Integrated Multi-Omics of the Longissimus Dorsal Muscle Transcriptomics and Metabolomics Reveals Intramuscular Fat Accumulation Mechanism with Diet Energy Differences in Yaks
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sample Collection
2.2. Longissimus Dorsal Muscle Quality Measurement and Hematoxylin–Eosin Staining (HE Staining)
2.3. RNA-Seq Library Construction and Sequencing
2.4. Lipid Extraction and Data Preprocessing
2.5. Real-Time Fluorescence Quantitative PCR (RT-qPCR)
2.6. Statistical Analysis
3. Results
3.1. Determination of pH, Color Difference, Shear, and Cooking Loss of the LD
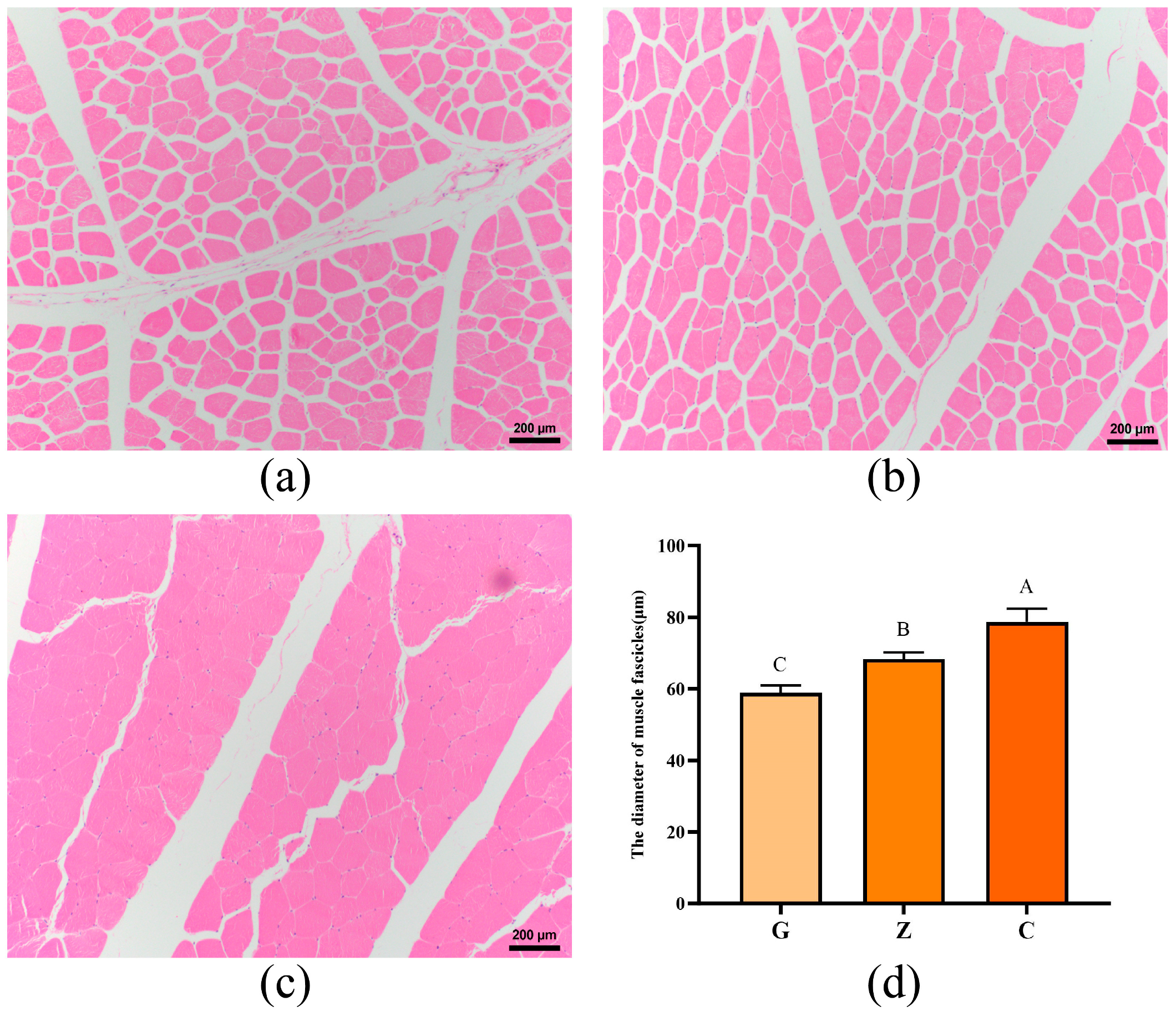
3.2. Histology of the Longissimus Dorsal Muscles
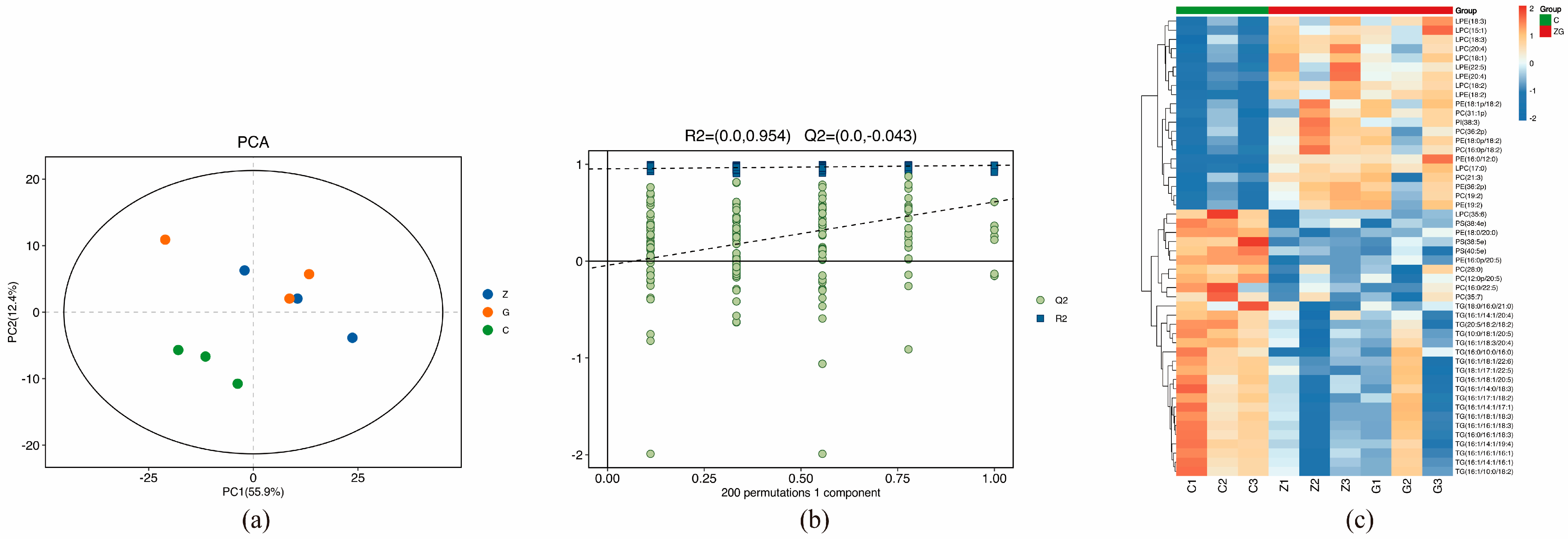
3.3. Lipidomics Results and Analysis
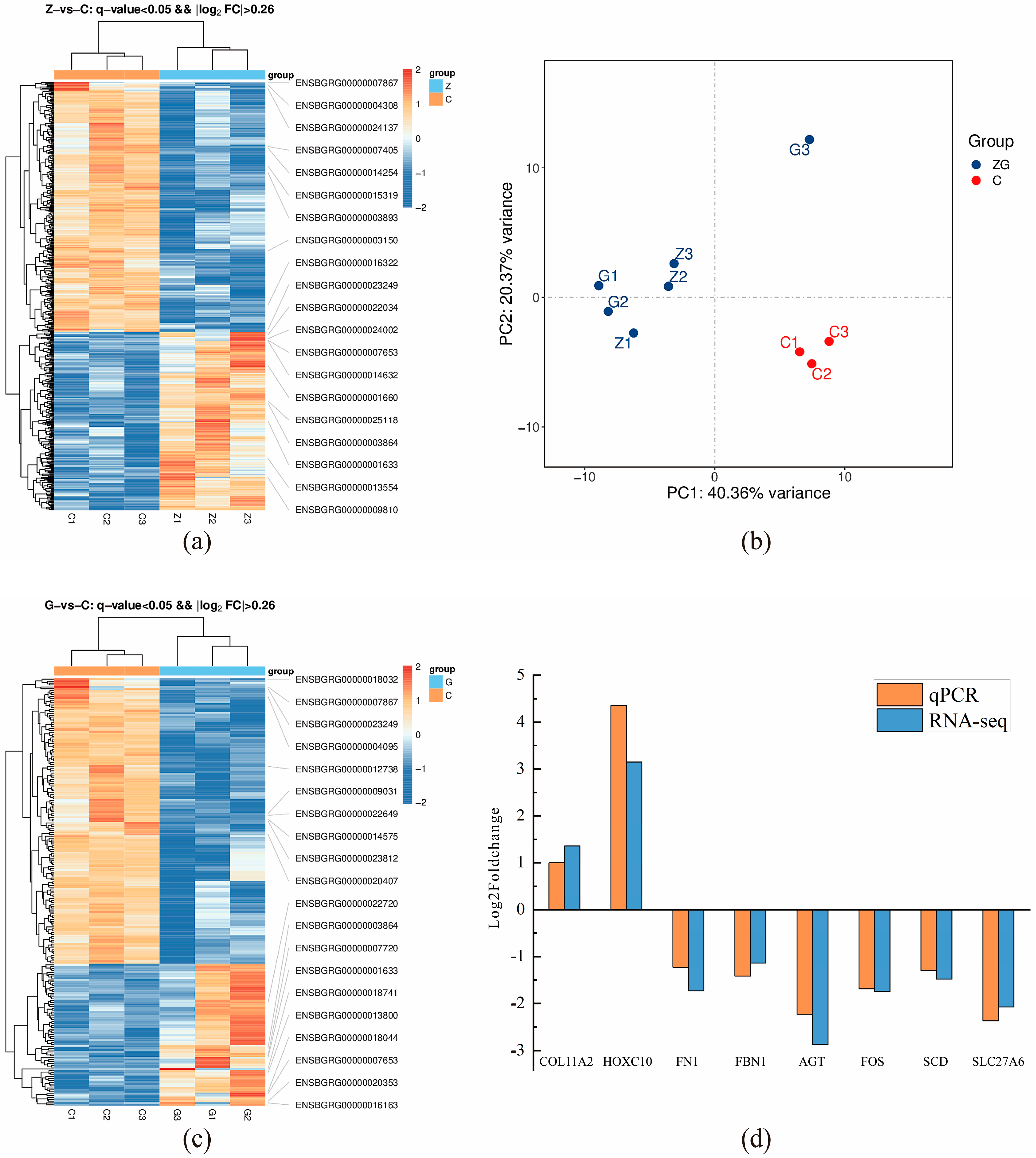
3.4. Transcriptomics Results and Analysis
3.5. Validation of mRNA Sequencing Using RT-qPCR
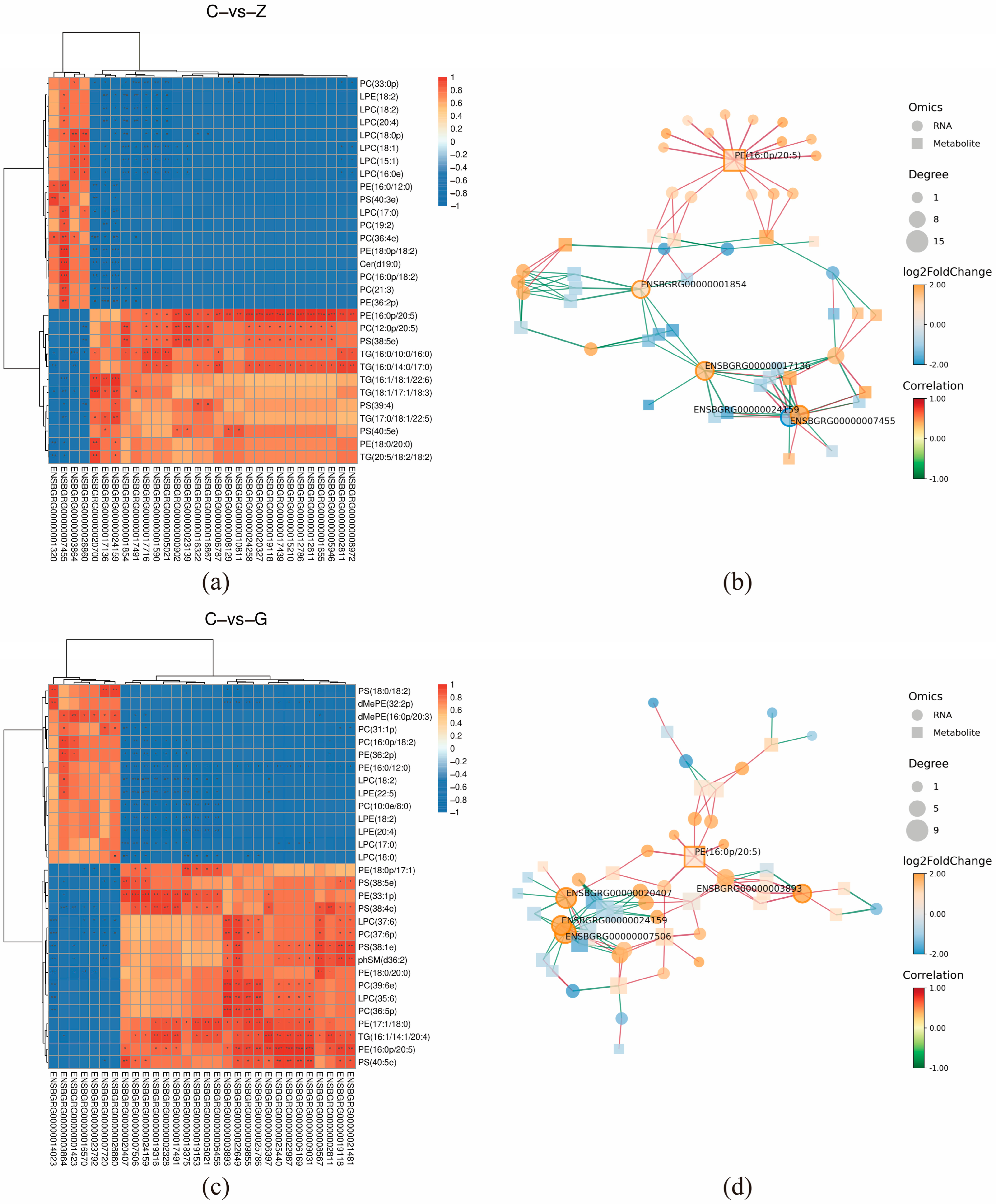
3.6. Correlation Analysis of Lipid Metabolomics and Transcriptomics
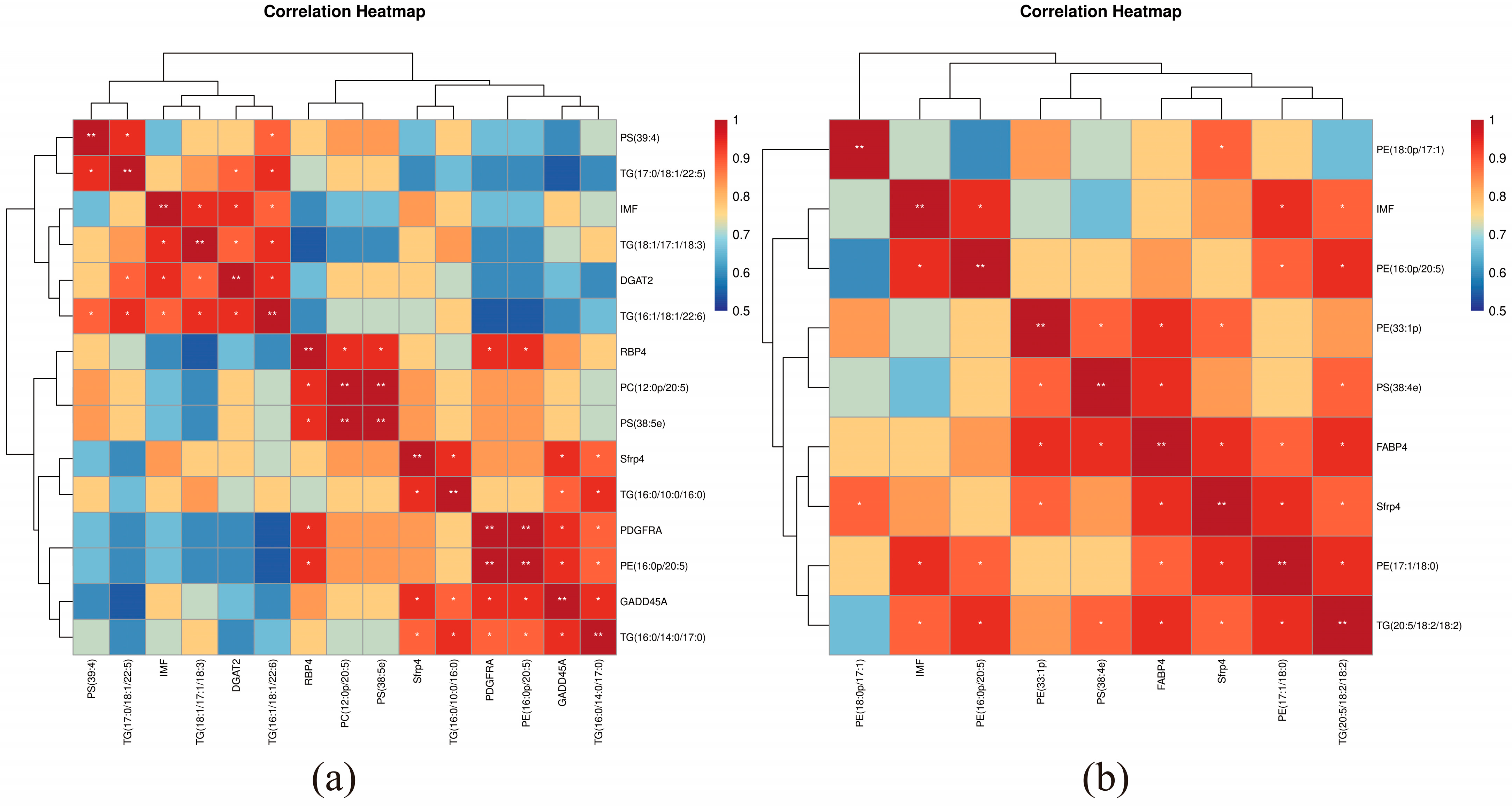
3.7. Correlation of IMF, DEG and SDL
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IMF | Intramuscular fat |
| LD | Longissimus dorsi muscle |
| DEG | Differentially expressed genes |
| SDL | Significant differential lipids |
| QC | Quality control |
| PCA | Principal component analysis |
| OPLS-DA | Orthogonal partial least squares discriminant analysis |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Jing, X.P.; Ding, L.M.; Zhou, J.W.; Huang, X.D.; Degen, A.; Long, R.J. The adaptive strategies of yaks to live in the Asian highlands. Anim. Nutr. 2002, 9, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zong, W.; Zhao, S.; Qie, M.; Yang, X.; Zhao, Y. Nutrition and edible characteristics, origin traceability and authenticity identification of yak meat and milk: A review. Trends Food Sci. Technol. 2023, 139, 104133. [Google Scholar] [CrossRef]
- Guo, X.; Wu, X.; Chu, M.; Bao, P.; Xiong, L.; Liang, C.; Ding, X.; Pei, J.; Yan, P. Characterization of the complete mitochondrial genome of the Pamir yak (Bos grunniens). Mitochondrial DNA B Resour. 2019, 4, 3165–3166. [Google Scholar] [CrossRef] [PubMed]
- Bao, P.J.; Ding, Z.Q.; Ma, X.M.; La, Y.F.; Guo, T.; Zheng, X.B.; Ma, J.E.D.; Dao, M.; Zhou, C.; Zuo, E.W.; et al. Study on Slaughter Performance and Meat Quality of Pamir Yak. China Herbiv. Sci. 2024, 44, 83–87. [Google Scholar]
- Zhong, J.C.; Wang, H.; Chai, Z.X.; Ma, Z.J. Exploitation and innovative utilization of yak germplasm resources. Chin. Livest. Poult. Breed. 2022, 18, 22–29. [Google Scholar]
- Zhao, J.; Xu, Z.; Chen, A.; You, X.; Zhao, Y.; He, W.; Zhao, L.; Yang, S. Identification of meat from yak and cattle using SNP markers with integrated allele-specific polymerase chain reaction-capillary electrophoresis method. Meat Sci. 2019, 148, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.L.; Gao, C.Q.; Wang, Q.; Zhang, Z.M.; Xu, Y.L.; Li, H.C.; Yan, H.C.; Wang, X.Q. Effects of pioglitazone hydrochloride and vitamin E on meat quality, antioxidant status and fatty acid profiles in finishing pigs. Meat Sci. 2018, 145, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Hou, B.; Zhao, Y.; He, P.; Xu, C.; Ma, P.; Lam, S.M.; Li, B.; Gil, V.; Shui, G.; Qiang, G.; et al. Targeted lipidomics and transcriptomics profiling reveal the heterogeneity of visceral and subcutaneous white adipose tissue. Life Sci. 2020, 245, 117352. [Google Scholar] [CrossRef] [PubMed]
- Dannenberger, D.; Nuernberg, G.; Scollan, N.; Ender, K.; Nuernberg, K. Diet alters the fatty acid composition of individual phospholipid classes in beef muscle. J. Agric. Food Chem. 2007, 55, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhong, J.; Zhang, C.; Chai, Z.; Cao, H.; Wang, J.; Zhu, J.; Wang, J.; Ji, Q. The whole-transcriptome landscape of muscle and adipose tissues reveals the ceRNA regulation network related to intramuscular fat deposition in yak. BMC Genom. 2020, 21, 347. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Hu, J.; Li, L.; Xue, L.; Tian, J.; Zhang, T.; Yang, L.; Gu, Y.; Zhang, J. Integrating lipidomics and metabolomics to reveal biomarkers of fat deposition in chicken meat. Food Chem. 2025, 464 Pt 2, 141732. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.C.; Bruce, H.L. Contribution of intramuscular connective tissue and its structural components on meat tenderness-revisited: A review. Crit. Rev. Food Sci. Nutr. 2024, 64, 9280–9310. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Lamas, L.; Barron, L.J.R.; Farmer, L.; Aldai, N. Fatty acid composition of intramuscular fat and odour-active compounds of lamb commercialized in northern Spain. Meat Sci. 2018, 139, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Igene, J.O.; Pearson, A.M. Role of phospholipids and triglycerides in warmed-over flavor development in meat model systems. J. Food Sci. 1979, 44, 1285–1290. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Y.; Hussain, T.; Dai, C.; Li, J.; Huang, P.; Li, Y.; Ding, X.; Huang, J.; Ji, F.; et al. Effects of dietary energy level on growth performance, blood parameters and meat quality in fattening male Hu lambs. J. Anim. Physiol. Anim. Nutr. 2020, 104, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Li, N.Y.; Yang, M.H.; Bi, B.L.; Kong, L.F.; Gao, S.Z.; Zhao, S.M.; Su, Z.F.; Huang, Y. Effect of Dietary Energy Levels on Gene Expression Related with Intramuscular Fat Deposition in Muscle Tissue of Wujin Pigs. Chin. J. Anim. Sci. 2014, 50, 41–45. [Google Scholar]
- Kang, K.; Ma, J.; Wang, H.; Wang, Z.; Peng, Q.; Hu, R.; Zou, H.; Bao, S.; Zhang, W.; Sun, B. High-energy diet improves growth performance, meat quality and gene expression related to intramuscular fat deposition in finishing yaks raised by barn feeding. Vet. Med. Sci. 2020, 6, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, H.; Wu, F.; Qiu, X.; Yu, Z.; Niu, W.; He, Y.; Su, H.; Cao, B. Effects of Dietary Energy on Growth Performance, Rumen Fermentation and Bacterial Community, and Meat Quality of Holstein-Friesians Bulls Slaughtered at Different Ages. Animals 2019, 9, 1123. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhu, Y.; Wang, X.; He, Y.; Cao, B. Effects of different dietary energy and protein levels and sex on growth performance, carcass characteristics and meat quality of F1 Angus × Chinese Xiangxi yellow cattle. J. Anim. Sci. Biotechnol. 2014, 5, 21. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Chen, Y. Transcriptomics: Advances and approaches. Sci. China Life Sci. 2013, 56, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Hatzirodos, N.; Irving-Rodgers, H.F.; Hummitzsch, K.; Harland, M.L.; Morris, S.E.; Rodgers, R.J. Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes. BMC Genom. 2014, 15, 24. [Google Scholar] [CrossRef] [PubMed]
- Idle, J.R.; Gonzalez, F.J. Metabolomics. Cell Metab. 2007, 6, 348–351. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Fischer, H.P. Mathematical modeling of complex biological systems: From parts lists to understanding systems behavior. Alcohol. Res. Health 2008, 31, 49–59. [Google Scholar] [PubMed]
- Du, L.; Chang, T.; An, B.; Liang, M.; Deng, T.; Li, K.; Cao, S.; Du, Y.; Gao, X.; Xu, L.; et al. Transcriptomics and Lipid Metabolomics Analysis of Subcutaneous, Visceral, and Abdominal Adipose Tissues of Beef Cattle. Genes 2022, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Zhang, X.; Wang, Z.; Li, F.; Yue, X. Integrating lipidomics and transcriptomics to reveal the heterogeneity of sheep adipose tissues. Food Biosci. 2024, 60, 104393. [Google Scholar] [CrossRef]
- Meyer, S.; Gessner, D.K.; Braune, M.S.; Friedhoff, T.; Most, E.; Höring, M.; Liebisch, G.; Zorn, H.; Eder, K.; Ringseis, R. Comprehensive evaluation of the metabolic effects of insect meal from Tenebrio molitor L. in growing pigs by transcriptomics, metabolomics and lipidomics. J. Anim. Sci. Biotechnol. 2020, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Lin, Z.; Peng, H.; Zhang, S.; Zhang, Z.; Zhang, X.; Nie, Q.; Luo, W. The transmembrane protein TMEM182 promotes fat deposition and alters metabolomics and lipidomics. Int. J. Biol. Macromol. 2024, 259 Pt 1, 129144. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Hu, H.; Lin, H.; Wang, C.; Wang, Y.; Wang, J. Muscle Transcriptome Analysis Reveals Potential Candidate Genes and Pathways Affecting Intramuscular Fat Content in Pigs. Front. Genet. 2020, 11, 877. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liao, Q.; Sun, Y.; Pan, T.; Liu, S.; Miao, W.; Li, Y.; Zhou, L.; Xu, G. Lipidomic and Transcriptomic Analysis of the Longissimus Muscle of Luchuan and Duroc Pigs. Front. Nutr. 2021, 8, 667622. [Google Scholar] [CrossRef] [PubMed]
- Bo, Q. Intramuscular Fat (IMF) Deposition Pattern and its FatMetabolism Mechanism in Yaks of Different Ages. Ph.D. Thesis, Southwest Minzu University, Chengdu, China, 2023. [Google Scholar]
- Peng, Q.H.; Wang, Z.S.; Tan, C.; Zhang, H.B.; Hu, Y.N.; Zou, H.W. Effects of different pomace and pulp dietary energy density on growth performance and intramuscular fat deposition relating mRNA expression in beef cattle. J. Food Agric. Environ. 2012, 10, 404–407. [Google Scholar]
- Smet, S.D.; Webb, E.C.; Claeys, E.; Uytterhaegen, L.; Demeyer, D.I. Effect of dietary energy and protein levels on fatty acid composition of intramuscular fat in double-muscled Belgian Blue bulls. Meat Sci. 2000, 56, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, Y.; Wu, J.; Qiao, M.; Xu, Z.; Peng, X.; Mei, S. Proteomic and lipidomic analyses reveal saturated fatty acids, phosphatidylinositol, phosphatidylserine, and associated proteins contributing to intramuscular fat deposition. J. Proteomics 2021, 241, 104235. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhu, M.; Chai, W.; Wang, Y.; Song, Y.; Liu, B.; Cai, C.; Song, Y.; Sun, X.; Xue, P.; et al. Determination of the Heterogeneity of Intramuscular Fat and Visceral Adipose Tissue from Dezhou Donkey by Lipidomics and Transcriptomics Profiling. Front. Nutr. 2021, 8, 746684. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, M.; Suh, J.M.; Hah, N.; Liddle, C.; Atkins, A.R.; Downes, M.; Evans, R.M. PPARγ signaling and metabolism: The good, the bad and the future. Nat. Med. 2013, 19, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.Y.; Kim, J.S.; Nguyen, T.H.; Lee, H.J.; Baik, M. Wnt/β-catenin signaling and adipogenic genes are associated with intramuscular fat content in the longissimus dorsi muscle of Korean cattle. Anim. Genet. 2013, 44, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Hörbelt, T.; Knebel, B.; Fahlbusch, P.; Barbosa, D.; de Wiza, D.H.; Van de Velde, F.; Van Nieuwenhove, Y.; Lapauw, B.; Thoresen, G.H.; Al-Hasani, H.; et al. The adipokine sFRP4 induces insulin resistance and lipogenesis in the liver. Biochim. Biophys. Acta Mol. Basis. Dis. 2019, 1865, 2671–2684. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Liang, J.; Yang, L.; Lan, G.; Wang, L.; Wang, L. Systematic Identification and Comparison of the Expressed Profiles of lncRNAs, miRNAs, circRNAs, and mRNAs with Associated Co-Expression Networks in Pigs with Low and High Intramuscular Fat. Animals 2021, 11, 3212. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Jiang, H. Molecular and Cellular Mechanisms of Intramuscular Fat Development and Growth in Cattle. Int. J. Mol. Sci. 2024, 25, 2520. [Google Scholar] [CrossRef] [PubMed]
- Goszczynski, D.E.; Papaleo-Mazzucco, J.; Ripoli, M.V.; Villarreal, E.L.; Rogberg-Muñoz, A.; Mezzadra, C.A.; Melucci, L.M.; Giovambattista, G. Genetic Variation in FABP4 and Evaluation of Its Effects on Beef Cattle Fat Content. Anim. Biotechnol. 2017, 28, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Cui, H.; Dong, N.; Zhu, X.; Li, S.; Ma, X.; Niu, D. Effects of phosphatidylethanolamine on intramuscular fat deposition and key gene identification by transcriptome sequencing in broiler chickens. Poult. Sci. 2025, 104, 104914. [Google Scholar] [CrossRef] [PubMed]
- Pannier, L.; Mullen, A.M.; Hamill, R.M.; Stapleton, P.C.; Sweeney, T. Association analysis of single nucleotide polymorphisms in DGAT1, TG and FABP4 genes and intramuscular fat in crossbred Bos taurus cattle. Meat Sci. 2010, 85, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhu, R.; Yang, X.; Feng, Y.; He, Q.; Wang, H.; Liu, Q.; Shi, D.; Huang, J. The role of lncFABP4 in modulating adipogenic differentiation in buffalo intramuscular preadipocytes. Anim. Sci. J. 2024, 95, e13951. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Shu, G.; Yuan, F.; Zhu, X.; Gao, P.; Wang, S.; Wang, L.; Xi, Q.; Zhang, S.; Zhang, Y.; et al. Fatty acid and transcriptome profiling of longissimus dorsi muscles between pig breeds differing in meat quality. Int. J. Biol. Sci. 2013, 9, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Sadkowski, T.; Ciecierska, A.; Majewska, A.; Oprządek, J.; Dasiewicz, K.; Ollik, M.; Wicik, Z.; Motyl, T. Transcriptional background of beef marbling—Novel genes implicated in intramuscular fat deposition. Meat Sci. 2014, 97, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.S.; Lee, K.T.; Choi, J.W.; Jeon, H.J.; Lee, S.W.; Cho, Y.M.; Kim, T.H. Novel SNPs in the growth arrest and DNA damage-inducible protein 45 alpha gene (GADD45A) associated with meat quality traits in Berkshire pigs. Genet. Mol. Res. 2015, 14, 8581–8588. [Google Scholar] [CrossRef] [PubMed]
- You, W.; Liu, S.; Ji, J.; Ling, D.; Tu, Y.; Zhou, Y.; Chen, W.; Valencak, T.G.; Wang, Y.; Shan, T. Growth arrest and DNA damage-inducible alpha regulates muscle repair and fat infiltration through ATP synthase F1 subunit alpha. J. Cachexia Sarcopenia Muscle 2023, 14, 326–341. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zhang, Z.; Yuan, Z.; Lo, L.J.; Chen, J.; Wang, Y.; Peng, J. Distinctive genes determine different intramuscular fat and muscle fiber ratios of the longissimus dorsi muscles in Jinhua and landrace pigs. PLoS ONE 2013, 8, e53181. [Google Scholar] [CrossRef] [PubMed]
- Bénézech, C.; Mader, E.; Desanti, G.; Khan, M.; Nakamura, K.; White, A.; Ware, C.F.; Anderson, G.; Caamaño, J.H. Lymphotoxin-β receptor signaling through NF-κB2-RelB pathway reprograms adipocyte precursors as lymph node stromal cells. Immunity 2012, 37, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.M.; Qin, J.; Liu, S.G.; Cai, R.; Chen, X.C.; Wang, X.M.; Pang, W.J. PDGFRα Regulated by miR-34a and FoxO1 Promotes Adipogenesis in Porcine Intramuscular Preadipocytes through Erk Signaling Pathway. Int. J. Mol. Sci. 2017, 18, 2424. [Google Scholar] [CrossRef] [PubMed]
- Martins, T.S.; Sanglard, L.M.; Silva, W.; Chizzotti, M.L.; Rennó, L.N.; Serão, N.V.; Silva, F.F.; Guimarães, S.E.; Ladeira, M.M.; Dodson, M.V.; et al. Molecular Factors Underlying the Deposition of Intramuscular Fat and Collagen in Skeletal Muscle of Nellore and Angus Cattle. PLoS ONE 2015, 10, e0139943. [Google Scholar] [CrossRef] [PubMed]
- Blaner, W.S. Retinol-binding protein: The serum transport protein for vitamin A. Endocr. Rev. 1989, 10, 308–316. [Google Scholar] [CrossRef] [PubMed]
- de Jager, N.; Hudson, N.J.; Reverter, A.; Barnard, R.; Cafe, L.M.; Greenwood, P.L.; Dalrymple, B.P. Gene expression phenotypes for lipid metabolism and intramuscular fat in skeletal muscle of cattle. J. Anim. Sci. 2013, 91, 1112–1128. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Albrecht, E.; Dannenberger, D.; Hammon, H.M.; Kuehn, C.; Sauerwein, H.; Yang, R.; Zhao, Z.; Maak, S. Retinol binding protein 4 abundance in plasma and tissues is related to body fat deposition in cattle. Sci. Rep. 2019, 9, 8056. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Yin, Z.; Wang, M.; Zhang, W.; Raza, S.H.A.; Althobaiti, F.; Qi, L.; Wang, J. Expression of DGAT2 Gene and Its Associations with Intramuscular Fat Content and Breast Muscle Fiber Characteristics in Domestic Pigeons (Columba livia). Front. Vet. Sci. 2022, 9, 847363. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.X.; Zeng, Y.Q.; Wang, H.; Chen, W.; Du, J.F.; Chen, Q.M.; Hu, Y.X.; Yang, L. The effects of DGAT1 and DGAT2 mRNA expression on fat deposition in fatty and lean breeds of pig. Livest. Sci. 2011, 140, 292–296. [Google Scholar] [CrossRef]
- Wang, H.; Wang, J.; Yang, D.D.; Liu, Z.L.; Zeng, Y.Q.; Chen, W. Expression of lipid metabolism genes provides new insights into intramuscular fat deposition in Laiwu pigs. Asian-Australas. J. Anim. Sci. 2020, 33, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Kwon, E.G.; Im, S.K.; Seo, K.S.; Baik, M. Expression of fat deposition and fat removal genes is associated with intramuscular fat content in longissimus dorsi muscle of Korean cattle steers. J. Anim. Sci. 2012, 90, 2044–2053. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shi, B.; Xie, J.; Zhou, H.; Wang, J.; Liu, X.; Li, S.; Zhao, Z.; Luo, Y. Tissue Expression and Variation of the DGAT2 Gene and Its Effect on Carcass and Meat Quality Traits in Yak. Animals 2019, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, Q.; Lin, Y.; Wang, Y.; Shi, H.; Xiang, H.; Zhu, J. Diacylglycerol acyltransferase 2 promotes the adipogenesis of intramuscular preadipocytes in goat. Anim. Biotechnol. 2023, 34, 2376–2383. [Google Scholar] [CrossRef] [PubMed]
- Nian, Z.; Sun, Z.; Yu, L.; Toh, S.Y.; Sang, J.; Li, P. Fat-specific protein 27 undergoes ubiquitin-dependent degradation regulated by triacylglycerol synthesis and lipid droplet formation. J. Biol. Chem. 2010, 285, 9604–9615. [Google Scholar] [CrossRef] [PubMed]
| Item | Low Energy Supply (C) | Medium Energy Supply (Z) | High Energy Supply (G) |
|---|---|---|---|
| Ingredient (%) | |||
| Corn | 37.4 | 59.5 | 68.8 |
| 46 Cotton meal | 0 | 9.2 | 10 |
| 43 Soybean meal | 0 | 12.1 | 13 |
| Soybean Oil | 0 | 0 | 1.2 |
| Bran (especially wheat) | 7.6 | 11.7 | 2 |
| Syrup | 0 | 2.5 | 0 |
| Premix ② | 5 | 5 | 5 |
| Total | 100.00 | 100.00 | 100.00 |
| Nutrient Level ① | |||
| Crude protein (%) | 9.95 | 19.38 | 19.17 |
| Net energy of weight Gain (MJ/Kg) | 0.75 | 1.38 | 1.53 |
| Gene | Primer Sequences (5′–3′) | Annealing Temperature (°C) |
|---|---|---|
| COL11A2 | F: ACCACCAAGACTTCACAGGC R: GGGGTGGGGTCCTGATAAT | 60.3 |
| HOXC10 | F: CGACAACGAAGCAAAAGAGGAG R: TCCAGCGTCTGGTGTTTAGT | 60.3 |
| FN1 | F: CCCTGGTGTCACAGAAGCTA R: CTGGGGGAGCTCATCTGTCTT | 60.3 |
| FBN1 | F: TGGCTCCAGATCCATCCAACA R: CCTTTCTGGCACAGACAGTGA | 60.3 |
| AGT | F: GACCCAAATCTCGCTGCTGA R: GAAGCCCCTCATCTTTCCTTGG | 60.3 |
| FOS | F: AGGGGCAAGGTAGAACAGTTG R: CTAGTTGGTCTGTCTCCGCTT | 60.3 |
| SCD | F: TCCCGACGTGGCTTTTTCTT R: CACCAGGTTTGTAGTACCTCCT | 60.3 |
| SLC27A6 | F: TGTGGTTGTGCCAGGTTATGA R: AAAACTGTGGACGAGCGTAAG | 60.3 |
| β-actin | F: GCAGGTCATCACCATCGG R: CCGTGTTGGCGTAGAGGT | 60.3 |
| Item | Group C | Group Z | Group G |
|---|---|---|---|
| pH*45min | 6.99 ± 0.00 a | 6.7 ± 0.02 b | 5.49 ± 0.03 c |
| L*45min | 9.25 ± 0.03 c | 9.48 ± 0.02 a | 9.32 ± 0.04 b |
| a*45min | 20.78 ± 0.03 c | 21.31 ± 0.22 b | 21.70 ± 0.11 a |
| b*45min | 3.39 ± 0.03 c | 3.54 ± 0.02 b | 3.82 ± 0.03 a |
| pH*24h | 6.42 ± 0.02 a | 5.64 ± 0.03 b | 5.28 ± 0.02 c |
| L*24h | 9.78 ± 0.06 b | 10.75 ± 0.10 a | 10.79 ± 0.05 a |
| a*24h | 21.39 ± 0.13 b | 21.51 ± 0.07 b | 23.49 ± 0.02 a |
| b*24h | 4.50 ± 0.12 b | 3.95 ± 0.06 c | 4.77 ± 0.00 a |
| Steaming loss (%) | 30.42 ± 1.15 | 29.71 ± 2.98 | 27.67 ± 2.86 |
| Shearing force (kg f) | 100.90 ± 0.61 a | 98.27 ± 1.78 a | 83.69 ± 1.89 b |
| Intramuscular fat (g/100 g) | 0.97 ± 0.02 b | 1.25 ± 0.07 b | 2.04 ± 0.42 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, J.; Bao, P.; Li, N.; Kong, S.; Wang, T.; Zhang, M.; Yu, Q.; Cao, X.; Jia, J.; Yan, P. Integrated Multi-Omics of the Longissimus Dorsal Muscle Transcriptomics and Metabolomics Reveals Intramuscular Fat Accumulation Mechanism with Diet Energy Differences in Yaks. Biomolecules 2025, 15, 1025. https://doi.org/10.3390/biom15071025
Deng J, Bao P, Li N, Kong S, Wang T, Zhang M, Yu Q, Cao X, Jia J, Yan P. Integrated Multi-Omics of the Longissimus Dorsal Muscle Transcriptomics and Metabolomics Reveals Intramuscular Fat Accumulation Mechanism with Diet Energy Differences in Yaks. Biomolecules. 2025; 15(7):1025. https://doi.org/10.3390/biom15071025
Chicago/Turabian StyleDeng, Jingying, Pengjia Bao, Ning Li, Siyuan Kong, Tong Wang, Minghao Zhang, Qinran Yu, Xinyu Cao, Jianlei Jia, and Ping Yan. 2025. "Integrated Multi-Omics of the Longissimus Dorsal Muscle Transcriptomics and Metabolomics Reveals Intramuscular Fat Accumulation Mechanism with Diet Energy Differences in Yaks" Biomolecules 15, no. 7: 1025. https://doi.org/10.3390/biom15071025
APA StyleDeng, J., Bao, P., Li, N., Kong, S., Wang, T., Zhang, M., Yu, Q., Cao, X., Jia, J., & Yan, P. (2025). Integrated Multi-Omics of the Longissimus Dorsal Muscle Transcriptomics and Metabolomics Reveals Intramuscular Fat Accumulation Mechanism with Diet Energy Differences in Yaks. Biomolecules, 15(7), 1025. https://doi.org/10.3390/biom15071025






