Quiescence Multiverse
Abstract
1. Introduction
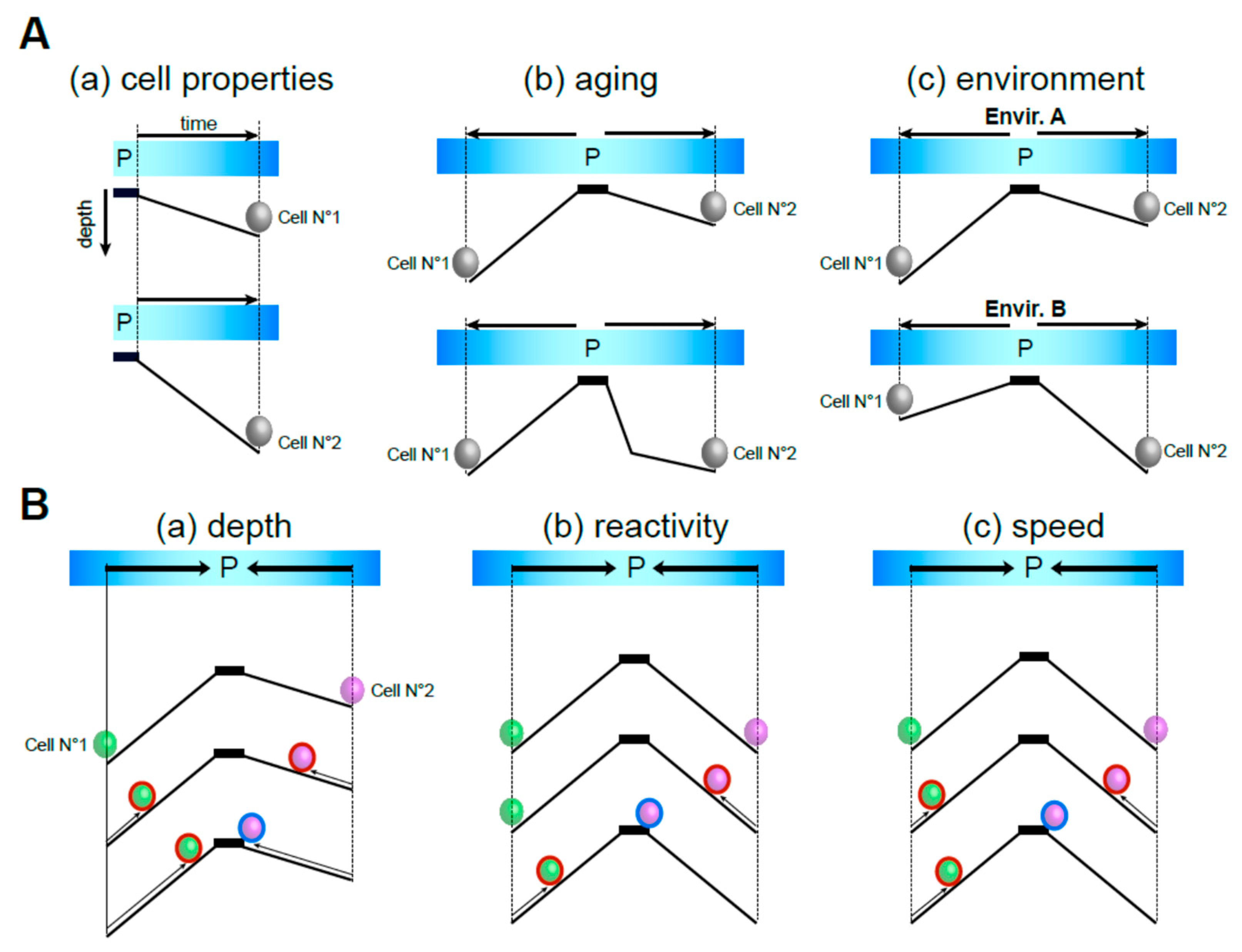
2. The Multiple Scales of Ageing
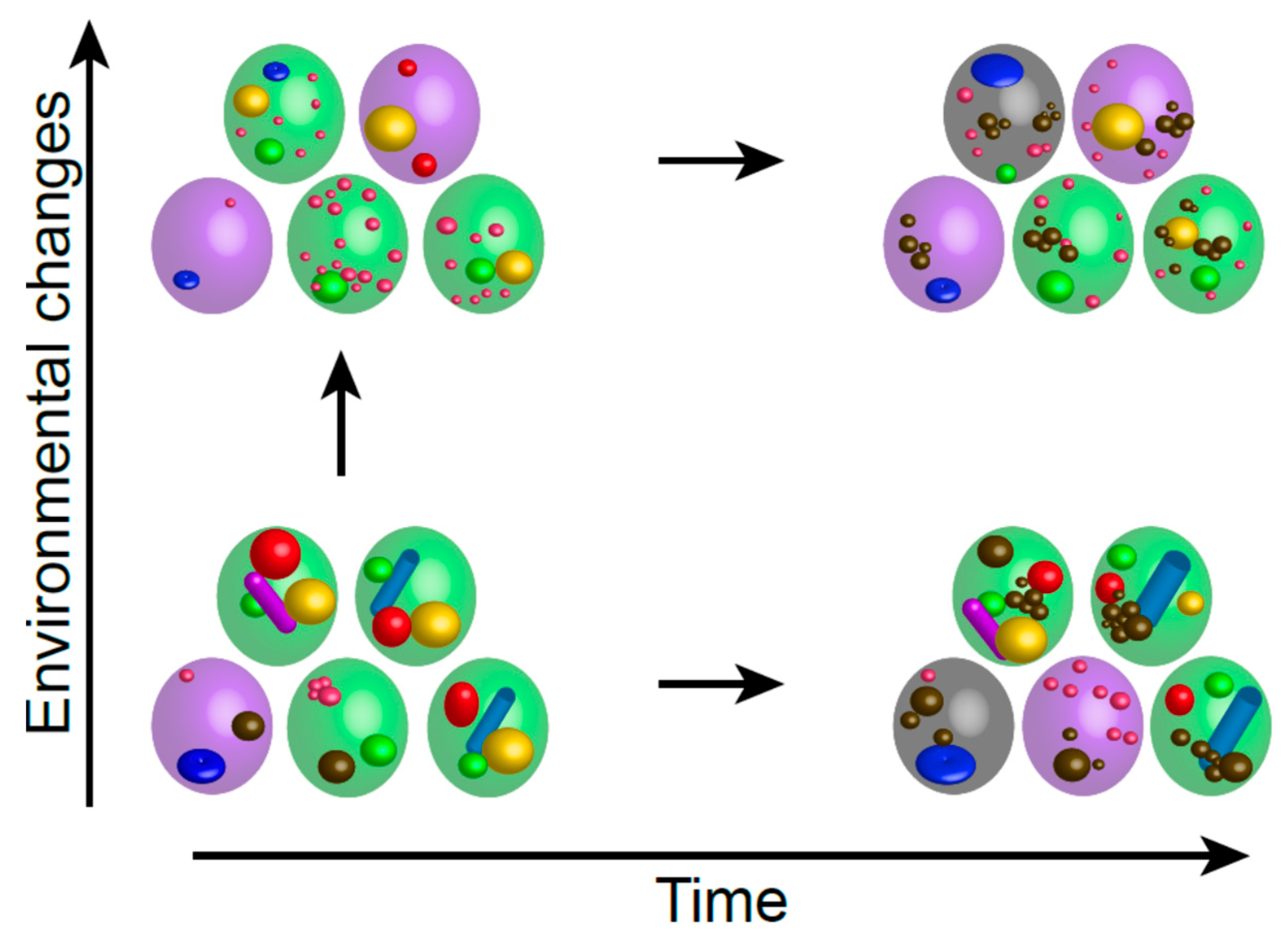
3. Time Generates Heterogeneity
4. Heterogeneity and Robustness in Facing Time
5. Quiescence Deepening
6. Quiescence Exit Efficiency
7. Conclusions and Model
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EGF | Epidermal Growth Factor |
| MuSC | Muscle Stem Cell |
| HSC | Hematopoietic Stem Cell |
| NSC | Neural Stem Cell |
References
- Sender, R.; Milo, R. The Distribution of Cellular Turnover in the Human Body. Nat. Med. 2021, 27, 45–48. [Google Scholar] [CrossRef] [PubMed]
- van Velthoven, C.T.J.; Rando, T.A. Stem Cell Quiescence: Dynamism, Restraint, and Cellular Idling. Cell Stem Cell 2019, 24, 213–225. [Google Scholar] [CrossRef]
- Marescal, O.; Cheeseman, I.M. Cellular Mechanisms and Regulation of Quiescence. Dev. Cell 2020, 55, 259–271. [Google Scholar] [CrossRef] [PubMed]
- de Morree, A.; Rando, T.A. Regulation of Adult Stem Cell Quiescence and Its Functions in the Maintenance of Tissue Integrity. Nat. Rev. Mol. Cell Biol. 2023, 24, 334–354. [Google Scholar] [CrossRef] [PubMed]
- Cheung, T.H.; Rando, T.A. Molecular Regulation of Stem Cell Quiescence. Nat. Rev. Mol. Cell Biol. 2013, 14, 329–340. [Google Scholar] [CrossRef]
- Mitra, M.; Batista, S.L.; Coller, H.A. Transcription Factor Networks in Cellular Quiescence. Nat. Cell Biol. 2025, 27, 14–27. [Google Scholar] [CrossRef]
- Sang, L.; Coller, H.A.; Roberts, J.M. Control of the Reversibility of Cellular Quiescence by the Transcriptional Repressor HES1. Science 2008, 321, 1095–1100. [Google Scholar] [CrossRef]
- De Virgilio, C. The Essence of Yeast Quiescence. FEMS Microbiol. Rev. 2012, 36, 306–339. [Google Scholar] [CrossRef]
- Gray, J.V.; Petsko, G.A.; Johnston, G.C.; Ringe, D.; Singer, R.A.; Werner-Washburne, M. “Sleeping Beauty”: Quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. MMBR 2004, 68, 187–206. [Google Scholar] [CrossRef]
- Daignan-Fornier, B.; Laporte, D.; Sagot, I. Quiescence Through the Prism of Evolution. Front. Cell Dev. Biol. 2021, 9, 745069. [Google Scholar] [CrossRef]
- Fiore, A.P.Z.P.; de Ribeiro, P.F.; Bruni-Cardoso, A. Sleeping Beauty and the Microenvironment Enchantment: Microenvironmental Regulation of the Proliferation-Quiescence Decision in Normal Tissues and in Cancer Development. Front. Cell Dev. Biol. 2018, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Boer, V.M.; Crutchfield, C.A.; Bradley, P.H.; Botstein, D.; Rabinowitz, J.D. Growth-Limiting Intracellular Metabolites in Yeast Growing under Diverse Nutrient Limitations. Mol. Biol. Cell 2010, 21, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Klosinska, M.M.; Crutchfield, C.A.; Bradley, P.H.; Rabinowitz, J.D.; Broach, J.R. Yeast Cells Can Access Distinct Quiescent States. Genes Dev. 2011, 25, 336–349. [Google Scholar] [CrossRef]
- Coller, H.A.; Sang, L.; Roberts, J.M. A New Description of Cellular Quiescence. PLoS Biol. 2006, 4, e83. [Google Scholar] [CrossRef] [PubMed]
- Coller, H.A. The Paradox of Metabolism in Quiescent Stem Cells. FEBS Lett. 2019, 593, 2817–2839. [Google Scholar] [CrossRef]
- Toynbee, A.J.; Markowitz, W.; Smart, J.J.C. Time and its role in the history of thought and action. Encyclopedia Britannica, 1 January 2025. [Google Scholar]
- Lemoine, M. Defining Aging. Biol. Philos. 2020, 35, 46. [Google Scholar] [CrossRef]
- Blasimme, A.; Boniolo, G.; Nathan, M.J. Rethinking Ageing: Introduction. Hist. Philos. Life Sci. 2021, 43, 95. [Google Scholar] [CrossRef]
- Saborido, C.; García-Barranquero, P. Is Aging a Disease? The Theoretical Definition of Aging in the Light of the Philosophy of Medicine. J. Med. Philos. Forum Bioeth. Philos. Med. 2022, 47, 770–783. [Google Scholar] [CrossRef]
- Harman, D. Aging: A Theory Based on Free Radical and Radiation Chemistry. J. Gerontol. 1956, 11, 298–300. [Google Scholar] [CrossRef]
- Kennedy, B.K.; Berger, S.L.; Brunet, A.; Campisi, J.; Cuervo, A.M.; Epel, E.S.; Franceschi, C.; Lithgow, G.J.; Morimoto, R.I.; Pessin, J.E.; et al. Geroscience: Linking Aging to Chronic Disease. Cell 2014, 159, 709–713. [Google Scholar] [CrossRef]
- Kroemer, G.; Maier, A.B.; Cuervo, A.M.; Gladyshev, V.N.; Ferrucci, L.; Gorbunova, V.; Kennedy, B.K.; Rando, T.A.; Seluanov, A.; Sierra, F.; et al. From Geroscience to Precision Geromedicine: Understanding and Managing Aging. Cell 2025, 188, 2043–2062. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Vershinina, O.; Bacalini, M.G.; Zaikin, A.; Franceschi, C.; Ivanchenko, M. Disentangling Age-Dependent DNA Methylation: Deterministic, Stochastic, and Nonlinear. Sci. Rep. 2021, 11, 9201. [Google Scholar] [CrossRef] [PubMed]
- Olecka, M.; van Bömmel, A.; Best, L.; Haase, M.; Foerste, S.; Riege, K.; Dost, T.; Flor, S.; Witte, O.W.; Franzenburg, S.; et al. Nonlinear DNA Methylation Trajectories in Aging Male Mice. Nat. Commun. 2024, 15, 3074. [Google Scholar] [CrossRef]
- Shen, X.; Wang, C.; Zhou, X.; Zhou, W.; Hornburg, D.; Wu, S.; Snyder, M.P. Nonlinear Dynamics of Multi-Omics Profiles during Human Aging. Nat. Aging 2024, 4, 1619–1634. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, J.; Oh, H.; Wyss-Coray, T. Measuring Biological Age Using Omics Data. Nat. Rev. Genet. 2022, 23, 715–727. [Google Scholar] [CrossRef]
- Vandereyken, K.; Sifrim, A.; Thienpont, B.; Voet, T. Methods and Applications for Single-Cell and Spatial Multi-Omics. Nat. Rev. Genet. 2023, 24, 494–515. [Google Scholar] [CrossRef]
- Schaum, N.; Lehallier, B.; Hahn, O.; Pálovics, R.; Hosseinzadeh, S.; Lee, S.E.; Sit, R.; Lee, D.P.; Losada, P.M.; Zardeneta, M.E.; et al. Ageing Hallmarks Exhibit Organ-Specific Temporal Signatures. Nature 2020, 583, 596–602. [Google Scholar] [CrossRef]
- Nie, C.; Li, Y.; Li, R.; Yan, Y.; Zhang, D.; Li, T.; Li, Z.; Sun, Y.; Zhen, H.; Ding, J.; et al. Distinct Biological Ages of Organs and Systems Identified from a Multi-Omics Study. Cell Rep. 2022, 38, 110459. [Google Scholar] [CrossRef]
- Izgi, H.; Han, D.; Isildak, U.; Huang, S.; Kocabiyik, E.; Khaitovich, P.; Somel, M.; Dönertaş, H.M. Inter-Tissue Convergence of Gene Expression during Ageing Suggests Age-Related Loss of Tissue and Cellular Identity. eLife 2022, 11, e68048. [Google Scholar] [CrossRef]
- Chien, J.-F.; Liu, H.; Wang, B.-A.; Luo, C.; Bartlett, A.; Castanon, R.; Johnson, N.D.; Nery, J.R.; Osteen, J.; Li, J.; et al. Cell-Type-Specific Effects of Age and Sex on Human Cortical Neurons. Neuron 2024, 112, 2524–2539.e5. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.-C.; Brbić, M.; Park, Y.-J.; Jackson, T.; Chen, J.; Kolluru, S.S.; Qi, Y.; Katheder, N.S.; Cai, X.T.; Lee, S.; et al. Aging Fly Cell Atlas Identifies Exhaustive Aging Features at Cellular Resolution. Science 2023, 380, eadg0934. [Google Scholar] [CrossRef] [PubMed]
- Hayflick, L.; Moorhead, P.S. The Serial Cultivation of Human Diploid Cell Strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.; Yuan, H.; Soifer, I.; Maile, T.M.; Wang, R.Y.; Ireland, A.; O’Brien, J.J.; Goudeau, J.; Chan, L.J.G.; Vijay, T.; et al. Novel Insights from a Multiomics Dissection of the Hayflick Limit. eLife 2022, 11, e70283. [Google Scholar] [CrossRef]
- Wright, W.E.; Shay, J.W. Historical Claims and Current Interpretations of Replicative Aging. Nat. Biotechnol. 2002, 20, 682–688. [Google Scholar] [CrossRef]
- Hormoz, S. Stem Cell Population Asymmetry Can Reduce Rate of Replicative Aging. J. Theor. Biol. 2013, 331, 19–27. [Google Scholar] [CrossRef]
- Sheldrake, A.R. Cellular Senescence, Rejuvenation and Potential Immortality. Proc. Biol. Sci. 2022, 289, 20212434. [Google Scholar] [CrossRef]
- Pardee, A.B. A Restriction Point for Control of Normal Animal Cell Proliferation. Proc. Natl. Acad. Sci. USA 1974, 71, 1286–1290. [Google Scholar] [CrossRef]
- Laporte, D.; Lebaudy, A.; Sahin, A.; Pinson, B.; Ceschin, J.; Daignan-Fornier, B.; Sagot, I. Metabolic Status Rather than Cell Cycle Signals Control Quiescence Entry and Exit. J. Cell Biol. 2011, 192, 949–957. [Google Scholar] [CrossRef]
- Otsuki, L.; Brand, A.H. Cell Cycle Heterogeneity Directs the Timing of Neural Stem Cell Activation from Quiescence. Science 2018, 360, 99–102. [Google Scholar] [CrossRef]
- Costello, G.; Rodgers, L.; Beach, D. Fission Yeast Enters the Stationary Phase G0 State from Either Mitotic G1 or G2. Curr. Genet. 1986, 11, 119–125. [Google Scholar] [CrossRef]
- Takeo, K.; Tanaka, R.; Miyaji, M.; Nishimura, K. Unbudded G2 as Well as G1 Arrest in the Stationary Phase of the Basidiomycetous Yeast Cryptococcus neoformans. FEMS Microbiol. Lett. 1995, 129, 231–235. [Google Scholar] [PubMed]
- Cameron, I.L.; Bols, N.C. Effect of Cell Population Density on G2 Arrest in Tetrahymena. J. Cell Biol. 1975, 67, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Conger, B.V.; Carabia, J.V. Microspectrophotometric Determination of the 2C and 4C Nuclear Complement in the Root and Shoot of the Dormant Maize Embryo. Environ. Exp. Bot. 1976, 16, 171–175. [Google Scholar] [CrossRef]
- McKeown, C.R.; Cline, H.T. Nutrient Restriction Causes Reversible G2 Arrest in Xenopus Neural Progenitors. Dev. Camb. Engl. 2019, 146, dev178871. [Google Scholar] [CrossRef]
- Tripathi, A.; Kumar, K.V.P.; Chaube, S.K. Meiotic Cell Cycle Arrest in Mammalian Oocytes. J. Cell. Physiol. 2010, 223, 592–600. [Google Scholar] [CrossRef]
- Cooper, S. On the Proposal of a G0 Phase and the Restriction Point. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 1998, 12, 367–373. [Google Scholar] [CrossRef][Green Version]
- Cooper, S. Reappraisal of Serum Starvation, the Restriction Point, G0, and G1 Phase Arrest Points. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2003, 17, 333–340. [Google Scholar] [CrossRef]
- Matson, J.P.; Cook, J.G. Cell Cycle Proliferation Decisions: The Impact of Single Cell Analyses. FEBS J. 2017, 284, 362–375. [Google Scholar] [CrossRef]
- Ogrodnik, M.; Salmonowicz, H.; Gladyshev, V.N. Integrating Cellular Senescence with the Concept of Damage Accumulation in Aging: Relevance for Clearance of Senescent Cells. Aging Cell 2019, 18, e12841. [Google Scholar] [CrossRef]
- Gallart-Palau, X.; Tan, L.M.; Serra, A.; Gao, Y.; Ho, H.H.; Richards, A.M.; Kandiah, N.; Chen, C.P.; Kalaria, R.N.; Sze, S.K. Degenerative Protein Modifications in the Aging Vasculature and Central Nervous System: A Problem Shared Is Not Always Halved. Ageing Res. Rev. 2019, 53, 100909. [Google Scholar] [CrossRef] [PubMed]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid Peroxidation: Production, Metabolism, and Signaling Mechanisms of Malondialdehyde and 4-Hydroxy-2-Nonenal. Oxid. Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef]
- Gille, B.; Galuska, C.E.; Fuchs, B.; Peleg, S. Recent Advances in Studying Age-Associated Lipids Alterations and Dietary Interventions in Mammals. Front. Aging 2021, 2, 773795. [Google Scholar] [CrossRef]
- Gęgotek, A.; Skrzydlewska, E. Biological Effect of Protein Modifications by Lipid Peroxidation Products. Chem. Phys. Lipids 2019, 221, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Brunet, A.; Goodell, M.A.; Rando, T.A. Ageing and Rejuvenation of Tissue Stem Cells and Their Niches. Nat. Rev. Mol. Cell Biol. 2023, 24, 45–62. [Google Scholar] [CrossRef]
- Malecki, K.M.C.; Andersen, J.K.; Geller, A.M.; Harry, G.J.; Jackson, C.L.; James, K.A.; Miller, G.W.; Ottinger, M.A. Integrating Environment and Aging Research: Opportunities for Synergy and Acceleration. Front. Aging Neurosci. 2022, 14, 824921. [Google Scholar] [CrossRef] [PubMed]
- Stockholm, D.; Benchaouir, R.; Picot, J.; Rameau, P.; Neildez, T.M.A.; Landini, G.; Laplace-Builhé, C.; Paldi, A. The Origin of Phenotypic Heterogeneity in a Clonal Cell Population in Vitro. PLoS ONE 2007, 2, e394. [Google Scholar] [CrossRef]
- Sutcu, H.H.; Ricchetti, M. Loss of Heterogeneity, Quiescence, and Differentiation in Muscle Stem Cells. Stem Cell Investig. 2018, 5, 9. [Google Scholar] [CrossRef]
- Laporte, D.; Courtout, F.; Salin, B.; Ceschin, J.; Sagot, I. An Array of Nuclear Microtubules Reorganizes the Budding Yeast Nucleus during Quiescence. J. Cell Biol. 2013, 203, 585–594. [Google Scholar] [CrossRef]
- Laporte, D.; Courtout, F.; Tollis, S.; Sagot, I. Quiescent Saccharomyces cerevisiae Forms Telomere Hyperclusters at the Nuclear Membrane Vicinity through a Multifaceted Mechanism Involving Esc1, the Sir Complex, and Chromatin Condensation. Mol. Biol. Cell 2016, 27, 1875–1884. [Google Scholar] [CrossRef]
- Laporte, D.; Sagot, I. Microtubules Move the Nucleus to Quiescence. Nucleus 2014, 5, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Guidi, M.; Ruault, M.; Marbouty, M.; Loïodice, I.; Cournac, A.; Billaudeau, C.; Hocher, A.; Mozziconacci, J.; Koszul, R.; Taddei, A. Spatial Reorganization of Telomeres in Long-Lived Quiescent Cells. Genome Biol. 2015, 16, 206. [Google Scholar] [CrossRef]
- Rutledge, M.T.; Russo, M.; Belton, J.-M.; Dekker, J.; Broach, J.R. The Yeast Genome Undergoes Significant Topological Reorganization in Quiescence. Nucleic Acids Res. 2015, 43, 8299–8313. [Google Scholar] [CrossRef]
- Sheth, U.; Parker, R. Decapping and Decay of Messenger RNA Occur in Cytoplasmic Processing Bodies. Science 2003, 300, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Laporte, D.; Salin, B.; Daignan-Fornier, B.; Sagot, I. Reversible Cytoplasmic Localization of the Proteasome in Quiescent Yeast Cells. J. Cell Biol. 2008, 181, 737–745. [Google Scholar] [CrossRef]
- Sagot, I.; Laporte, D. The Cell Biology of Quiescent Yeast—A Diversity of Individual Scenarios. J. Cell Sci. 2019, 132, jcs213025. [Google Scholar] [CrossRef] [PubMed]
- Laporte, D.; Massoni-Laporte, A.; Lefranc, C.; Dompierre, J.; Mauboules, D.; Nsamba, E.T.; Royou, A.; Gal, L.; Schuldiner, M.; Gupta, M.L.; et al. A Stable Microtubule Bundle Formed through an Orchestrated Multistep Process Controls Quiescence Exit. eLife 2024, 12, RP89958. [Google Scholar] [CrossRef]
- Laporte, D.; Sagot, I. Microtubule Reorganization and Quiescence: An Intertwined Relationship. Physiol. Bethesda Md. 2025, 40, 145–157. [Google Scholar] [CrossRef]
- Laporte, D.; Gouleme, L.; Jimenez, L.; Khemiri, I.; Sagot, I. Mitochondria Reorganization upon Proliferation Arrest Predicts Individual Yeast Cell Fate. eLife 2018, 7, e35685. [Google Scholar] [CrossRef]
- Niu, H.; Gu, J.; Zhang, Y. Bacterial Persisters: Molecular Mechanisms and Therapeutic Development. Signal Transduct. Target. Ther. 2024, 9, 174. [Google Scholar] [CrossRef]
- Yu, J.; Liu, Y.; Yin, H.; Chang, Z. Regrowth-Delay Body as a Bacterial Subcellular Structure Marking Multidrug-Tolerant Persisters. Cell Discov. 2019, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.M.; Isberg, R.R. Defining Heterogeneity within Bacterial Populations via Single Cell Approaches. BioEssays News Rev. Mol. Cell. Dev. Biol. 2016, 38, 782–790. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.F.; Herrero, M.A.; Acosta, F.J.; Fernandez-Arias, C. Population Mechanics: A Mathematical Framework to Study T Cell Homeostasis. Sci. Rep. 2017, 7, 9511. [Google Scholar] [CrossRef] [PubMed]
- Vartanian, A.D.; Quétin, M.; Michineau, S.; Auradé, F.; Hayashi, S.; Dubois, C.; Rocancourt, D.; Drayton-Libotte, B.; Szegedi, A.; Buckingham, M.; et al. PAX3 Confers Functional Heterogeneity in Skeletal Muscle Stem Cell Responses to Environmental Stress. Cell Stem Cell 2019, 24, 958–973.e9. [Google Scholar] [CrossRef]
- Bucher, N.L.R. Regeneration of mammalian liver. Int. Rev. Cytol. 1963, 15, 245–300. [Google Scholar] [CrossRef]
- Augenlicht, L.H.; Baserga, R. Changes in the G0 State of WI-38 Fibroblasts at Different Times after Confluence. Exp. Cell Res. 1974, 89, 255–262. [Google Scholar] [CrossRef]
- Owen, T.A.; Soprano, D.R.; Soprano, K.J. Analysis of the Growth Factor Requirements for Stimulation of WI-38 Cells after Extended Periods of Density-Dependent Growth Arrest. J. Cell. Physiol. 1989, 139, 424–431. [Google Scholar] [CrossRef]
- Owen, T.A.; Cosenza, S.C.; Soprano, D.R.; Soprano, K.J. Time of C-Fos and c-Myc Expression in Human Diploid Fibroblasts Stimulated to Proliferate after Prolonged Periods in Quiescence. J. Biol. Chem. 1987, 262, 15111–15117. [Google Scholar] [CrossRef]
- Rossini, M.; Lin, J.-C.; Baserga, R. Effects of Prolonged Quiescence on Nuclei and Chromatin of WI-38 Fibroblasts. J. Cell. Physiol. 1976, 88, 1–11. [Google Scholar] [CrossRef]
- Yanez, I.; O’Farrell, M. Variation in the Length of the Lag Phase Following Serum Restimulation of Mouse 3T3 Cells. Cell Biol. Int. Rep. 1989, 13, 453–462. [Google Scholar] [CrossRef]
- Levin-Reisman, I.; Gefen, O.; Fridman, O.; Ronin, I.; Shwa, D.; Sheftel, H.; Balaban, N.Q. Automated Imaging with ScanLag Reveals Previously Undetectable Bacterial Growth Phenotypes. Nat. Methods 2010, 7, 737–739. [Google Scholar] [CrossRef] [PubMed]
- Laporte, D.; Jimenez, L.; Gouleme, L.; Sagot, I. Yeast Quiescence Exit Swiftness Is Influenced by Cell Volume and Chronological Age. Microb. Cell 2017, 5, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Sheng, N.; Zhang, Z.; He, C.; Zhao, Y.; Sun, H.; Chen, J.; Yang, X.; Tang, C. Initial Nutrient Condition Determines the Recovery Speed of Quiescent Cells in Fission Yeast. Heliyon 2024, 10, e26558. [Google Scholar] [CrossRef] [PubMed]
- Olmedo, M.; Mata-Cabana, A.; Rodríguez-Palero, M.J.; García-Sánchez, S.; Fernández-Yañez, A.; Merrow, M.; Artal-Sanz, M. Prolonged Quiescence Delays Somatic Stem Cell-like Divisions in Caenorhabditis Elegans and Is Controlled by Insulin Signaling. Aging Cell 2020, 19, e13085. [Google Scholar] [CrossRef]
- Kwon, J.S.; Everetts, N.J.; Wang, X.; Wang, W.; Della Croce, K.; Xing, J.; Yao, G. Controlling Depth of Cellular Quiescence by an Rb-E2F Network Switch. Cell Rep. 2017, 20, 3223–3235. [Google Scholar] [CrossRef]
- Urbán, N.; Cheung, T.H. Stem Cell Quiescence: The Challenging Path to Activation. Development 2021, 148, dev165084. [Google Scholar] [CrossRef]
- Cabezas-Wallscheid, N.; Buettner, F.; Sommerkamp, P.; Klimmeck, D.; Ladel, L.; Thalheimer, F.B.; Pastor-Flores, D.; Roma, L.P.; Renders, S.; Zeisberger, P.; et al. Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell 2017, 169, 807–823.e19. [Google Scholar] [CrossRef]
- Fujimaki, K.; Li, R.; Chen, H.; Della Croce, K.; Zhang, H.H.; Xing, J.; Bai, F.; Yao, G. Graded Regulation of Cellular Quiescence Depth between Proliferation and Senescence by a Lysosomal Dimmer Switch. Proc. Natl. Acad. Sci. USA 2019, 116, 22624–22634. [Google Scholar] [CrossRef]
- Audesse, A.J.; Webb, A.E. Mechanisms of Enhanced Quiescence in Neural Stem Cell Aging. Mech. Ageing Dev. 2020, 191, 111323. [Google Scholar] [CrossRef]
- Donigan, A.M.; Cavalli, R.C.; Pena, A.A.; Savage, C.R.; Soprano, D.R.; Soprano, K.J. Epidermal Growth Factor Receptors Lose Ligand Binding Ability as WI-38 Cells Progress from Short-Term to Long-Term Quiescence. J. Cell. Physiol. 1993, 155, 164–170. [Google Scholar] [CrossRef]
- Soprano, K.J. WI-38 Cell Long-Term Quiescence Model System: A Valuable Tool to Study Molecular Events That Regulate Growth. J. Cell. Biochem. 1994, 54, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Yao, G. Quiescence-Origin Senescence: A New Paradigm in Cellular Aging. Biomedicines 2024, 12, 1837. [Google Scholar] [CrossRef]
- Wei, M.Y.; Yao, G. Modeling the Depth of Cellular Dormancy from RNA-Sequencing Data. Methods Mol. Biol. 2024, 2811, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Murley, A.; Popovici, A.C.; Hu, X.S.; Lund, A.; Wickham, K.; Durieux, J.; Joe, L.; Koronyo, E.; Zhang, H.; Genuth, N.R.; et al. Quiescent Cell Re-Entry Is Limited by Macroautophagy-Induced Lysosomal Damage. Cell 2025, 188, 2670–2686.e14. [Google Scholar] [CrossRef]
- Murley, A.; Dillin, A. Macroautophagy in Quiescent and Senescent Cells: A Pathway to Longevity? Trends Cell Biol. 2023, 33, 495–504. [Google Scholar] [CrossRef]
- Fujimaki, K.; Yao, G. Cell Dormancy Plasticity: Quiescence Deepens into Senescence through a Dimmer Switch. Physiol. Genom. 2020, 52, 558–562. [Google Scholar] [CrossRef]
- Leeman, D.S.; Hebestreit, K.; Ruetz, T.; Webb, A.E.; McKay, A.; Pollina, E.A.; Dulken, B.W.; Zhao, X.; Yeo, R.W.; Ho, T.T.; et al. Lysosome Activation Clears Aggregates and Enhances Quiescent Neural Stem Cell Activation during Aging. Science 2018, 359, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, B.; Pan, Q.; Kwon, J.S.; Miller, M.A.; Croce, K.D.; Yao, G. Circadian Proteins Cry and Rev-Erb Converge to Deepen Cellular Quiescence by Downregulating Cyclin D and Cdk4,6. bioRxiv 2021. [Google Scholar] [CrossRef]
- Mira, H.; Andreu, Z.; Suh, H.; Lie, D.C.; Jessberger, S.; Consiglio, A.; San Emeterio, J.; Hortigüela, R.; Marqués-Torrejón, M.A.; Nakashima, K.; et al. Signaling through BMPR-IA Regulates Quiescence and Long-Term Activity of Neural Stem Cells in the Adult Hippocampus. Cell Stem Cell 2010, 7, 78–89. [Google Scholar] [CrossRef]
- Mira, H.; Morante, J. Neurogenesis From Embryo to Adult—Lessons From Flies and Mice. Front. Cell Dev. Biol. 2020, 8, 533. [Google Scholar] [CrossRef]
- Elkin, A.M.; Robbins, S.; Barros, C.S.; Bossing, T. The Critical Balance Between Quiescence and Reactivation of Neural Stem Cells. Biomolecules 2025, 15, 672. [Google Scholar] [CrossRef] [PubMed]
- Eames, A.; Chandrasekaran, S. Leveraging Metabolic Modeling and Machine Learning to Uncover Modulators of Quiescence Depth. PNAS Nexus 2024, 3, pgae013. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Segura, A.; de Jong, T.V.; Melov, S.; Guryev, V.; Campisi, J.; Demaria, M. Unmasking Transcriptional Heterogeneity in Senescent Cells. Curr. Biol. CB 2017, 27, 2652–2660.e4. [Google Scholar] [CrossRef]
- Goel, A.J.; Rieder, M.-K.; Arnold, H.-H.; Radice, G.L.; Krauss, R.S. Niche Cadherins Control the Quiescence-to-Activation Transition in Muscle Stem Cells. Cell Rep. 2017, 21, 2236–2250. [Google Scholar] [CrossRef] [PubMed]
- Guilhot, C.; Catenacci, M.; Lofaro, S.; Rudnicki, M.A. The Satellite Cell in Skeletal Muscle: A Story of Heterogeneity. Curr. Top. Dev. Biol. 2024, 158, 15–51. [Google Scholar] [CrossRef]
- Kann, A.P.; Hung, M.; Wang, W.; Nguyen, J.; Gilbert, P.M.; Wu, Z.; Krauss, R.S. An Injury-Responsive Rac-to-Rho GTPase Switch Drives Activation of Muscle Stem Cells through Rapid Cytoskeletal Remodeling. Cell Stem Cell 2022, 29, 933–947.e6. [Google Scholar] [CrossRef]
- Rodgers, J.T.; King, K.Y.; Brett, J.O.; Cromie, M.J.; Charville, G.W.; Maguire, K.K.; Brunson, C.; Mastey, N.; Liu, L.; Tsai, C.-R.; et al. mTORC1 Controls the Adaptive Transition of Quiescent Stem Cells from G0 to G(Alert). Nature 2014, 510, 393–396. [Google Scholar] [CrossRef]
- Ancel, S.; Michaud, J.; Sizzano, F.; Tauzin, L.; Oliveira, M.; Migliavacca, E.; Schüler, S.C.; Raja, S.; Dammone, G.; Karaz, S.; et al. A Dual-Color PAX7 and MYF5 in Vivo Reporter to Investigate Muscle Stem Cell Heterogeneity in Regeneration and Aging. Stem Cell Rep. 2024, 19, 1024–1040. [Google Scholar] [CrossRef]
- Wilson, A.; Laurenti, E.; Oser, G.; van der Wath, R.C.; Blanco-Bose, W.; Jaworski, M.; Offner, S.; Dunant, C.F.; Eshkind, L.; Bockamp, E.; et al. Hematopoietic Stem Cells Reversibly Switch from Dormancy to Self-Renewal during Homeostasis and Repair. Cell 2008, 135, 1118–1129. [Google Scholar] [CrossRef]
- Laurenti, E.; Frelin, C.; Xie, S.; Ferrari, R.; Dunant, C.F.; Zandi, S.; Neumann, A.; Plumb, I.; Doulatov, S.; Chen, J.; et al. CDK6 Levels Regulate Quiescence Exit in Human Hematopoietic Stem Cells. Cell Stem Cell 2015, 16, 302–313. [Google Scholar] [CrossRef]
- Ibneeva, L.; Singh, S.P.; Sinha, A.; Eski, S.E.; Wehner, R.; Rupp, L.; Kovtun, I.; Pérez-Valencia, J.A.; Gerbaulet, A.; Reinhardt, S.; et al. CD38 Promotes Hematopoietic Stem Cell Dormancy. PLoS Biol. 2024, 22, e3002517. [Google Scholar] [CrossRef] [PubMed]
- Mann, Z.; Sengar, M.; Verma, Y.K.; Rajalingam, R.; Raghav, P.K. Hematopoietic Stem Cell Factors: Their Functional Role in Self-Renewal and Clinical Aspects. Front. Cell Dev. Biol. 2022, 10, 664261. [Google Scholar] [CrossRef] [PubMed]
- Ruffinatto, L.; Groult, Y.; Iacono, J.; Sarrazin, S.; de Laval, B. Hematopoietic Stem Cell a Reservoir of Innate Immune Memory. Front. Immunol. 2024, 15, 1491729. [Google Scholar] [CrossRef] [PubMed]
- Marqués-Torrejón, M.Á.; Williams, C.A.C.; Southgate, B.; Alfazema, N.; Clements, M.P.; Garcia-Diaz, C.; Blin, C.; Arranz-Emparan, N.; Fraser, J.; Gammoh, N.; et al. LRIG1 Is a Gatekeeper to Exit from Quiescence in Adult Neural Stem Cells. Nat. Commun. 2021, 12, 2594. [Google Scholar] [CrossRef]
- Meng, H.; Huan, Y.; Zhang, K.; Yi, X.; Meng, X.; Kang, E.; Wu, S.; Deng, W.; Wang, Y. Quiescent Adult Neural Stem Cells: Developmental Origin and Regulatory Mechanisms. Neurosci. Bull. 2024, 40, 1353–1363. [Google Scholar] [CrossRef]
- Llorens-Bobadilla, E.; Zhao, S.; Baser, A.; Saiz-Castro, G.; Zwadlo, K.; Martin-Villalba, A. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells That Become Activated upon Brain Injury. Cell Stem Cell 2015, 17, 329–340. [Google Scholar] [CrossRef]
- Kalamakis, G.; Brüne, D.; Ravichandran, S.; Bolz, J.; Fan, W.; Ziebell, F.; Stiehl, T.; Catalá-Martinez, F.; Kupke, J.; Zhao, S.; et al. Quiescence Modulates Stem Cell Maintenance and Regenerative Capacity in the Aging Brain. Cell 2019, 176, 1407–1419.e14. [Google Scholar] [CrossRef]
- Wang, X.; Fujimaki, K.; Mitchell, G.C.; Kwon, J.S.; Della Croce, K.; Langsdorf, C.; Zhang, H.H.; Yao, G. Exit from Quiescence Displays a Memory of Cell Growth and Division. Nat. Commun. 2017, 8, 321. [Google Scholar] [CrossRef]
- de Jong, I.G.; Haccou, P.; Kuipers, O.P. Bet Hedging or Not? A Guide to Proper Classification of Microbial Survival Strategies. BioEssays 2011, 33, 215–223. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laporte, D.; Sagot, I. Quiescence Multiverse. Biomolecules 2025, 15, 960. https://doi.org/10.3390/biom15070960
Laporte D, Sagot I. Quiescence Multiverse. Biomolecules. 2025; 15(7):960. https://doi.org/10.3390/biom15070960
Chicago/Turabian StyleLaporte, Damien, and Isabelle Sagot. 2025. "Quiescence Multiverse" Biomolecules 15, no. 7: 960. https://doi.org/10.3390/biom15070960
APA StyleLaporte, D., & Sagot, I. (2025). Quiescence Multiverse. Biomolecules, 15(7), 960. https://doi.org/10.3390/biom15070960





