New Insights into the Sex Chromosome Evolution of the Common Barker Frog Species Complex (Anura, Leptodactylidae) Inferred from Its Satellite DNA Content
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens
2.2. Whole Genome Sequencing and Primary Analysis
2.3. RepeatExplorer Analysis
2.4. RepeatMasker Analysis
2.5. Criteria Used for satDNA Designation and Variants Delimitation
2.6. Chromosome Mapping of satDNAs by Fluorescent In Situ Hybridization
3. Results
3.1. Characterization of the Ph. ephippifer Satellitome
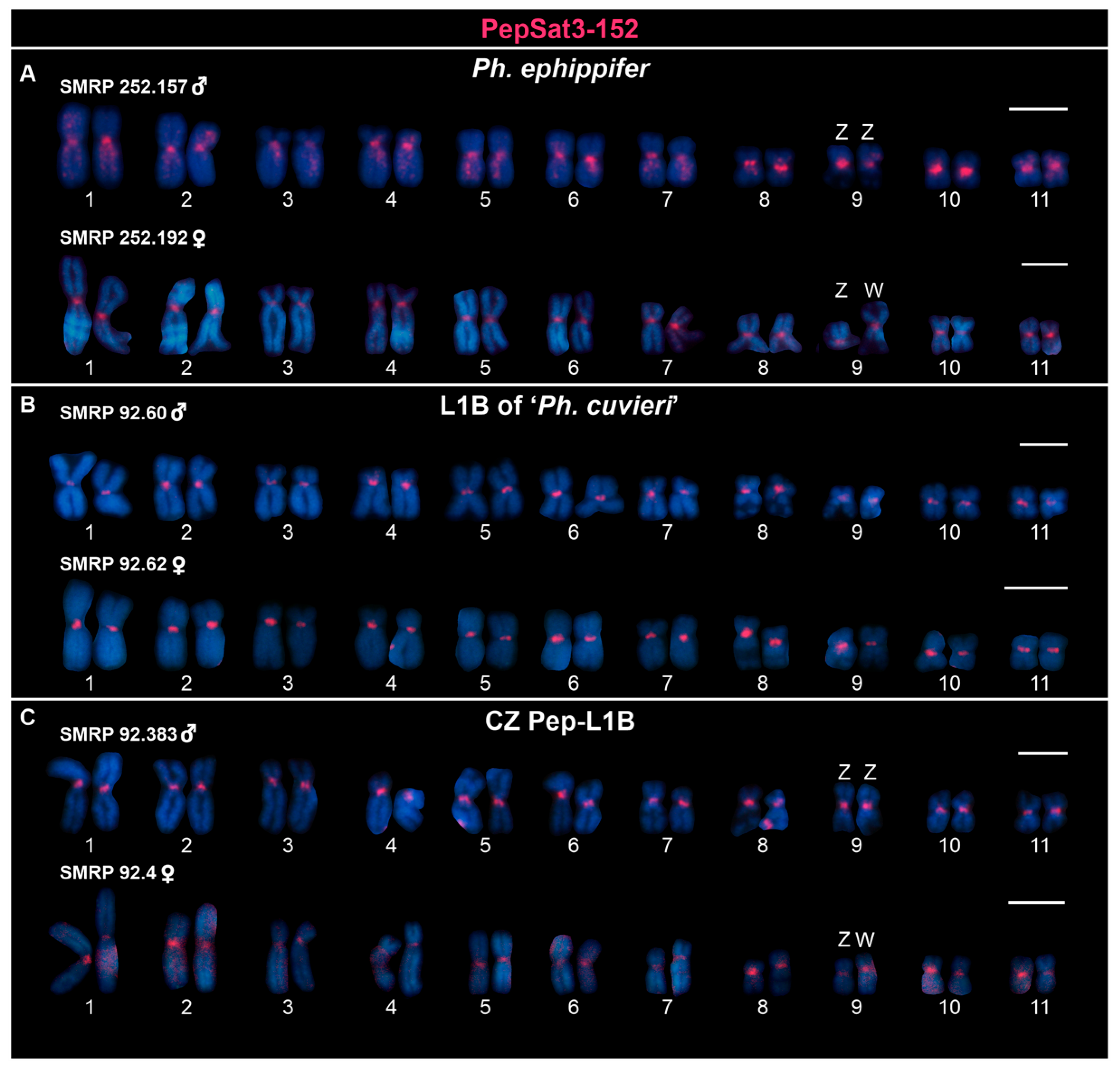
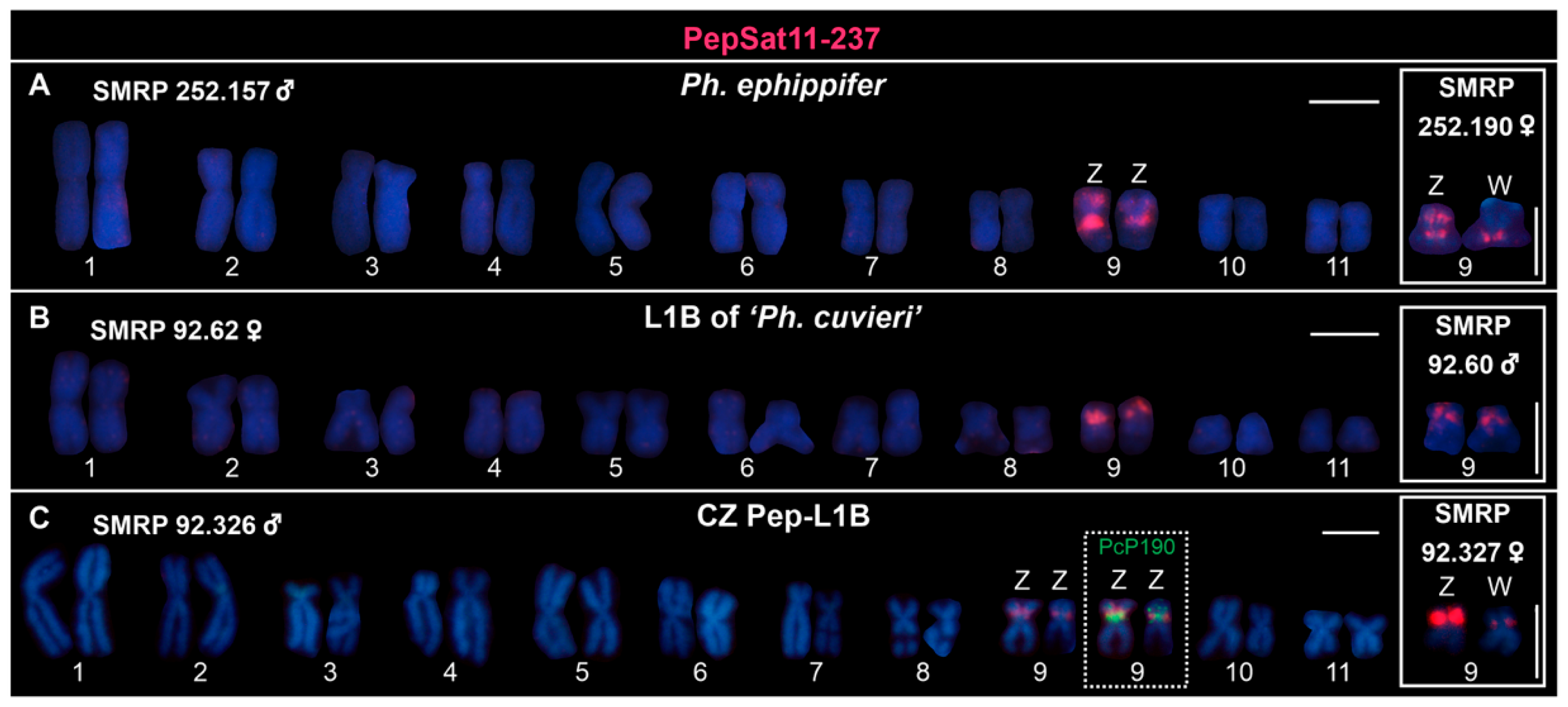
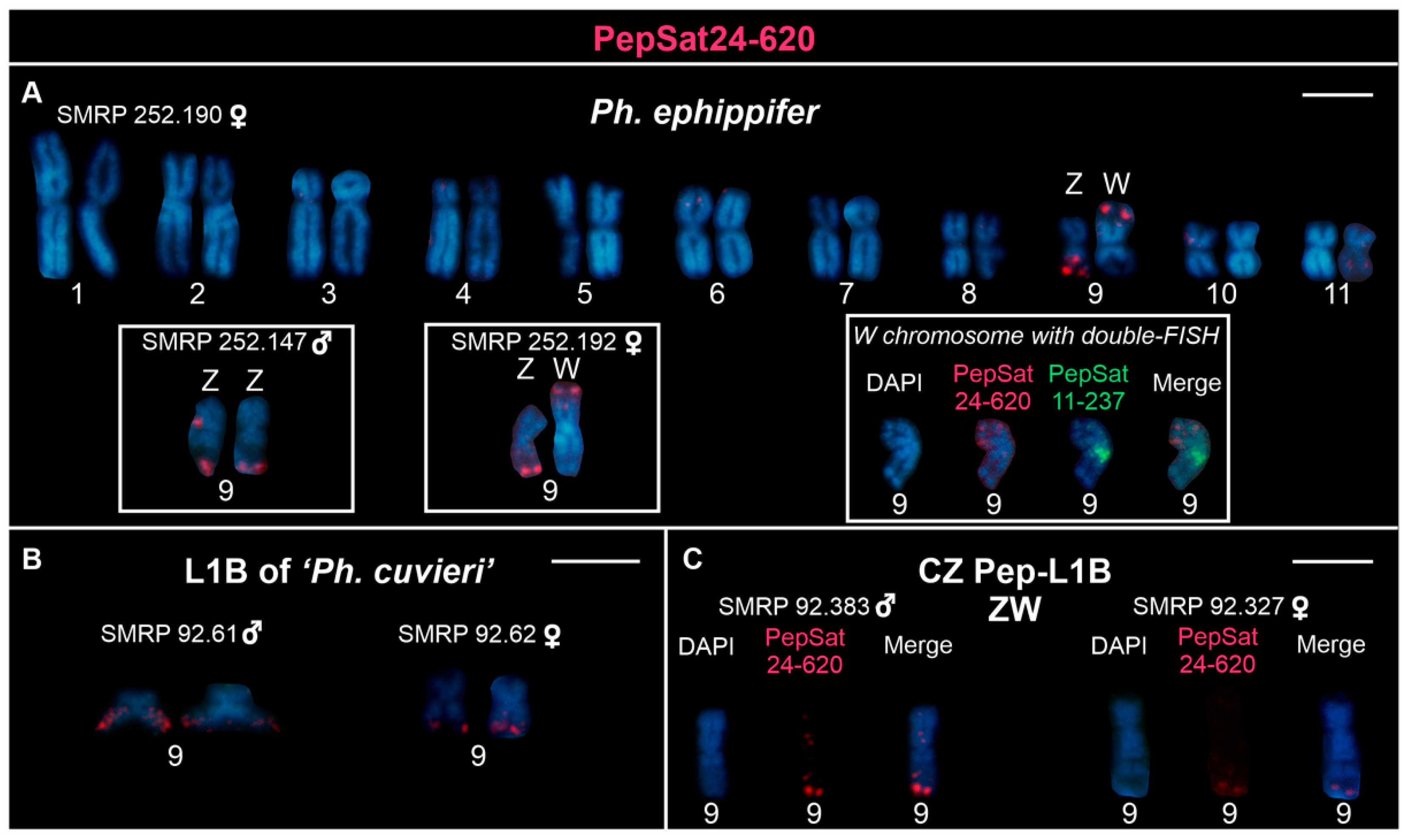
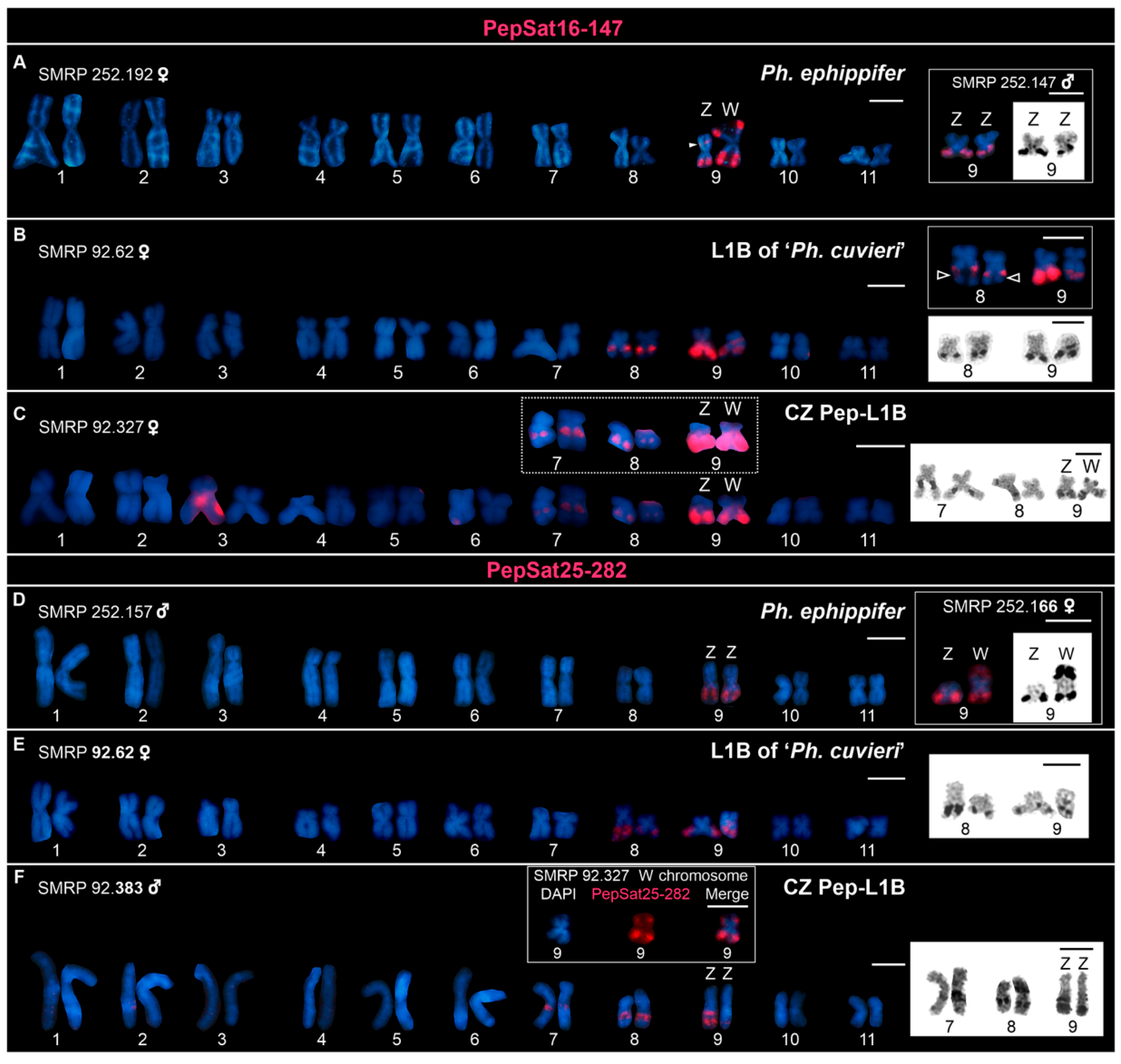
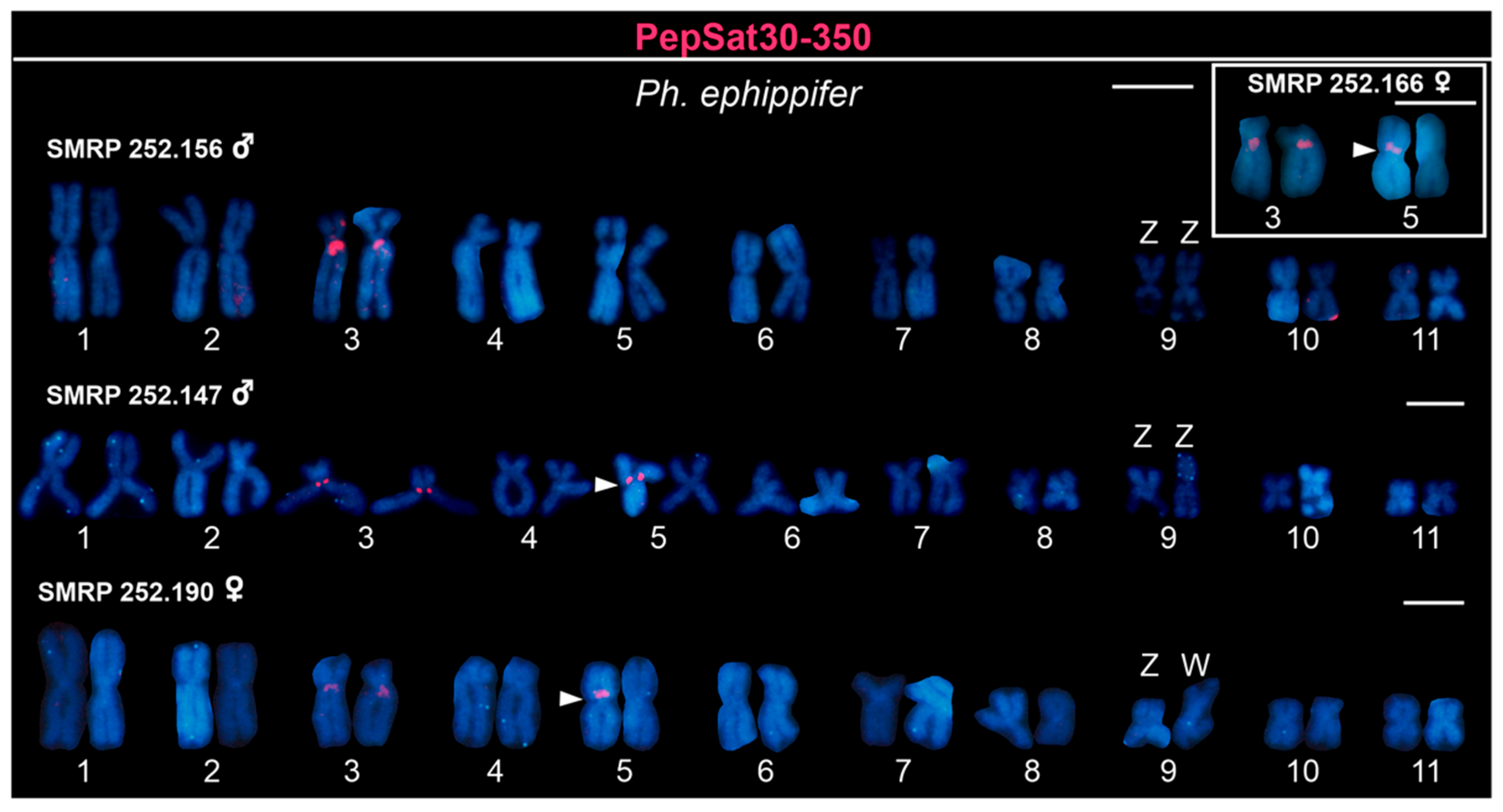
3.2. Chromosome Mapping of Satellite DNAs in the Clade Ph. ephippifer–CZ Pep-L1B–L1B of ‘Ph. cuvieri’
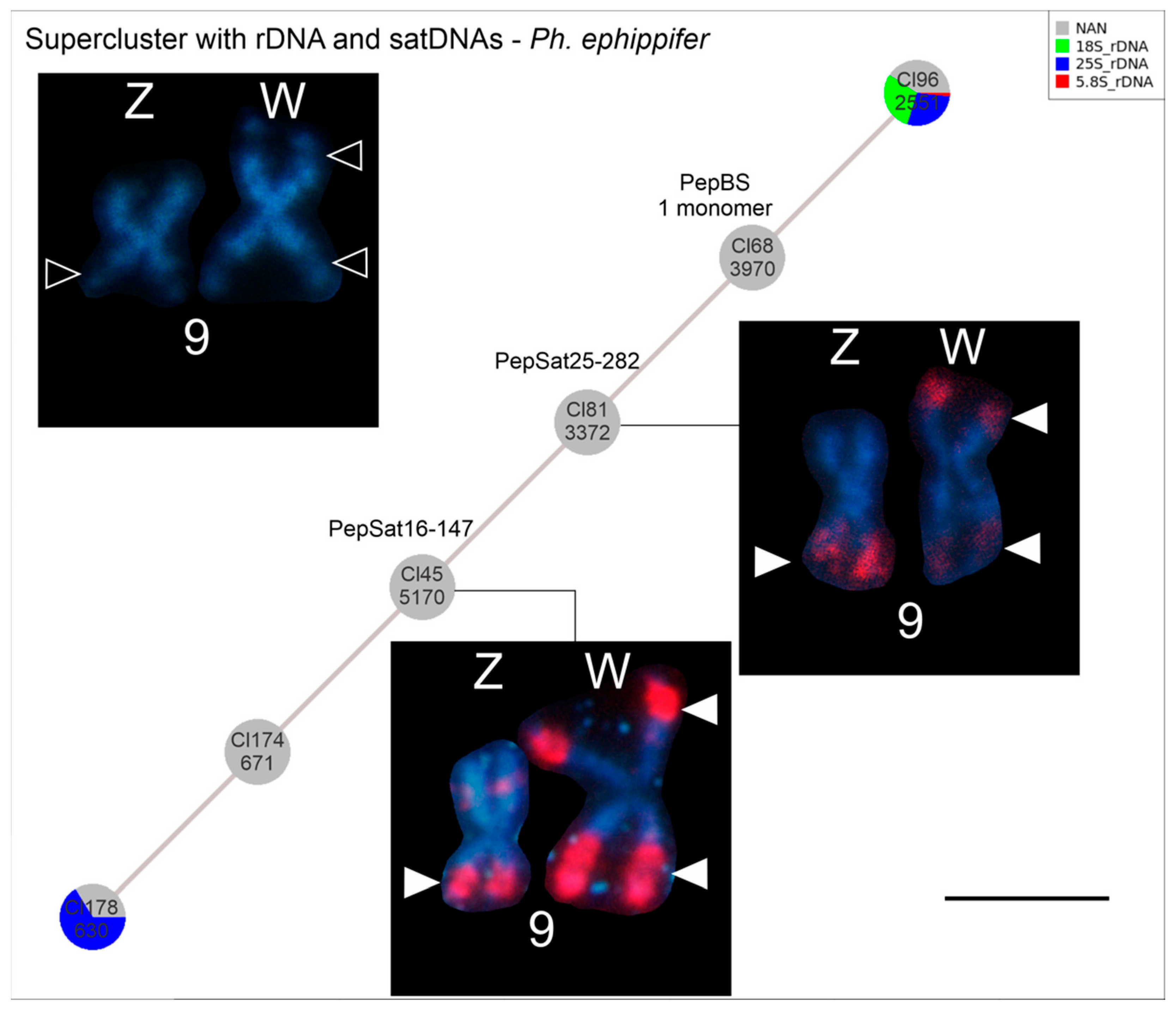
3.3. Supercluster Formed by rDNA and Satellite DNAs
4. Discussion
4.1. Genome Size Estimation
4.2. General Aspects of the Satellitome of Ph. ephippifer
4.3. Common Challenges in the Study of Satellite DNA Revealed by the Analysis of Ph. ephippifer
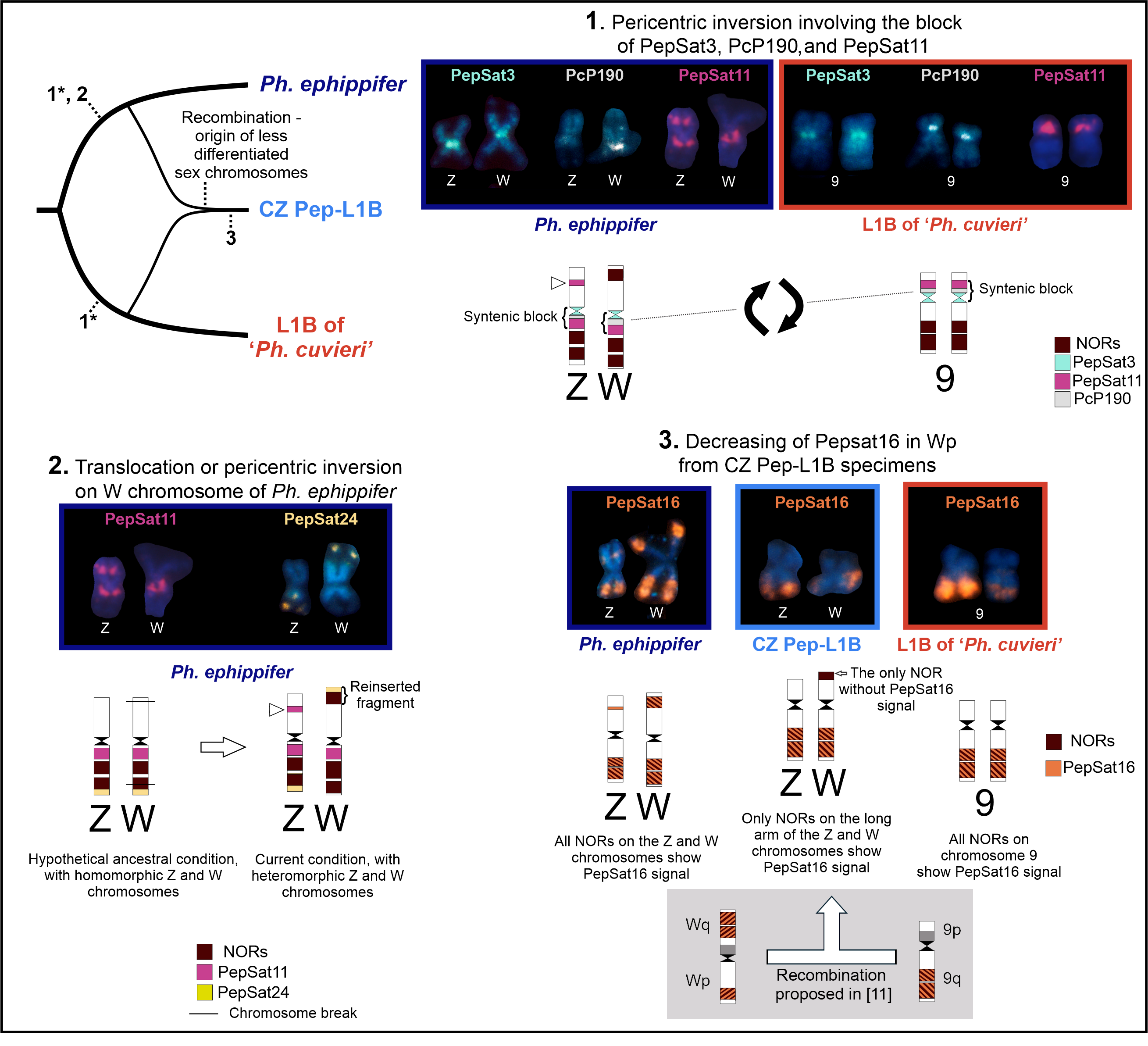
4.4. Chromosome Mapping of Satellite DNAs Is Useful for the Comparative Analysis of Physalaemus ephippifer, L1B, and CZ Pep-L1B
4.4.1. Centromeric Satellite DNA
4.4.2. Satellite DNAs Associated with rDNA
4.4.3. Satellite DNAs Associated with Sex Chromosomes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| satDNA | Satellite DNA |
| L1B | Lineage 1B of ‘Physalaemus cuvieri’ |
| NOR | Nucleolar organizer region |
| TE | Transposable elements |
| rDNA | Ribosomal DNA |
| snRNA | Small nuclear RNA |
| SPAB | São Pedro da Água Branca, Maranhão state, Brazil |
| VNM | Vila Nova dos Martírios, Maranhão state, Brazil |
| TS | Trecho Seco—São Francisco do Brejão, Maranhão state, Brazil |
| Imp | Imperatriz, Maranhão state, Brazil |
| DE | Dom Eliseu, Pará state, Brazil |
| CZ Pep-L1B | Lineage originated from secondary contact between P. ephippifer and L1B |
References
- Biscotti, M.A.; Canapa, A.; Forconi, M.; Olmo, E.; Barucca, M. Transcription of Tandemly Repetitive DNA: Functional Roles. Chromosome Res. 2015, 23, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Ramos, M. Satellite DNA: An Evolving Topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- López-Flores, I.; Garrido-Ramos, M. The Repetitive DNA Content of Eukaryotic Genomes. In Repetitive DNA; Karger Publishers: Basel, Switzerland, 2012; Volume 7, pp. 1–28. [Google Scholar]
- Plohl, M.; Meštrović, N.; Mravinac, B. Satellite DNA Evolution. In Genome Dynamics; Garrido-Ramos, M.A., Ed.; S. Karger AG: Basel, Switzerland, 2012; Volume 7, pp. 126–152. ISBN 978-3-318-02149-3. [Google Scholar]
- Ugarković, Ð.; Plohl, M. Variation in Satellite DNA Profiles—Causes and Effects. EMBO J. 2002, 21, 5955–5959. [Google Scholar] [CrossRef] [PubMed]
- Dover, G.A. Molecular Drive in Multigene Families: How Biological Novelties Arise, Spread and Are Assimilated. Trends Genet. 1986, 2, 159–165. [Google Scholar] [CrossRef]
- Dover, G. Molecular Drive: A Cohesive Mode of Species Evolution. Nature 1982, 299, 111–117. [Google Scholar] [CrossRef]
- Ferree, P.M.; Prasad, S. How Can Satellite DNA Divergence Cause Reproductive Isolation? Let Us Count the Chromosomal Ways. Genet. Res. Int. 2012, 2012, 430136. [Google Scholar] [CrossRef]
- Plohl, M.; Meštrović, N.; Mravinac, B. Centromere Identity from the DNA Point of View. Chromosoma 2014, 123, 313–325. [Google Scholar] [CrossRef]
- Camacho, J.P.M.; Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J. Satellite DNA Is an Inseparable Fellow Traveler of B Chromosomes. In Satellite DNAs in Physiology and Evolution; Ðurðica, U., Ed.; Springer Nature: Geneva, Switzerland, 2021; pp. 85–102. [Google Scholar]
- Silva, D.M.Z.D.A.; Utsunomia, R.; Ruiz-Ruano, F.J.; Daniel, S.N.; Porto-Foresti, F.; Hashimoto, D.T.; Oliveira, C.; Camacho, J.P.M.; Foresti, F. High-Throughput Analysis Unveils a Highly Shared Satellite DNA Library among Three Species of Fish Genus Astyanax. Sci. Rep. 2017, 7, 12726. [Google Scholar] [CrossRef]
- Stornioli, J.H.F.; Goes, C.A.G.; Calegari, R.M.; Dos Santos, R.Z.; Giglio, L.M.; Foresti, F.; Oliveira, C.; Penitente, M.; Porto-Foresti, F.; Utsunomia, R. The B Chromosomes of Prochilodus Lineatus (Teleostei, Characiformes) Are Highly Enriched in Satellite DNAs. Cells 2021, 10, 1527. [Google Scholar] [CrossRef]
- Ferretti, A.B.S.M.; Milani, D.; Palacios-Gimenez, O.M.; Ruiz-Ruano, F.J.; Cabral-de-Mello, D.C. High Dynamism for Neo-Sex Chromosomes: Satellite DNAs Reveal Complex Evolution in a Grasshopper. Heredity 2020, 125, 124–137. [Google Scholar] [CrossRef]
- Kretschmer, R.; Goes, C.A.G.; Bertollo, L.A.C.; Ezaz, T.; Porto-Foresti, F.; Toma, G.A.; Utsunomia, R.; De Bello Cioffi, M. Satellitome Analysis Illuminates the Evolution of ZW Sex Chromosomes of Triportheidae Fishes (Teleostei: Characiformes). Chromosoma 2022, 131, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Dias, G.B.; De Lima, L.G.; Kuhn, G.C.E.S.; Ramos, É.; Martins, C.; Cabral-de-Mello, D.C. High-Throughput Analysis of the Satellitome Revealed Enormous Diversity of Satellite DNAs in the Neo-Y Chromosome of the Cricket Eneoptera surinamensis. Sci. Rep. 2017, 7, 6422. [Google Scholar] [CrossRef] [PubMed]
- Utsunomia, R.; Silva, D.M.Z.D.A.; Ruiz-Ruano, F.J.; Goes, C.A.G.; Melo, S.; Ramos, L.P.; Oliveira, C.; Porto-Foresti, F.; Foresti, F.; Hashimoto, D.T. Satellitome Landscape Analysis of Megaleporinus Macrocephalus (Teleostei, Anostomidae) Reveals Intense Accumulation of Satellite Sequences on the Heteromorphic Sex Chromosome. Sci. Rep. 2019, 9, 5856. [Google Scholar] [CrossRef]
- Navarro-Domínguez, B.; Cabrero, J.; López-León, M.D.; Ruiz-Ruano, F.J.; Pita, M.; Bella, J.L.; Camacho, J.P.M. Tandem Repeat DNA Provides Many Cytological Markers for Hybrid Zone Analysis in Two Subspecies of the Grasshopper Chorthippus parallelus. Genes 2023, 14, 397. [Google Scholar] [CrossRef]
- Souza, L.H.B.; Ferro, J.M.; Gatto, K.P.; De Sá, F.P.; Haddad, C.F.B.; Lourenço, L.B. Clinal Variation in Autosomal Satellite DNA Clusters across a Contact Zone in Barker Frogs. J. Evol. Biol. 2025, 38, 167–179. [Google Scholar] [CrossRef]
- Lourenço, L.B.; Targueta, C.P.; Baldo, D.; Nascimento, J.; Garcia, P.C.A.; Andrade, G.V.; Haddad, C.F.B.; Recco-Pimentel, S.M. Phylogeny of Frogs from the Genus Physalaemus (Anura, Leptodactylidae) Inferred from Mitochondrial and Nuclear Gene Sequences. Mol. Phylogenet. Evol. 2015, 92, 204–216. [Google Scholar] [CrossRef]
- Nascimento, J.; Lima, J.D.; Suárez, P.; Baldo, D.; Andrade, G.V.; Pierson, T.W.; Fitzpatrick, B.M.; Haddad, C.F.B.; Recco-Pimentel, S.M.; Lourenço, L.B. Extensive Cryptic Diversity within the Physalaemus cuvieri–Physalaemus ephippifer Species Complex (Amphibia, Anura) Revealed by Cytogenetic, Mitochondrial, and Genomic Markers. Front. Genet. 2019, 10, 719. [Google Scholar] [CrossRef]
- Souza, L.H.B.; Pierson, T.W.; Tenório, R.O.; Ferro, J.M.; Gatto, K.P.; Silva, B.C.; De Andrade, G.V.; Suárez, P.; Haddad, C.F.B.; Lourenço, L.B. Multiple Contact Zones and Karyotypic Evolution in a Neotropical Frog Species Complex. Sci. Rep. 2024, 14, 1119. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, J.; Quinderé, Y.R.S.D.; Recco-Pimentel, S.M.; Lima, J.R.F.; Lourenço, L.B. Heteromorphic Z and W Sex Chromosomes in Physalaemus ephippifer (Steindachner, 1864) (Anura, Leiuperidae). Genetica 2010, 138, 1127–1132. [Google Scholar] [CrossRef]
- Quinderé, Y.R.S.D.; Lourenço, L.B.; Andrade, G.V.; Tomatis, C.; Baldo, D.; Recco-Pimentel, S.M. Polytypic and Polymorphic Cytogenetic Variations in the Widespread Anuran Physalaemus cuvieri (Anura, Leiuperidae) with Emphasis on Nucleolar Organizing Regions. Biol. Res. 2009, 42, 79–92. [Google Scholar] [CrossRef]
- Targueta, C.P.; Gatto, K.P.; Vittorazzi, S.E.; Recco-Pimentel, S.M.; Lourenço, L.B. High Diversity of 5S Ribosomal DNA and Evidence of Recombination with the Satellite DNA PcP190 in Frogs. Gene 2023, 851, 147015. [Google Scholar] [CrossRef] [PubMed]
- Vittorazzi, S.E.; Lourenço, L.B.; Del-Grande, M.L.; Recco-Pimentel, S.M. Satellite DNA Derived from 5S rDNA in Physalaemus cuvieri (Anura, Leiuperidae). Cytogenet. Genome Res. 2011, 134, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Vittorazzi, S.E.; Lourenço, L.B.; Recco-Pimentel, S.M. Long-Time Evolution and Highly Dynamic Satellite DNA in Leptodactylid and Hylodid Frogs. BMC Genet. 2014, 15, 111. [Google Scholar] [CrossRef] [PubMed]
- Gatto, K.P.; Souza, L.H.B.; Nascimento, J.; Suárez, P.; Lourenço, L.B. Comparative Mapping of a New Repetitive DNA Sequence and Chromosome Region-Specific Probes Unveiling Rearrangements in an Amazonian Frog Complex. Genome 2021, 64, 857–868. [Google Scholar] [CrossRef]
- Souza, L.H.B.; Silva, B.C.; Pompeo, J.N.; Gatto, K.P.; Lourenço, L.B. Chromosome Homologies and Polymorphisms in a Neotropical Species Complex of Frogs Revealed by the U2 snRNA Gene. Genome 2025, 68, 1–11. [Google Scholar] [CrossRef]
- Graves, J.A.M. Weird Animal Genomes and the Evolution of Vertebrate Sex and Sex Chromosomes. Annu. Rev. Genet. 2008, 42, 565–586. [Google Scholar] [CrossRef]
- Kratochvíl, L.; Stöck, M.; Rovatsos, M.; Bullejos, M.; Herpin, A.; Jeffries, D.L.; Peichel, C.L.; Perrin, N.; Valenzuela, N.; Pokorná, M.J. Expanding the Classical Paradigm: What We Have Learnt from Vertebrates about Sex Chromosome Evolution. Phil. Trans. R. Soc. B 2021, 376, 20200097. [Google Scholar] [CrossRef]
- Ma, W.-J.; Veltsos, P. The Diversity and Evolution of Sex Chromosomes in Frogs. Genes 2021, 12, 483. [Google Scholar] [CrossRef]
- Schartl, M. Sex Chromosome Evolution in Non-Mammalian Vertebrates. Cur. Op. Genet. Dev. 2004, 14, 634–641. [Google Scholar] [CrossRef]
- King, M.; Rofe, R. Karyotypic Variation in the Australian Gekko Phyllodactylus marmoratus (Gray) (Gekkonidae: Reptilia). Chromosoma 1976, 54, 75–87. [Google Scholar] [CrossRef]
- Gatto, K.P.; Mattos, J.V.; Seger, K.R.; Lourenço, L.B. Sex Chromosome Differentiation in the Frog Genus Pseudis Involves Satellite DNA and Chromosome Rearrangements. Front. Genet. 2018, 9, 301. [Google Scholar] [CrossRef] [PubMed]
- Rhie, A.; Walenz, B.P.; Koren, S.; Phillippy, A.M. Merqury: Reference-Free Quality, Completeness, and Phasing Assessment for Genome Assemblies. Genome Biol. 2020, 21, 245. [Google Scholar] [CrossRef] [PubMed]
- Ranallo-Benavidez, T.R.; Jaron, K.S.; Schatz, M.C. GenomeScope 2.0 and Smudgeplot for Reference-Free Profiling of Polyploid Genomes. Nat. Commun. 2020, 11, 1432. [Google Scholar] [CrossRef]
- Abueg, L.A.L.; Afgan, E.; Allart, O.; Awan, A.H.; Bacon, W.A.; Baker, D.; Bassetti, M.; Batut, B.; Bernt, M.; The Galaxy Community; et al. The Galaxy Platform for Accessible, Reproducible, and Collaborative Data Analyses: 2024 Update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Macas, J. Global Analysis of Repetitive DNA from Unassembled Sequence Reads Using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Macas, J. Graph-Based Clustering and Characterization of Repetitive Sequences in next-Generation Sequencing Data. BMC Bioinform. 2010, 11, 378. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy-Based Web Server for Genome-Wide Characterization of Eukaryotic Repetitive Elements from next-Generation Sequence Reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. In Gene Prediction. Methods in Molecular Biology; Kollmar, M., Ed.; Humana: New York, NY, USA, 2019; Volume 1962, pp. 1–14. [Google Scholar]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-Line: Integrating Search and Context for Analysis of Transfer RNA Genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. Available online: http://www.repeatmasker.org (accessed on 20 January 2025).
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-Throughput Analysis of the Satellitome Illuminates Satellite DNA Evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Negm, S.; Greenberg, A.; Larracuente, A.M.; Sproul, J.S. RepeatProfiler: A Pipeline for Visualization and Comparative Analysis of Repetitive DNA Profiles. Mol. Ecol. Resour. 2021, 21, 969–981. [Google Scholar] [CrossRef] [PubMed]
- Koressaar, T.; Remm, M. Enhancements and Modifications of Primer Design Program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Viegas-Péquignot, E. In Situ Hybridization to Chromosomes with Biotinylated Probes. In In situ Hyridization, A Practical Approach; Wilkinson, D., Ed.; Oxford University Press: Oxford, UK, 1992; pp. 137–158. [Google Scholar]
- Howell, W.M.; Black, D.A. Controlled Silver-Staining of Nucleolus Organizer Regions with a Protective Colloidal Developer: A 1-Step Method. Experientia 1980, 36, 1014–1015. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Da Silva, M.J.; Gazoni, T.; Haddad, C.F.B.; Parise-Maltempi, P.P. Analysis in Proceratophrys Boiei Genome Illuminates the Satellite DNA Content in a Frog from the Brazilian Atlantic Forest. Front. Genet. 2023, 14, 1101397. [Google Scholar] [CrossRef]
- Da Silva, M.J.; Fogarin Destro, R.; Gazoni, T.; Narimatsu, H.; Pereira Dos Santos, P.S.; Haddad, C.F.B.; Parise-Maltempi, P.P. Great Abundance of Satellite DNA in Proceratophrys (Anura, Odontophrynidae) Revealed by Genome Sequencing. Cytogenet. Genome Res. 2020, 160, 141–147. [Google Scholar] [CrossRef]
- Simandle, E.T.; Peacock, M.M.; Zirelli, L.; Tracy, C.R. Sixteen Microsatellite Loci for the Bufo Boreas Group. Mol. Ecol. Notes 2006, 6, 116–119. [Google Scholar] [CrossRef]
- Gregory, T.R. Animal Genome Size Database. 2025. Available online: http://www.genomesize.com (accessed on 9 May 2025).
- Boštjančić, L.L.; Bonassin, L.; Anušić, L.; Lovrenčić, L.; Besendorfer, V.; Maguire, I.; Grandjean, F.; Austin, C.M.; Greve, C.; Hamadou, A.B.; et al. The Pontastacus leptodactylus (Astacidae) Repeatome Provides Insight into Genome Evolution and Reveals Remarkable Diversity of Satellite DNA. Front. Genet. 2021, 11, 611745. [Google Scholar] [CrossRef]
- Rico-Porras, J.M.; Mora, P.; Palomeque, T.; Montiel, E.E.; Cabral-de-Mello, D.C.; Lorite, P. Heterochromatin Is Not the Only Place for satDNAs: The High Diversity of satDNAs in the Euchromatin of the Beetle Chrysolina americana (Coleoptera, Chrysomelidae). Genes 2024, 15, 395. [Google Scholar] [CrossRef]
- Sales-Oliveira, V.C.; Dos Santos, R.Z.; Goes, C.A.G.; Calegari, R.M.; Garrido-Ramos, M.A.; Altmanová, M.; Ezaz, T.; Liehr, T.; Porto-Foresti, F.; Utsunomia, R.; et al. Evolution of Ancient Satellite DNAs in Extant Alligators and Caimans (Crocodylia, Reptilia). BMC Biol. 2024, 22, 47. [Google Scholar] [CrossRef]
- Portik, D.M.; Streicher, J.W.; Wiens, J.J. Frog Phylogeny: A Time-Calibrated, Species-Level Tree Based on Hundreds of Loci and 5,242 Species. Mol. Phylogenet. Evol. 2023, 188, 107907. [Google Scholar] [CrossRef] [PubMed]
- Guzmán, K.; Roco, Á.S.; Stöck, M.; Ruiz-García, A.; García-Muñoz, E.; Bullejos, M. Identification and Characterization of a New Family of Long Satellite DNA, Specific of True Toads (Anura, Amphibia, Bufonidae). Sci. Rep. 2022, 12, 13960. [Google Scholar] [CrossRef]
- Fry, K.; Salser, W. Nucleotide Sequences of HS-α Satellite DNA from Kangaroo Rat Dipodomys ordii and Characterization of Similar Sequences in Other Rodents. Cell 1977, 12, 1069–1084. [Google Scholar] [CrossRef]
- Camacho, J.P.M.; Cabrero, J.; López-León, M.D.; Martín-Peciña, M.; Perfectti, F.; Garrido-Ramos, M.A.; Ruiz-Ruano, F.J. Satellitome Comparison of Two Oedipodine Grasshoppers Highlights the Contingent Nature of Satellite DNA Evolution. BMC Biol. 2022, 20, 36. [Google Scholar] [CrossRef]
- De Lima, L.G.; Ruiz-Ruano, F.J. In-Depth Satellitome Analyses of 37 Drosophila Species Illuminate Repetitive DNA Evolution in the Drosophila Genus. Genome Biol. Evol. 2022, 14, evac064. [Google Scholar] [CrossRef] [PubMed]
- Pita, S.; Panzera, F.; Mora, P.; Vela, J.; Cuadrado, Á.; Sánchez, A.; Palomeque, T.; Lorite, P. Comparative Repeatome Analysis on Triatoma infestans Andean and Non-Andean Lineages, Main Vector of Chagas Disease. PLoS ONE 2017, 12, e0181635. [Google Scholar] [CrossRef]
- Mora, P.; Vela, J.; Ruiz-Ruano, F.J.; Ruiz-Mena, A.; Montiel, E.E.; Palomeque, T.; Lorite, P. Satellitome Analysis in the Ladybird Beetle Hippodamia variegata (Coleoptera, Coccinellidae). Genes 2020, 11, 783. [Google Scholar] [CrossRef]
- Montiel, E.E.; Mora, P.; Rico-Porras, J.M.; Palomeque, T.; Lorite, P. Satellitome of the Red Palm Weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae), the Most Diverse among Insects. Front. Ecol. Evol. 2022, 10, 826808. [Google Scholar] [CrossRef]
- Utsunomia, R.; Ruiz-Ruano, F.J.; Silva, D.M.Z.A.; Serrano, É.A.; Rosa, I.F.; Scudeler, P.E.S.; Hashimoto, D.T.; Oliveira, C.; Camacho, J.P.M.; Foresti, F. A Glimpse into the Satellite DNA Library in Characidae Fish (Teleostei, Characiformes). Front. Genet. 2017, 8, 103. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; Navarro-Domínguez, B.; Camacho, J.P.M.; Garrido-Ramos, M.A. Characterization of the Satellitome in Lower Vascular Plants: The Case of the Endangered Fern Vandenboschia speciosa. Ann. Bot. 2019, 123, 587–599. [Google Scholar] [CrossRef]
- Van-Lume, B.; Mata-Sucre, Y.; Báez, M.; Ribeiro, T.; Huettel, B.; Gagnon, E.; Leitch, I.J.; Pedrosa-Harand, A.; Lewis, G.P.; Souza, G. Evolutionary Convergence or Homology? Comparative Cytogenomics of Caesalpinia Group Species (Leguminosae) Reveals Diversification in the Pericentromeric Heterochromatic Composition. Planta 2019, 250, 2173–2186. [Google Scholar] [CrossRef]
- Sader, M.; Vaio, M.; Cauz-Santos, L.A.; Dornelas, M.C.; Vieira, M.L.C.; Melo, N.; Pedrosa-Harand, A. Large vs Small Genomes in Passiflora: The Influence of the Mobilome and the Satellitome. Planta 2021, 253, 86. [Google Scholar] [CrossRef]
- Ribeiro, T.; Vasconcelos, E.; Dos Santos, K.G.B.; Vaio, M.; Brasileiro-Vidal, A.C.; Pedrosa-Harand, A. Diversity of Repetitive Sequences within Compact Genomes of Phaseolus L. Beans and Allied Genera Cajanus L. and Vigna Savi. Chromosome Res. 2020, 28, 139–153. [Google Scholar] [CrossRef]
- Šatović-Vukšić, E.; Plohl, M. Classification Problems of Repetitive DNA Sequences. DNA 2021, 1, 84–90. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; Mora, P.; Rico-Porras, J.M.; Ferretti, A.B.S.M.; Palomeque, T.; Lorite, P. The Spread of Satellite DNAs in Euchromatin and Insights into the Multiple Sex Chromosome Evolution in Hemiptera Revealed by Repeatome Analysis of the Bug Oxycarenus hyalinipennis. Insect Mol. Biol. 2023, 32, 725–737. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Zhang, J.; Feng, Y.; Chen, Q.; Liu, Z.-S.; Liu, C.-L.; He, W.; Wang, H.; Yang, S.-F.; et al. Comparative Analysis of Transposable Elements and the Identification of Candidate Centromeric Elements in the Prunus Subgenus Cerasus and Its Relatives. Genes 2022, 13, 641. [Google Scholar] [CrossRef]
- Hartley, G.A.; Okhovat, M.; Hoyt, S.J.; Fuller, E.; Pauloski, N.; Alexandre, N.; Alexandrov, I.; Drennan, R.; Dubocanin, D.; Gilbert, D.M.; et al. Centromeric Transposable Elements and Epigenetic Status Drive Karyotypic Variation in the Eastern Hoolock Gibbon. Cell Genom. 2025, 5, 100808. [Google Scholar] [CrossRef]
- Piras, F.M.; Nergadze, S.G.; Magnani, E.; Bertoni, L.; Attolini, C.; Khoriauli, L.; Raimondi, E.; Giulotto, E. Uncoupling of Satellite DNA and Centromeric Function in the Genus Equus. PLoS Genet. 2010, 6, e1000845. [Google Scholar] [CrossRef]
- Da Silva, M.J.; Destro, R.F.; Gazoni, T.; Parise-Maltempi, P.P. Interspecific Cytogenomic Comparison Reveals a Potential Chromosomal Centromeric Marker in Proceratophrys Frog Species. Chromosoma 2024, 133, 195–202. [Google Scholar] [CrossRef]
- De Lucchini, S.; Andronico, F.; Andreazzoli, M.; Giuliani, M.; Savino, R.; Nardi, I. Extra-Ribosomal Spacer Sequences in Triturus. J. Mol. Biol. 1988, 204, 805–813. [Google Scholar] [CrossRef]
- Schmidt, E.R.; Godwin, E.A.; Keyl, H.-G.; Israelewski, N. Cloning and Analysis of Ribosomal DNA of Chironomus thummi piger and Chironomus thummi thummi. Chromosoma 1982, 87, 389–407. [Google Scholar] [CrossRef]
- Stupar, R.M.; Song, J.; Tek, A.L.; Cheng, Z.; Dong, F.; Jiang, J. Highly Condensed Potato Pericentromeric Heterochromatin Contains rDNA-Related Tandem Repeats. Genetics 2002, 162, 1435–1444. [Google Scholar] [CrossRef]
- Jo, S.-H.; Koo, D.-H.; Kim, J.F.; Hur, C.-G.; Lee, S.; Yang, T.; Kwon, S.-Y.; Choi, D. Evolution of Ribosomal DNA-Derived Satellite Repeat in Tomato Genome. BMC Plant. Biol. 2009, 9, 42. [Google Scholar] [CrossRef]
- Dalíková, M.; Provazníková, I.; Provazník, J.; Grof-Tisza, P.; Pepi, A.; Nguyen, P. The Role of Repetitive Sequences in Repatterning of Major Ribosomal DNA Clusters in Lepidoptera. Genome Biol. Evol. 2023, 15, evad090. [Google Scholar] [CrossRef]
- Almeida, C.; Fonsêca, A.; Dos Santos, K.G.B.; Mosiolek, M.; Pedrosa-Harand, A. Contrasting Evolution of a Satellite DNA and Its Ancestral IGS rDNA in Phaseolus (Fabaceae). Genome 2012, 55, 683–689. [Google Scholar] [CrossRef]
- Cardone, M.F.; Ballarati, L.; Ventura, M.; Rocchi, M.; Marozzi, A.; Ginelli, E.; Meneveri, R. Evolution of Beta Satellite DNA Sequences: Evidence for Duplication-Mediated Repeat Amplification and Spreading. Mol. Biol. Evol. 2004, 21, 1792–1799. [Google Scholar] [CrossRef]
- Barth, A.; Solé, M.; Costa, M.A. Chromosome Polymorphism in Phyllomedusa Rohdei Populations (Anura: Hylidae). J. Herpetol. 2009, 43, 676–679. [Google Scholar] [CrossRef]
- Castro, J.; Rodríguez, S.; Pardo, B.G.; Sánchez, L.; Martínez, P. Population Analysis of an Unusual NOR-Site Polymorphism in Brown Trout (Salmo trutta L.). Heredity 2001, 86, 291–302. [Google Scholar] [CrossRef]
- Medeiros, L.R. Chromosomal Differentiation of Hyla nana and Hyla sanborni (Anura, Hylidae) with a Description of NOR Polymorphism in H. Nana. J. Hered. 2003, 94, 149–154. [Google Scholar] [CrossRef][Green Version]
- Schmid, M. Chromosome Banding in Amphibia: VII. Analysis of the Structure and Variability of NORs in Anura. Chromosoma 1982, 87, 327–344. [Google Scholar] [CrossRef]
- Plohl, M.; Luchetti, A.; Meštrović, N.; Mantovani, B. Satellite DNAs between Selfishness and Functionality: Structure, Genomics and Evolution of Tandem Repeats in Centromeric (Hetero)Chromatin. Gene 2008, 409, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Kratochvíl, L.; Gamble, T.; Rovatsos, M. Sex Chromosome Evolution among Amniotes: Is the Origin of Sex Chromosomes Non-Random? Phil. Trans. R. Soc. B 2021, 376, 20200108. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B.; Charlesworth, D. The Degeneration of Y Chromosomes. Phil. Trans. R. Soc. Lond. B 2000, 355, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
| Group | Locality (City-State) | Number of Specimens | Accession Number in Zoological Collection | Accession Number in SMRP Collection |
|---|---|---|---|---|
| Physalaemus ephippifer | Santa Bárbara-PA | 4 males/3 females | ZUEC 24907; CFBH 46955; ZUEC 24908; CFBH 46958/CFBH 46750, 46782, 46784 | 252.147; 252.153; 252.156; 252.157/252.166; 252.190; 252.192 |
| CZ Pep-L1B | Dom Eliseu-PA | 2 males/- | CFBH 46703, 46711 | 92.383; 92.391 |
| São Pedro da Água Branca-PA | 1 male/2 females | HUFMA 2295/MNRJ 24258; HUFMA 2290 | 92.332/92.4; 92.327 | |
| Lineage 1B | Riachão-MA 1 | 1 male/- | CFBH 46777 | 92.421 |
| Urbano Santos-MA | 2 males/1 female | ZUEC 13362, 13363/ZUEC 13364 | 92.60; 92.61/92.62 |
| satDNA | Monomer Size (bp) | Abundance (%) | Average Divergence (%) Between Total Monomers | A+T Percentage *2 | GenBank Accession Number |
|---|---|---|---|---|---|
| PepSat1-21 | 21 | 1.55 | 9.58 | 61.9 | PV463924 |
| PepSat2-97 | 97 | 0.96 | 7.25 | 55.67 | PV463925 |
| PepSat3-152 * | 152 | 0.86 | 1.58 | 59.21 | PV463926 |
| PepSat4-149 * | 149 | 0.71 | 9.03 | 61.07 | PV463927 |
| PepSat5-1310 | 1310 | 0.45 | 15.18 | 54.73 | PV463928 |
| PepSat6-1546 | 1546 | 0.40 | 10.25 | 52.78 | PV463929 |
| PepSat7-902 | 902 | 0.36 | 13.3 | 48 | PV463930 |
| PepSat8-36 | 36 | 0.28 | 10.88 | 52.78 | PV463931 |
| PepSat9-49 | 49 | 0.25 | 17.33 | 53.06 | PV463932 |
| PboSat8-92 1-v-Pep | 95 | 0.249 | 9.98 | 55.79 | PV463933 |
| PepSat11-237 * | 237 | 0.242 | 9.16 | 40.93 | PV463934 |
| PepSat12-73 | 73 | 0.23 | 10.52 | 65.75 | PV463935 |
| PepSat13-49 | 49 | 0.219 | 7.9 | 55.10 | PV463936 |
| PepSat14-100 | 100 | 0.21 | 5.38 | 57 | PV463937 |
| PboSat05-35 1,2-v-Pep | 36 | 0.193 | 3.43 | 63.89 | PV463938 |
| PepSat16-147 * | 147 | 0.192 | 7.9 | 45.58 | PV463939 |
| PepSat17-162 | 162 | 0.19 | 10.8 | 53.09 | PV463940 |
| PepSat18-1073 | 1073 | 0.177 | 4.26 | 52.1 | PV463941 |
| PepSat19-629 | 629 | 0.172 | 7.6 | 56.92 | PV463942 |
| PepSat20-22 | 22 | 0.16 | 9.83 | 54.55 | PV463943 |
| PepSat21-49 | 49 | 0.14 | 16.37 | 55.1 | PV463944 |
| PcP190 1a 3,4 | 190 | 0.13 | 2.08 | 52.11 | PV463945 |
| PboSat36-39 1-v-Pep | 39 | 0.129 | 9.31 | 56.41 | PV463946 |
| PepSat24-620 * | 620 | 0.122 | 7.98 | 56.13 | PV463947 |
| PepSat25-282 * | 282 | 0.11 | 4.61 | 45.39 | PV463948 |
| PepSat26-908 | 908 | 0.097 | 9.84 | 61.45 | PV463949 |
| PepSat27-280 | 280 | 0.092 | 2.11 | 62.5 | PV463950 |
| PboSat22-90 1-v-Pep | 31 | 0.08 | 5.45 | 58.06 | PV463951 |
| PboSat11-39 1,2-v-Pep | 38 | 0.076 | 3.6 | 52.63 | PV463952 |
| PepSat30-350 * | 350 | 0.075 | 4.13 | 59.71 | PV463953 |
| PepSat31-709 | 709 | 0.057 | 6.04 | 59.1 | PV463954 |
| BBR86 v-Pep 5 | 41 | 0.05 | 8.18 | 56.1 | PV463955 |
| PepSat33-30 | 30 | 0.049 | 16.45 | 56.67 | PV463956 |
| PepSat34-57 | 57 | 0.048 | 17.57 | 45.61 | PV463957 |
| PepSat35-28 | 28 | 0.047 | 12.43 | 42.86 | PV463958 |
| PepSat36-739 | 739 | 0.04 | 5.38 | 61.3 | PV463959 |
| PepSat37-101 | 101 | 0.04 | 9.96 | 60.4 | PV463960 |
| PepSat38-39 | 39 | 0.039 | 5.63 | 41.03 | PV463961 |
| PepSat39-90 | 90 | 0.038 | 3.24 | 57.78 | PV463962 |
| PboSat12-91 1,2-v-Pep | 45 | 0.03 | 4.33 | 64.44 | PV463963 |
| PboSat17-93 1,2-v-Pep | 93 | 0.027 | 5.67 | 63.44 | PV463964 |
| PepSat42-41 | 41 | 0.026 | 9.94 | 46.34 | PV463965 |
| PepSat43-54 | 54 | 0.024 | 7.31 | 38.89 | PV463966 |
| PepSat44-16 | 16 | 0.023 | 12.42 | 37.5 | PV463967 |
| PepSat45-33 | 33 | 0.022 | 3.18 | 66.67 | PV463968 |
| PepSat46-47 | 47 | 0.02 | 9.78 | 53.19 | PV463969 |
| PepSat47-74 | 74 | 0.019 | 2.83 | 43.24 | PV463970 |
| PepSat48-27 | 27 | 0.019 | 4.02 | 48.15 | PV463971 |
| PepSat49-68 | 68 | 0.018 | 5.55 | 60.29 | PV463972 |
| PepSat50-32 | 32 | 0.017 | 5.19 | 65.63 | PV463973 |
| PepSat51-109 | 109 | 0.015 | 11.5 | 60.55 | PV463974 |
| PepSat52-30 | 30 | 0.014 | 5.19 | 56.67 | PV463975 |
| PepSat53-643 | 643 | 0.013 | 2.36 | 57.7 | PV463976 |
| PepSat54-31 | 31 | 0.012 | 10.08 | 48.39 | PV463977 |
| PepSat55-33 | 33 | 0.011 | 16.07 | 48.48 | PV463978 |
| PepSat56-31 | 31 | 0.0092 | 5.25 | 61.29 | PV463979 |
| PepSat57-158 | 158 | 0.009 | 8.29 | 60.76 | PV463980 |
| PepSat58-45 | 45 | 0.009 | 4.75 | 46.67 | PV463981 |
| PepSat59-30 | 30 | 0.009 | 5.56 | 36.67 | PV463982 |
| PepSat60-110 | 110 | 0.009 | 5.98 | 51.82 | PV463983 |
| PepSat61-23 | 23 | 0.008 | 4.93 | 43.48 | PV463984 |
| PepSat62-56 | 56 | 0.004 | 3.64 | 35.71 | PV463985 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Souza, L.H.B.; Ferro, J.M.; Milanez, H.M.; Haddad, C.F.B.; Lourenço, L.B. New Insights into the Sex Chromosome Evolution of the Common Barker Frog Species Complex (Anura, Leptodactylidae) Inferred from Its Satellite DNA Content. Biomolecules 2025, 15, 876. https://doi.org/10.3390/biom15060876
Souza LHB, Ferro JM, Milanez HM, Haddad CFB, Lourenço LB. New Insights into the Sex Chromosome Evolution of the Common Barker Frog Species Complex (Anura, Leptodactylidae) Inferred from Its Satellite DNA Content. Biomolecules. 2025; 15(6):876. https://doi.org/10.3390/biom15060876
Chicago/Turabian StyleSouza, Lucas H. B., Juan M. Ferro, Helena M. Milanez, Célio F. B. Haddad, and Luciana B. Lourenço. 2025. "New Insights into the Sex Chromosome Evolution of the Common Barker Frog Species Complex (Anura, Leptodactylidae) Inferred from Its Satellite DNA Content" Biomolecules 15, no. 6: 876. https://doi.org/10.3390/biom15060876
APA StyleSouza, L. H. B., Ferro, J. M., Milanez, H. M., Haddad, C. F. B., & Lourenço, L. B. (2025). New Insights into the Sex Chromosome Evolution of the Common Barker Frog Species Complex (Anura, Leptodactylidae) Inferred from Its Satellite DNA Content. Biomolecules, 15(6), 876. https://doi.org/10.3390/biom15060876







