR-1238 gene polymorphisms and risk of vitiligo. Arch. Dermatol. Res. 2024, 316, 118. [Google Scholar] [CrossRef]
- Dai, X.; Li, H.; Chen, Y.; Fan, L.; Geng, H.; Li, S.; Qu, J.; Hou, L. Regulation of pigmentation by microRNAs: MITF-dependent microRNA-211 targets TGF-β receptor 2. Pigment. Cell Melanoma Res. 2015, 28, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Xue, V.W.; Chung, J.Y.; Córdoba, C.A.G.; Cheung, A.H.; Kang, W.; Lam, E.W.; Leung, K.T.; To, K.F.; Lan, H.Y.; Tang, P.M. Transforming Growth Factor-β: A Multifunctional Regulator of Cancer Immunity. Cancers 2020, 12, 3099. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J.; Sheppard, D. TGF-β signaling in health and disease. Cell 2023, 186, 4007–4037. [Google Scholar] [CrossRef]
- TGF-β Signaling in Health, Disease and Therapeutics|Signal Transduction and Targeted Therapy. Available online: https://www.nature.com/articles/s41392-024-01764-w (accessed on 1 June 2025).
- TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis—PMC. Available online: https://pmc.ncbi.nlm.nih.gov/articles/PMC6600375/ (accessed on 1 June 2025).
- Sanjabi, S.; Oh, S.A.; Li, M.O. Regulation of the Immune Response by TGF-β: From Conception to Autoimmunity and Infection. Cold Spring Harb. Perspect. Biol. 2017, 9, a022236. [Google Scholar] [CrossRef]
- Kubiczkova, L.; Sedlarikova, L.; Hajek, R.; Sevcikova, S. TGF-β—An excellent servant but a bad master. J. Transl. Med. 2012, 10, 183. [Google Scholar] [CrossRef] [PubMed]
- Batlle, E.; Massagué, J. Transforming Growth Factor-β Signaling in Immunity and Cancer. Immunity 2019, 50, 924–940. [Google Scholar] [CrossRef]
- Song, G.-q.; Zhao, Y. MicroRNA-211, a direct negative regulator of CDC25B expression, inhibits triple-negative breast cancer cells’ growth and migration. Tumor Biol. 2015, 36, 5001–5009. [Google Scholar] [CrossRef]
- Ostrowski, S.M.; Fisher, D.E. The melanocyte lineage factor miR-211 promotes BRAFV600E inhibitor resistance. Cancer Res. 2021, 141, 250–252. [Google Scholar] [CrossRef]
- Qin, X.; Zhang, J.; Lin, Y.; Sun, X.M.; Zhang, J.N.; Cheng, Z.Q. Identification of MiR-211-5p as a tumor suppressor by targeting ACSL4 in Hepatocellular Carcinoma. J. Transl. Med. 2020, 18, 326. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Huang, P.; Xie, J.; Li, R. microRNA-211 suppresses the growth and metastasis of cervical cancer by directly targeting ZEB1. Mol. Med. Rep. 2018, 17, 1275–1282. [Google Scholar] [CrossRef]
- Wang, K.; Jin, W.; Jin, P.; Fei, X.; Wang, X.; Chen, X. miR-211-5p Suppresses Metastatic Behavior by Targeting SNAI1 in Renal Cancer. Mol. Cancer Res. 2017, 15, 448–456. [Google Scholar] [CrossRef]
- Wang, W.; Liu, Z.; Zhang, X.; Liu, J.; Gui, J.; Cui, M.; Li, Y. miR-211-5p is down-regulated and a prognostic marker in bladder cancer. J. Gene Med. 2020, 22, e3270. [Google Scholar] [CrossRef]
- Melanoma Cell Invasiveness is Regulated by miR-211 Suppression of the BRN2 Transcription Factor—Boyle—2011—Pigment Cell & Melanoma Research—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/full/10.1111/j.1755-148X.2011.00849.x (accessed on 1 June 2025).
- Yang, C.-J.; Shen, W.G.; Liu, C.J.; Chen, Y.W.; Lu, H.H.; Tsai, M.M.; Lin, S.C. miR-221 and miR-222 expression increased the growth and tumorigenesis of oral carcinoma cells: miR-221 and miR-222 in OSCC. J. Oral Pathol. Med. 2011, 40, 560–566. [Google Scholar] [CrossRef]
- Yang, J.; Li, X.; Wei, S.; Peng, L.; Sang, H.; Jin, D.; Chen, M.; Dang, Y.; Zhang, G. Evaluation of the Diagnostic Potential of a Plasma Exosomal miRNAs Panel for Gastric Cancer. Front. Oncol. 2021, 11, 683465. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Kunhiraman, H.; Perera, R.J. The Paradoxical Behavior of microRNA-211 in Melanomas and Other Human Cancers. Front. Oncol. 2021, 10, 628367. [Google Scholar] [CrossRef]
- Wang, T.; Hao, D.; Yang, S.; Wang, Y.; Liu, J.; Wang, X.; Ma, J.; Xi, Z.; Yang, Y.; Qu, C. miR-211 facilitates platinum chemosensitivity by blocking the DNA damage response (DDR) in ovarian cancer. Cell Death Dis. 2019, 10, 495. [Google Scholar] [CrossRef]
- Foster, H.M.; Carle, M.N.; Jira, L.R.; Koh, D.W. TRPM2 Channels: A Potential Therapeutic Target in Melanoma? Int. J. Mol. Sci. 2023, 24, 10437. [Google Scholar] [CrossRef]
- Balzano, F.; Deiana, M.; Giudici, S.D.; Oggiano, A.; Baralla, A.; Pasella, S.; Mannu, A.; Pescatori, M.; Porcu, B.; Fanciulli, G.; et al. miRNA Stability in Frozen Plasma Samples. Molecules 2015, 20, 19030–19040. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.; Lu, H.; Wang, X.; Jin, H. MicroRNAs as potential biomarkers in cancer: Opportunities and challenges. Biomed Res. Int. 2015, 2015, 125094. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. microRNA as a new immune-regulatory agent in breast milk. Silence 2010, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E. Stem-Loop qRT-PCR for the Detection of Plant microRNAs. In Plant Epigenetics; Kovalchuk, I., Ed.; Springer: Boston, MA, USA, 2017; Volume 1456, pp. 163–175. [Google Scholar]
- Katsushima, K.; Lee, B.; Yuan, M.; Kunhiraman, H.; Stapleton, S.; Jallo, G.; Raabe, E.; Eberhart, C.; Perera, R. microRNA 211, a potential therapeutic agent for Group 3 medulloblastoma in children. Neuro-Oncol. 2021, 23 (Suppl. 6), vi40. [Google Scholar] [CrossRef]
- Lim, S.Y.; Boyd, S.C.; Diefenbach, R.J.; Rizos, H. Circulating microRNAs: Functional biomarkers for melanoma prognosis and treatment. Mol. Cancer 2025, 24, 99. [Google Scholar] [CrossRef] [PubMed]
- Pagoni, M.; Cava, C.; Sideris, D.C.; Avgeris, M.; Zoumpourlis, V.; Michalopoulos, I.; Drakoulis, N. miRNA-Based Technologies in Cancer Therapy. J. Pers. Med. 2023, 13, 1586. [Google Scholar] [CrossRef]
- Trang, P.; Wiggins, J.F.; Daige, C.L.; Cho, C.; Omotola, M.; Brown, D.; Weidhaas, J.B.; Bader, A.G.; Slack, F.J. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 2011, 19, 1116–1122. [Google Scholar] [CrossRef]
- Lee, S.W.L.; Paoletti, C.; Campisi, M.; Osaki, T.; Adriani, G.; Kamm, R.D.; Mattu, C.; Chiono, V. MicroRNA delivery through nanoparticles. J. Control. Release 2019, 313, 80–95. [Google Scholar] [CrossRef]
- Gareev, I.; Beylerli, O.; Tamrazov, R.; Ilyasova, T.; Shumadalova, A.; Du, W.; Yang, B. Methods of miRNA delivery and possibilities of their application in neuro-oncology. Non-Coding RNA Res. 2023, 8, 661–674. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.-K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.-L.; Kim, T.-Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Lee, H.; Lee, S.; Bae, H.; Kang, H.S.; Kim, S.J. Genome-wide identification of target genes for miR-204 and miR-211 identifies their proliferation stimulatory role in breast cancer cells. Sci. Rep. 2016, 6, 25287. [Google Scholar] [CrossRef] [PubMed]
- Ala, U. Competing Endogenous RNAs, Non-Coding RNAs and Diseases: An Intertwined Story. Cells 2020, 9, 1574. [Google Scholar] [CrossRef] [PubMed]
- Chiarella, A.M.; Lu, D.; Hathaway, N.A. Epigenetic Control of a Local Chromatin Landscape. Int. J. Mol. Sci. 2020, 21, 943. [Google Scholar] [CrossRef]
- Chen, S.; Sun, H.; Mookhtiar, A.K.; Chintala, P.K. Recent Advances in Non-Viral Delivery Systems for CRISPR/Cas-Based Genome Editing. Int. J. Mol. Sci. 2024, 25, 5462. [Google Scholar] [CrossRef]
- Di Martino, M.T.; Tagliaferri, P.; Tassone, P. MicroRNA in cancer therapy: Breakthroughs and challenges in early clinical applications. J. Exp. Clin. Cancer Res. 2025, 44, 126. [Google Scholar] [CrossRef]
- Alahverdi, M.; Dadmehr, M.; Sahebkar, A. Nanocarriers for microRNA delivery: A review of applied platforms and perspectives. Int. J. Biol. Macromol. 2025, 319, 145463. [Google Scholar] [CrossRef]
- Chen, R.; Bhavsar, C.; Lourie, R.; Li, S.; Wu, S.Y. Development of an innovative extracellular vesicle mimetic delivery platform for efficient miRNA delivery to tumours. Biomaterials 2025, 321, 123282. [Google Scholar] [CrossRef]
- Kumar, M.A.; Baba, S.K.; Sadida, H.Q.; Al Marzooqi, S.; Jerobin, J.; Altemani, F.H.; Algehainy, N.; Alanazi, M.A.; Abou-Samra, A.-B.; Kumar, R.; et al. Extracellular vesicles as tools and targets in therapy for diseases. Signal Transduct. Target. Ther. 2024, 9, 27. [Google Scholar] [CrossRef]
- Pottash, A.E.; Levy, D.; Jeyaram, A.; Kuo, L.; Kronstadt, S.M.; Chao, W.; Jay, S.M. Combinatorial microRNA Loading into Extracellular Vesicles for Increased Anti-Inflammatory Efficacy. Non-Coding RNA 2022, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Wang, Z.; Xing, H.; Huang, Y.; Lu, M. Extracellular vesicle-based targeted RNA therapies against cancer. J. Extracell Vesicles. 2025, 6, 100083. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rayo Parra, J.; Grand, Z.; Gonzalez, G.; Perera, R.; Pandeya, D.; Weiler, T.; Chapagain, P. The Multifaceted Role of miR-211 in Health and Disease. Biomolecules 2025, 15, 1109. https://doi.org/10.3390/biom15081109
Rayo Parra J, Grand Z, Gonzalez G, Perera R, Pandeya D, Weiler T, Chapagain P. The Multifaceted Role of miR-211 in Health and Disease. Biomolecules. 2025; 15(8):1109. https://doi.org/10.3390/biom15081109
Chicago/Turabian StyleRayo Parra, Juan, Zachary Grand, Gabriel Gonzalez, Ranjan Perera, Dipendra Pandeya, Tracey Weiler, and Prem Chapagain. 2025. "The Multifaceted Role of miR-211 in Health and Disease" Biomolecules 15, no. 8: 1109. https://doi.org/10.3390/biom15081109
APA StyleRayo Parra, J., Grand, Z., Gonzalez, G., Perera, R., Pandeya, D., Weiler, T., & Chapagain, P. (2025). The Multifaceted Role of miR-211 in Health and Disease. Biomolecules, 15(8), 1109. https://doi.org/10.3390/biom15081109






