Harnessing Microbiome, Bacterial Extracellular Vesicle, and Artificial Intelligence for Polycystic Ovary Syndrome Diagnosis and Management
Abstract
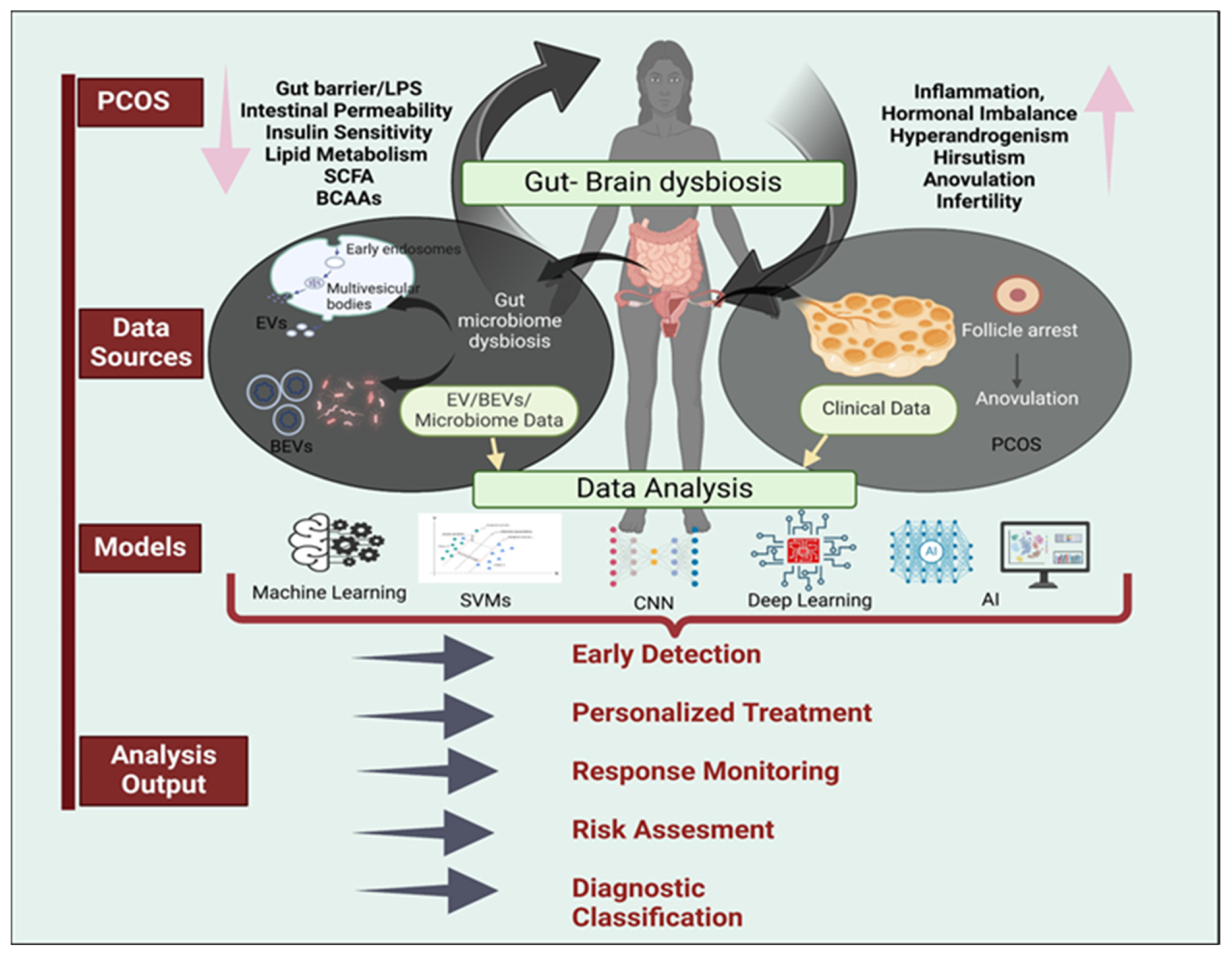
1. Introduction
2. Methods
3. Current Diagnostic Approaches for PCOS and Limitations
| Diagnostic Approach | Key Biomarkers | Specific Values/Thresholds | Strengths | Limitations | Performance | References |
|---|---|---|---|---|---|---|
| Traditional Approaches | ||||||
| NIH criteria (1992) | Hyperandrogenism: Elevated circulating androgens above 95th percentile of healthy controls OR clinical signs (hirsutism, acne, alopecia). Measured via total testosterone, free testosterone. Ovulatory dysfunction: Irregular or absent ovulation with menstrual irregularity, assessed through menstrual cycle patterns and ovulation markers. | Total testosterone: >88 ng/dL (>2.4 nmol/L); Free testosterone: >0.75 ng/dL; Oligomenorrhea: ≤8 cycles/year; Cycle length: >35 days or <21 days | High specificity (100%); Focus on reproductive-endocrine disorder components; Well-defined androgen thresholds | Fails to recognize metabolic components; Narrower phenotypic presentation; Limited sensitivity | Sensitivity: 60% Specificity: 100% | [57,75] |
| Rotterdam criteria (2003/updated 2023) | Oligo/anovulation: ≤8 menstrual cycles per year, assessed through cycle frequency and ovulation markers. Clinical hyperandrogenism: Hirsutism, acne, or androgenic alopecia measured by Ferriman–Gallwey score. Biochemical hyperandrogenism: Elevated testosterone or androstenedione levels. Polycystic ovaries: Increased follicle number (≥20) or ovarian volume (≥10 mL) on ultrasound using modern technology, assessed via follicle count and ovarian volume measurements. | Oligomenorrhea: ≤8 cycles/year; LH/FSH ratio: Often >2:1, Ferriman-Gallwey score: ≥8 (varies by ethnicity); Total testosterone: Variable by assay method, Updated criteria (2023); Follicle count: ≥20 per ovary (8 MHz transducer); Ovarian volume: ≥10 mL (either ovary), Previous: ≥12 follicles per ovary | Widely adopted in clinical practice; Updated follicle thresholds reflect improved imaging technology | Original 12-follicle threshold now considered too low; Inter-observer variability in ultrasound assessment; No incorporation of metabolic parameters | Original criteria: Sensitivity: 75% Specificity: 99% Updated follicle threshold reduces false positives | [4,76,77,78,79] |
| AE-PCOS Society criteria | Hyperandrogenism: Central diagnostic feature that must be present either clinically or biochemically, assessed through clinical manifestations and biochemical markers. Ovarian dysfunction: Either oligo/anovulation OR polycystic ovaries on ultrasound, evaluated via menstrual irregularity and polycystic ovaries assessment. | Clinical hyperandrogenism: Present; Biochemical hyperandrogenism: Method-dependent thresholds; Oligo/anovulation: Present; Polycystic ovarian morphology: As per updated criteria | Emphasizes hyperandrogenism as core feature; Better identification of women with metabolic risks | More restrictive than Rotterdam; Excludes some milder phenotypes; Implementation challenges | Performance metrics not extensively validated in large studies | [9,18] |
| Emerging Approaches | ||||||
| Microbiome Analysis | Gut dysbiosis: Altered microbiota composition characterized by reduced diversity and specific bacterial imbalances associated with metabolic dysfunction. Assessed via Firmicutes/Bacteroidetes ratio, specific bacterial genera (Escherichia-Shigella, Proteobacteria), alpha diversity measures, and beta diversity patterns. Microbiome–PCOS axis: Gut bacteria influence host metabolism, inflammation, and hormone regulation. | PCOS vs. Controls: Decreased Firmicutes/Bacteroidetes ratio; Increased Proteobacteria abundance; Increased Escherichia-Shigella: Variable but often elevated; Decreased overall alpha diversity Note: Specific thresholds vary significantly between studies and populations | Insights into pathogenesis; Potential therapeutic targets through microbiome modulation; Non-invasive sample collection | High inter-individual variability; Lack of standardized collection/analysis methods; Confounding by diet and lifestyle; Limited clinical validation | Machine learning classification accuracy varies widely; No consistent diagnostic thresholds established | [20,23,80,81,82] |
| Bacterial Extracellular Vesicles (BEVs) | EV dysregulation: Altered bacterial and cellular extracellular vesicle cargo reflecting systemic inflammation and metabolic dysfunction. Measured via various miRNA species, protein cargo markers, and cytokine profiles in EVs. Intercellular communication: EVs carry regulatory molecules between cells and tissues, serving as biomarkers for disease state. | Research-stage biomarkers: miRNA expression patterns: Study-dependent fold changes; EV concentration: Often elevated in PCOS; Inflammatory protein cargo: Variable across studies Note: Specific diagnostic thresholds not established | Potential for multi-parameter biomarker panels; Reflects systemic pathophysiology; Stable in circulation | Primarily used in base research; Standardization of isolation methods needed; Limited clinical validation studies; High technical complexity | Research-stage metrics: Various AUC values reported (0.8–0.95) in preliminary studies | [83,84,85] |
| Artificial Intelligence—Clinical Data | Machine learning classification: Algorithmic integration of multiple clinical parameters to generate diagnostic probability scores using various ML techniques (SVM, Random Forest, etc.). Input features include clinical features, laboratory values (BMI, testosterone levels, cycle regularity, LH/FSH ratios), and anthropometric measures. Output includes probability scores and classification decisions. | Algorithm performance varies: Feature combinations: Study-dependent; Probability thresholds: Typically > 0.5 for positive classification; Cross-validation: k-fold approaches Common features: BMI, testosterone levels, cycle regularity, LH/FSH ratios | High diagnostic accuracy; Integration of multiple data types; Objective decision making; Potential for clinical decision support | Need for large, diverse training datasets; Potential algorithmic bias; Model interpretability challenges; Validation across populations needed | Overall Performance: AUC: 73–100% Accuracy: 89–100% Sensitivity: 41–100% Specificity: 75–100% Standardized criteria studies: AUC: 80–100% Accuracy: 89–100% | [43,44,86,87] |
| Deep Learning—Ultrasound Image Analysis | Automated image analysis: Computer vision algorithms for objective ultrasound interpretation with automated feature extraction and pattern recognition. CNN-based features include automated follicle detection, ovarian morphology analysis, and texture and pattern recognition. Deep feature learning: CNNs learn hierarchical representations directly from image data without manual feature engineering, processing pixel-level analysis and feature extraction. | Technical specifications: Input image resolution: Typically 224 × 224 pixels; Follicle detection: Automated counting and sizing; CNN architectures: VGG16, ResNet, Inception V3, custom designs performance thresholds: Classification confidence: >0.5 probability; Image quality requirements: Variable by study | Reduced inter-observer variability; Objective measurements; Potential for real-time diagnosis; Automated follicle counting; Reduced dependency on operator expertise | Computational requirements; Need for large, annotated datasets; Model generalizability across different ultrasound systems; Black box interpretability | Individual studies: VGG16+XGBoost: 99.89% accuracy (Suha & Islam, 2022); Various CNN models: 82.6–99% accuracy; Sensitivity: 85–100%; Specificity: 80–94%; Precision: 82.6–97% | [88,89,90,91,92] |
| Integrated Multi-omics AI | Precision medicine approach: Integration of genetic, molecular, clinical, and imaging data for comprehensive phenotyping and personalized risk assessment. Input data includes genomic variants, clinical phenotypes, laboratory biomarkers, and imaging data. Systems biology: Understanding PCOS as a complex multi-system disorder with individualized presentations through multi-modal integration and pathway analysis. | Complex feature integration: SNP risk scores: Population-dependent; Multi-omics data fusion: Study-specific approaches; Ensemble methods: Combined algorithm outputs; Personalized risk stratification: Individual-based thresholds | Comprehensive molecular profiling; Individual risk stratification; Potential for personalized treatment; Integration of diverse data types | High cost and complexity; Data privacy concerns; Limited clinical accessibility; Standardization challenges; Requires specialized infrastructure | Research-stage metrics: Limited large-scale validation studies available; Promising preliminary results in small cohorts | [93] |
4. Microbiome Analysis in PCOS
5. EV and BEVs Analysis in PCOS
6. Artificial Intelligence and Machine Learning in Medical Diagnostics of PCOS
6.1. Random Forest
6.2. Support Vector Machines (SVMs)
6.3. Deep Learning for Image Analysis
- (a)
- Convolutional neural networks (CNNs): The CNN model, based on the ResNet-50 architecture, achieved 92.3% accuracy (95% CI: 90.1–94.5%), 91.4% sensitivity (95% CI: 88.7–94.1%), and 93.1% specificity (95% CI: 90.6–5.6%) in identifying polycystic ovary morphology, significantly outperforming traditional manual assessments [108,120,124,132,142,143,144,145,146,147,148]. In a two-phase approach, Gülhan et al. optimized follicle detection in ultrasound images through several preprocessing methods, followed by a CNN-based classification of ovarian images. The technique discriminated between normal and PCOS images, with accuracies of 65.81% for raw images and 77.81% for preprocessed images [122]. Similarly, Sumathi et al. used CNN-based image processing to classify ovarian cysts, achieving 85% accuracy [121]. Overall, these studies suggest that CNN-based approaches, particularly when combined with optimized preprocessing methods, offer promising potential for automated PCOS detection through ultrasound image analysis.
- (b)
- Advanced CNN architectures: Suha and Islam combined the CNN architecture for feature extraction and a stacking ensemble method for classification [123]. Compared to existing machine learning methods, this approach improved the accuracy and reduced training time, resulting in 99.89% classification accuracy. Furthermore, Garzia et al. investigated predictors of metformin treatment effectiveness in PCOS patients using artificial neural networks (ANNs), specifically focusing on weight loss and androgen level reduction outcomes. Using Auto-CM, a fourth-generation ANN, the authors developed semantic connectivity maps (SCMs) to correlate baseline clinical characteristics with treatment outcomes. The ANN analysis revealed that patients with oligo-amenorrhea and hyperandrogenemia at baseline were most likely to respond positively to metformin treatment, whereas lower baseline testosterone levels was a significant predictor of treatment discontinuation [131].
6.4. Integrated Approaches
7. Combining AI with Microbiome and BEVs Analysis for PCOS Diagnosis
7.1. Data Collection and Preprocessing
7.2. Feature Selection and Model Development
7.3. AI-Enabled PCOS Subtyping
8. Towards Personalized Treatment of PCOS
9. Ethical Challenges in AI and Microbiome-Based Approaches
9.1. Data Privacy and Security
9.2. AI Bias
9.3. Regulatory Frameworks
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| AI/MLSaMD | Artificial Intelligence and Machine Learning Software as a Medical Device |
| AMH | Anti-Müllerian Hormone |
| ANN | Artificial Neural Network |
| AUC | Area Under the Curve |
| Auto-CM | Auto-Contractive Map |
| BCAA | Branched-Chain Amino Acid |
| BEV | Bacterial Extracellular Vesicle |
| BMI | Body Mass Index |
| CNN | Convolutional Neural Network |
| CREB1 | cAMP Responsive Element Binding Protein 1 |
| DOGMA | Dysbiosis Of Gut Microbiota |
| EV | Extracellular Vesicle |
| FDA | Food and Drug Administration |
| FFEV | Follicular Fluid Extracellular Vesicle |
| FOSL2 | FOS Like 2, AP-1 Transcription Factor Subunit |
| FXR | Farnesoid X Receptor |
| GDCA | Glycodeoxycholic acid |
| HIF-1α | Hypoxia Inducible Factor 1 Alpha |
| HOMA-IR | Homeostatic Model Assessment for Insulin Resistance |
| IL | Interleukin |
| IRS-1 | Insulin Receptor Substrate 1 |
| LDHA | Lactate Dehydrogenase A |
| LPS | Lipopolysaccharide |
| M1/M2 | Macrophage phenotypes (M1: pro-inflammatory, M2: anti-inflammatory) |
| MAPK | Mitogen-Activated Protein Kinase |
| METTL3 | Methyltransferase Like 3 |
| MIF | Macrophage Migration Inhibitory Factor |
| miRNA | microRNA |
| MSC-EV | Mesenchymal Stem Cell-Derived Extracellular Vesicle |
| mTOR | Mammalian Target Of Rapamycin |
| NF-kB | Nuclear Factor Kappa-Light-Chain-Enhancer of Activated B Cells |
| NIH | National Institutes of Health |
| OUT | Operational Taxonomic Unit |
| PCOS | Polycystic Ovary Syndrome |
| PI3K-Akt | Phosphoinositide 3-kinase/Protein kinase B pathway |
| ROC | Receiver-Operating Characteristic |
| S6K1 | Protein S6 Kinase 1 |
| SCFA | Short-Chain Fatty Acid |
| SCM | Semantic Connectivity Map |
| sEV | Small Extracellular Vesicle |
| SIRT1 | Sirtuin 1 |
| SMAD5 | SMAD Family Member 5 |
| STAT1/STAT3 | Signal Transducer and Activator of Transcription 1/3 |
| SVM | Support Vector Machine |
| TLR2 | Toll-Like Receptor 2 |
| TNF-α | Tumor Necrosis Factor alpha |
| TUDCA | Tauroursodeoxycholic Acid |
| WNT | Wingless-Related Integration Site |
References
- Lizneva, D.; Suturina, L.; Walker, W.; Brakta, S.; Gavrilova-Jordan, L.; Azziz, R. Criteria, Prevalence, and Phenotypes of Polycystic Ovary Syndrome. Fertil Steril 2016, 106, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Azziz, R.; Carmina, E.; Chen, Z.; Dunaif, A.; Laven, J.S.; Legro, R.S.; Lizneva, D.; Natterson-Horowtiz, B.; Teede, H.J.; Yildiz, B.O. Polycystic Ovary Syndrome. Nat. Rev. Dis. Primers 2016, 2, 16057. [Google Scholar] [CrossRef] [PubMed]
- National Institutes of Health Evidence-Based Methodology Workshop on Polycystic Ovary Syndrome. Final Report. Exec. Summary. Bethesda Md. 2012, 1, 1–14.
- Rotterdam ESHRE/ASRM-Sponsored PCOS consensus workshop group Revised 2003 Consensus on Diagnostic Criteria and Long-Term Health Risks Related to Polycystic Ovary Syndrome (PCOS). Hum. Reprod. 2004, 19, 41–47. [CrossRef] [PubMed]
- Bozdag, G.; Mumusoglu, S.; Zengin, D.; Karabulut, E.; Yildiz, B.O. The Prevalence and Phenotypic Features of Polycystic Ovary Syndrome: A Systematic Review and Meta-Analysis. Hum. Reprod. 2016, 31, 2841–2855. [Google Scholar] [CrossRef]
- Rosenfield, R.L.; Ehrmann, D.A. The Pathogenesis of Polycystic Ovary Syndrome (PCOS): The Hypothesis of PCOS as Functional Ovarian Hyperandrogenism Revisited. Endocr. Rev. 2016, 37, 467–520. [Google Scholar] [CrossRef]
- Goodman, N.F.; Cobin, R.H.; Futterweit, W.; Glueck, J.S.; Legro, R.S.; Carmina, E.; American Association of Clinical Endocrinologists (AACE); American College of Endocrinology (ACE); Androgen Excess and PCOS Society (AES). AMERICAN ASSOCIATION OF CLINICAL ENDOCRINOLOGISTS, AMERICAN COLLEGE OF ENDOCRINOLOGY, AND ANDROGEN EXCESS AND PCOS SOCIETY DISEASE STATE CLINICAL REVIEW: GUIDE TO THE BEST PRACTICES IN THE EVALUATION AND TREATMENT OF POLYCYSTIC OVARY SYNDROME--PART 1. Endocr Pr. 2015, 21, 1291–1300. [Google Scholar] [CrossRef]
- Azziz, R. New Insights into the Genetics of Polycystic Ovary Syndrome. Nat. Rev. Endocrinol. 2016, 12, 74–75. [Google Scholar] [CrossRef]
- Azziz, R. Diagnosis of Polycystic Ovarian Syndrome: The Rotterdam Criteria Are Premature. J. Clin. Endocrinol. Metab. 2006, 91, 781–785. [Google Scholar] [CrossRef]
- Rosenfield, R.L.; Barnes, R.B.; Ehrmann, D.A. Hyperandrogenism, Hirsutism, and Polycystic Ovary Syndrome. Endocrinol. Adult Pediatr. 2016, 2275–2296.e6. [Google Scholar] [CrossRef]
- Legro, R.S.; Arslanian, S.A.; Ehrmann, D.A.; Hoeger, K.M.; Murad, M.H.; Pasquali, R.; Welt, C.K. Endocrine Society Diagnosis and Treatment of Polycystic Ovary Syndrome: An Endocrine Society Clinical Practice Guideline. J. Clin. Endocrinol. Metab. 2013, 98, 4565–4592. [Google Scholar] [CrossRef] [PubMed]
- DeUgarte, C.M.; Bartolucci, A.A.; Azziz, R. Prevalence of Insulin Resistance in the Polycystic Ovary Syndrome Using the Homeostasis Model Assessment. Fertil Steril 2005, 83, 1454–1460. [Google Scholar] [CrossRef] [PubMed]
- Legro, R.S. Evaluation and Treatment of Polycystic Ovary Syndrome. In Endotext; Feingold, K.R., Ahmed, S.F., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Harris, H.R.; Terry, K.L. Polycystic Ovary Syndrome and Risk of Endometrial, Ovarian, and Breast Cancer: A Systematic Review. Fertil. Res. Pract. 2016, 2, 14. [Google Scholar] [CrossRef]
- Daan, N.M.P.; Louwers, Y.V.; Koster, M.P.H.; Eijkemans, M.J.C.; de Rijke, Y.B.; Lentjes, E.W.G.; Fauser, B.C.J.M.; Laven, J.S.E. Cardiovascular and Metabolic Profiles amongst Different Polycystic Ovary Syndrome Phenotypes: Who Is Really at Risk? Fertil Steril 2014, 102, 1444–1451.e3. [Google Scholar] [CrossRef]
- Oishi, S.; Mekaru, K.; Tanaka, S.E.; Arai, W.; Ashikawa, K.; Sakuraba, Y.; Nishioka, M.; Nakamura, R.; Miyagi, M.; Akamine, K.; et al. Microbiome Analysis in Women with Endometriosis: Does a Microbiome Exist in Peritoneal Fluid and Ovarian Cystic Fluid? Reprod. Med. Biol. 2022, 21, e12441. [Google Scholar] [CrossRef] [PubMed]
- Kostroun, K.E.; Goldrick, K.; Mondshine, J.N.; Robinson, R.D.; Mankus, E.; Reddy, S.; Wang, Z.; Song, X.; Knudtson, J.F. Impact of Updated International Diagnostic Criteria for the Diagnosis of Polycystic Ovary Syndrome. FS Rep. 2023, 4, 173–178. [Google Scholar] [CrossRef]
- Azziz, R.; Carmina, E.; Dewailly, D.; Diamanti-Kandarakis, E.; Escobar-Morreale, H.F.; Futterweit, W.; Janssen, O.E.; Legro, R.S.; Norman, R.J.; Taylor, A.E.; et al. The Androgen Excess and PCOS Society Criteria for the Polycystic Ovary Syndrome: The Complete Task Force Report. Fertil Steril 2009, 91, 456–488. [Google Scholar] [CrossRef]
- Tremellen, K.; Pearce, K. Dysbiosis of Gut Microbiota (DOGMA)--a Novel Theory for the Development of Polycystic Ovarian Syndrome. Med. Hypotheses 2012, 79, 104–112. [Google Scholar] [CrossRef]
- Lindheim, L.; Bashir, M.; Münzker, J.; Trummer, C.; Zachhuber, V.; Leber, B.; Horvath, A.; Pieber, T.R.; Gorkiewicz, G.; Stadlbauer, V.; et al. Alterations in Gut Microbiome Composition and Barrier Function Are Associated with Reproductive and Metabolic Defects in Women with Polycystic Ovary Syndrome (PCOS): A Pilot Study. PLoS ONE 2017, 12, e0168390. [Google Scholar] [CrossRef]
- Insenser, M.; Murri, M.; del Campo, R.; Martínez-García, M.Á.; Fernández-Durán, E.; Escobar-Morreale, H.F. Gut Microbiota and the Polycystic Ovary Syndrome: Influence of Sex, Sex Hormones, and Obesity. J. Clin. Endocrinol. Metab. 2018, 103, 2552–2562. [Google Scholar] [CrossRef]
- Torres, P.J.; Siakowska, M.; Banaszewska, B.; Pawelczyk, L.; Duleba, A.J.; Kelley, S.T.; Thackray, V.G. Gut Microbial Diversity in Women With Polycystic Ovary Syndrome Correlates With Hyperandrogenism. J. Clin. Endocrinol. Metab. 2018, 103, 1502–1511. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Yun, C.; Sun, L.; Xia, J.; Wu, Q.; Wang, Y.; Wang, L.; Zhang, Y.; Liang, X.; Wang, L.; et al. Gut Microbiota–Bile Acid–Interleukin-22 Axis Orchestrates Polycystic Ovary Syndrome. Nat. Med. 2019, 25, 1225–1233. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Lai, Z.; Sun, L.; Zhang, Z.; Yang, J.; Li, Z.; Lin, J.; Zhang, Z. Structural and Functional Profiles of the Gut Microbial Community in Polycystic Ovary Syndrome with Insulin Resistance (IR-PCOS): A Pilot Study. Res. Microbiol. 2019, 170, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Lindheim, L.; Bashir, M.; Münzker, J.; Trummer, C.; Zachhuber, V.; Pieber, T.R.; Gorkiewicz, G.; Obermayer-Pietsch, B. The Salivary Microbiome in Polycystic Ovary Syndrome (PCOS) and Its Association with Disease-Related Parameters: A Pilot Study. Front. Microbiol. 2016, 7, 1270. [Google Scholar] [CrossRef]
- Torres, P.J.; Ho, B.S.; Arroyo, P.; Sau, L.; Chen, A.; Kelley, S.T.; Thackray, V.G. Exposure to a Healthy Gut Microbiome Protects Against Reproductive and Metabolic Dysregulation in a PCOS Mouse Model. Endocrinology 2019, 160, 1193–1204. [Google Scholar] [CrossRef]
- Ou, Z.; Situ, B.; Huang, X.; Xue, Y.; He, X.; Li, Q.; Ou, D.; He, B.; Chen, J.; Huang, Y.; et al. Single-particle Analysis of Circulating Bacterial Extracellular Vesicles Reveals Their Biogenesis, Changes in Blood and Links to Intestinal Barrier. J. Extracell Vesicles 2023, 12, 12395. [Google Scholar] [CrossRef]
- Liu, C.; Yazdani, N.; Moran, C.S.; Salomon, C.; Seneviratne, C.J.; Ivanovski, S.; Han, P. Unveiling Clinical Applications of Bacterial Extracellular Vesicles as Natural Nanomaterials in Disease Diagnosis and Therapeutics. Acta Biomater. 2024, 180, 18–45. [Google Scholar] [CrossRef]
- Yang, L.; Liu, T.; Liao, Y.; Ren, Y.; Zheng, Z.; Zhang, M.; Yu, Y.; Liu, C.; Wang, C.; Chen, T.; et al. Potential Therapeutic Application and Mechanism of Gut Microbiota-Derived Extracellular Vesicles in Polycystic Ovary Syndrome. Biomed. Pharmacother. 2024, 180, 117504. [Google Scholar] [CrossRef]
- Sun, D.; Chen, P.; Xi, Y.; Sheng, J. From Trash to Treasure: The Role of Bacterial Extracellular Vesicles in Gut Health and Disease. Front. Immunol. 2023, 14, 1274295. [Google Scholar] [CrossRef]
- Joseph, A.; Anton, L.; Guan, Y.; Ferguson, B.; Mirro, I.; Meng, N.; France, M.; Ravel, J.; Elovitz, M.A. Extracellular Vesicles from Vaginal Gardnerella Vaginalis and Mobiluncus Mulieris Contain Distinct Proteomic Cargo and Induce Inflammatory Pathways. NPJ Biofilms Microbiomes 2024, 10, 28. [Google Scholar] [CrossRef]
- Wagner, M.; Hicks, C.; El-Omar, E.; Combes, V.; El-Assaad, F. The Critical Role of Host and Bacterial Extracellular Vesicles in Endometriosis. Biomedicines 2024, 12, 2585. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Shin, T.-S.; Kim, J.S.; Jee, Y.-K.; Kim, Y.-K. A New Horizon of Precision Medicine: Combination of the Microbiome and Extracellular Vesicles. Exp. Mol. Med. 2022, 54, 466–482. [Google Scholar] [CrossRef]
- Xie, J.; Haesebrouck, F.; Van Hoecke, L.; Vandenbroucke, R.E. Bacterial Extracellular Vesicles: An Emerging Avenue to Tackle Diseases. Trends Microbiol. 2023, 31, 1206–1224. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Jian, T.; Gai, Y.; Chen, J. Microbiota and Plant-Derived Vesicles That Serve as Therapeutic Agents and Delivery Carriers to Regulate Metabolic Syndrome. Adv. Drug Deliv. Rev. 2023, 196, 114774. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Xing, J.; Zhao, S.; Chen, H.; Yin, X.; Zhu, X. Engineered Extracellular Vesicles in Female Reproductive Disorders. Biomed. Pharmacother. 2023, 166, 115284. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Q.; Zhao, L.; Bin, Y.; Wang, L.; Wang, L.; Zhang, K.; Li, Q. Blood Bacterial 16S rRNA Gene Alterations in Women With Polycystic Ovary Syndrome. Front. Endocrinol. 2022, 13, 814520. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, J.; Gober, H.-J.; Leung, W.T.; Huang, Z.; Pan, X.; Li, C.; Zhang, N.; Wang, L. Alterations in the Intestinal Microbiome Associated with PCOS Affect the Clinical Phenotype. Biomed. Pharmacother. 2021, 133, 110958. [Google Scholar] [CrossRef]
- Corrie, L.; Awasthi, A.; Kaur, J.; Vishwas, S.; Gulati, M.; Kaur, I.P.; Gupta, G.; Kommineni, N.; Dua, K.; Singh, S.K. Interplay of Gut Microbiota in Polycystic Ovarian Syndrome: Role of Gut Microbiota, Mechanistic Pathways and Potential Treatment Strategies. Pharmaceuticals 2023, 16, 197. [Google Scholar] [CrossRef]
- Batra, M.; Bhatnager, R.; Kumar, A.; Suneja, P.; Dang, A.S. Interplay between PCOS and Microbiome: The Road Less Travelled. Am. J. Reprod. Immunol. 2022, 88, e13580. [Google Scholar] [CrossRef]
- Sun, Y.; Gao, S.; Ye, C.; Zhao, W. Gut Microbiota Dysbiosis in Polycystic Ovary Syndrome: Mechanisms of Progression and Clinical Applications. Front Cell Infect Microbiol 2023, 13, 1142041. [Google Scholar] [CrossRef]
- Yin, H.; Xie, J.; Xing, S.; Lu, X.; Yu, Y.; Ren, Y.; Tao, J.; He, G.; Zhang, L.; Yuan, X.; et al. Machine Learning-Based Analysis Identifies and Validates Serum Exosomal Proteomic Signatures for the Diagnosis of Colorectal Cancer. CR Med. 2024, 5, 101689. [Google Scholar] [CrossRef] [PubMed]
- Barrera, F.J.; Brown, E.D.L.; Rojo, A.; Obeso, J.; Plata, H.; Lincango, E.P.; Terry, N.; Rodríguez-Gutiérrez, R.; Hall, J.E.; Shekhar, S. Application of Machine Learning and Artificial Intelligence in the Diagnosis and Classification of Polycystic Ovarian Syndrome: A Systematic Review. Front. Endocrinol. 2023, 14, 1106625. [Google Scholar] [CrossRef] [PubMed]
- Elmannai, H.; El-Rashidy, N.; Mashal, I.; Alohali, M.A.; Farag, S.; El-Sappagh, S.; Saleh, H. Polycystic Ovary Syndrome Detection Machine Learning Model Based on Optimized Feature Selection and Explainable Artificial Intelligence. Diagnostics 2023, 13, 1506. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial Intelligence in Healthcare: Past, Present and Future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Guixue, G.; Yifu, P.; Yuan, G.; Xialei, L.; Fan, S.; Qian, S.; Jinjin, X.; Linna, Z.; Xiaozuo, Z.; Wen, F.; et al. Progress of the Application Clinical Prediction Model in Polycystic Ovary Syndrome. J. Ovarian Res. 2023, 16, 230. [Google Scholar] [CrossRef]
- Mubasher Hassan, M.; Mirza, T. Comparative Analysis of Machine Learning Algorithms in Diagnosis of Polycystic Ovarian Syndrome. IJCA 2020, 175, 42–53. [Google Scholar] [CrossRef]
- Wang, D.-D.; Li, Y.-F.; Mao, Y.-Z.; He, S.-M.; Zhu, P.; Wei, Q.-L. A Machine-Learning Approach for Predicting the Effect of Carnitine Supplementation on Body Weight in Patients with Polycystic Ovary Syndrome. Front. Nutr. 2022, 9. [Google Scholar] [CrossRef]
- Verma, P.; Maan, P.; Gautam, R.; Arora, T. Unveiling the Role of Artificial Intelligence (AI) in Polycystic Ovary Syndrome (PCOS) Diagnosis: A Comprehensive Review. Reprod. Sci. 2024, 31, 2901–2915. [Google Scholar] [CrossRef]
- Sinthia, G.; Poovizhi, T.; Khilar, R. Analysis on Polycystic Ovarian Syndrome and Comparative Study of Different Machine Learning Algorithms. In Proceedings of the Advances in Intelligent Computing and Communication; Mohanty, M.N., Das, S., Eds.; Springer Nature: Singapore, 2022; pp. 191–196. [Google Scholar]
- Ahmad, R.; Maghrabi, L.A.; Khaja, I.A.; Maghrabi, L.A.; Ahmad, M. SMOTE-Based Automated PCOS Prediction Using Lightweight Deep Learning Models. Diagnostics 2024, 14, 2225. [Google Scholar] [CrossRef]
- Faris, N.N.; Miften, F.S. An Intelligence Model for Detection of PCOS Based on K-Means Coupled with LS-SVM. Concurr. Comput. Pract. Exp. 2022, 34, e7139. [Google Scholar] [CrossRef]
- Gianfrancesco, M.A.; Tamang, S.; Yazdany, J.; Schmajuk, G. Potential Biases in Machine Learning Algorithms Using Electronic Health Record Data. JAMA Intern Med 2018, 178, 1544–1547. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y. Ethical Issues of Human Microbiome Research. Encycl. Life Sci. 2021, 1–9. [Google Scholar] [CrossRef]
- Chuong, K.H.; Hwang, D.M.; Tullis, D.E.; Waters, V.J.; Yau, Y.C.W.; Guttman, D.S.; O’Doherty, K.C. Navigating Social and Ethical Challenges of Biobanking for Human Microbiome Research. BMC Med. Ethics 2017, 18, 1. [Google Scholar] [CrossRef] [PubMed]
- Chuong, K.H.; Mack, D.R.; Stintzi, A.; O’Doherty, K.C. Human Microbiome and Learning Healthcare Systems: Integrating Research and Precision Medicine for Inflammatory Bowel Disease. OMICS 2018, 22, 119–126. [Google Scholar] [CrossRef]
- Zawadri, J. Diagnostic Criteria for Polycystic Ovary Syndrome: Towards a Rational Approach. In Polycystic Ovary Syndrome; Dunaif, A., Givens, J.R., Haseltine, F., Eds.; Blackwell Scientific: Boston, MA, USA, 1992; pp. 377–384. [Google Scholar]
- Lujan, M.E.; Chizen, D.R.; Pierson, R.A. Diagnostic Criteria for Polycystic Ovary Syndrome: Pitfalls and Controversies. J. Obstet. Gynaecol. Can. 2008, 30, 671–679. [Google Scholar] [CrossRef]
- Franks, S. Controversy in Clinical Endocrinology: Diagnosis of Polycystic Ovarian Syndrome: In Defense of the Rotterdam Criteria. J Clin Endocrinol Metab 2006, 91, 786–789. [Google Scholar] [CrossRef]
- Amer, S.a.K.S.; Li, T.C.; Bygrave, C.; Sprigg, A.; Saravelos, H.; Cooke, I.D. An Evaluation of the Inter-Observer and Intra-Observer Variability of the Ultrasound Diagnosis of Polycystic Ovaries. Hum. Reprod. 2002, 17, 1616–1622. [Google Scholar] [CrossRef]
- Ezeh, U.; Pall, M.; Mathur, R.; Azziz, R. Association of Fat to Lean Mass Ratio with Metabolic Dysfunction in Women with Polycystic Ovary Syndrome. Hum. Reprod. 2014, 29, 1508–1517. [Google Scholar] [CrossRef]
- Amiri, M.; Hatoum, S.; Hopkins, D.; Buyalos, R.P.; Ezeh, U.; Pace, L.A.; Bril, F.; Sheidaei, A.; Azziz, R. The Association Between Obesity and Polycystic Ovary Syndrome: An Epidemiologic Study of Observational Data. J. Clin. Endocrinol. Metab. 2024, 109, 2640–2657. [Google Scholar] [CrossRef]
- Lee, I.T.-L.; Sansone, S.; Irfan, M.; Copp, T.; Beidas, R.; Dokras, A. Implementation of International Guidelines for Polycystic Ovary Syndrome: Barriers and Facilitators among Gynecologists and Primary Care Providers. FS Rep. 2022, 3, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Joham, A.E.; Piltonen, T.; Lujan, M.E.; Kiconco, S.; Tay, C.T. Challenges in Diagnosis and Understanding of Natural History of Polycystic Ovary Syndrome. Clin. Endocrinol. 2022, 97, 165–173. [Google Scholar] [CrossRef]
- Rosner, W.; Auchus, R.J.; Azziz, R.; Sluss, P.M.; Raff, H. Utility, Limitations, and Pitfalls in Measuring Testosterone: An Endocrine Society Position Statement. J. Clin. Endocrinol. Metab. 2007, 92, 405–413. [Google Scholar] [CrossRef]
- Handelsman, D.J.; Teede, H.J.; Desai, R.; Norman, R.J.; Moran, L.J. Performance of Mass Spectrometry Steroid Profiling for Diagnosis of Polycystic Ovary Syndrome. Hum. Reprod 2017, 32, 418–422. [Google Scholar] [CrossRef]
- Yildiz, B.O.; Bolour, S.; Woods, K.; Moore, A.; Azziz, R. Visually Scoring Hirsutism. Hum. Reprod Update 2010, 16, 51–64. [Google Scholar] [CrossRef]
- Unfer, V.; Kandaraki, E.; Pkhaladze, L.; Roseff, S.; Vazquez-Levin, M.H.; Laganà, A.S.; Shiao-Yng, C.; Yap-Garcia, M.I.M.; Greene, N.D.E.; Soulage, C.O.; et al. When One Size Does Not Fit All: Reconsidering PCOS Etiology, Diagnosis, Clinical Subgroups, and Subgroup-Specific Treatments. Endocr. Metab. Sci. 2024, 14, 100159. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, J.; Cheng, X.; Nie, X.; He, B. Insulin Resistance in Polycystic Ovary Syndrome across Various Tissues: An Updated Review of Pathogenesis, Evaluation, and Treatment. J. Ovarian Res. 2023, 16, 9. [Google Scholar] [CrossRef]
- Zhao, Y.; Pan, S.; Li, Y.; Wu, X. Exosomal miR-143-3p Derived from Follicular Fluid Promotes Granulosa Cell Apoptosis by Targeting BMPR1A in Polycystic Ovary Syndrome. Sci. Rep. 2022, 12, 4359. [Google Scholar] [CrossRef]
- Jonard, S.; Robert, Y.; Cortet-Rudelli, C.; Pigny, P.; Decanter, C.; Dewailly, D. Ultrasound Examination of Polycystic Ovaries: Is It Worth Counting the Follicles? Hum. Reprod. 2003, 18, 598–603. [Google Scholar] [CrossRef]
- Kim, J.J.; Hwang, K.R.; Chae, S.J.; Yoon, S.H.; Choi, Y.M. Impact of the Newly Recommended Antral Follicle Count Cutoff for Polycystic Ovary in Adult Women with Polycystic Ovary Syndrome. Hum Reprod 2020, 35, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.; Han, Q.; Xu, J.; Wang, J.; Sun, Y.; Li, W.; Chen, Z.-J.; Du, Y. Metagenomic Analysis Identified Microbiome Alterations and Pathological Association between Intestinal Microbiota and Polycystic Ovary Syndrome. Fertil. Steril. 2020, 113, 1286–1298.e4. [Google Scholar] [CrossRef] [PubMed]
- Di Michele, S.; Fulghesu, A.M.; Pittui, E.; Cordella, M.; Sicilia, G.; Mandurino, G.; D’Alterio, M.N.; Vitale, S.G.; Angioni, S. Ultrasound Assessment in Polycystic Ovary Syndrome Diagnosis: From Origins to Future Perspectives—A Comprehensive Review. Biomedicines 2025, 13, 453. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, C.; Shi, Y.; Zhang, F.; Li, L.; Wang, X.; Ling, Y.; Fu, H.; Dong, W.; Shen, J.; et al. Dysbiosis of Gut Microbiota Associated with Clinical Parameters in Polycystic Ovary Syndrome. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E. Short-Chain Fatty Acids in Control of Body Weight and Insulin Sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Acharya, A.; Shetty, S.S.; Kumari N, S. Role of Gut Microbiota Derived Short Chain Fatty Acid Metabolites in Modulating Female Reproductive Health. Hum. Nutr. Metab. 2024, 36, 200256. [Google Scholar] [CrossRef]
- McNabney, S.M.; Henagan, T.M. Short Chain Fatty Acids in the Colon and Peripheral Tissues: A Focus on Butyrate, Colon Cancer, Obesity and Insulin Resistance. Nutrients 2017, 9, 1348. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A Double-Edged Sword for Health? Adv. Nutr. 2018, 9, 21–29. [Google Scholar] [CrossRef]
- Liu, K.; He, X.; Huang, J.; Yu, S.; Cui, M.; Gao, M.; Liu, L.; Qian, Y.; Xie, Y.; Hui, M.; et al. Short-Chain Fatty Acid-Butyric Acid Ameliorates Granulosa Cells Inflammation through Regulating METTL3-Mediated N6-Methyladenosine Modification of FOSL2 in Polycystic Ovarian Syndrome. Clin. Epigenetics 2023, 15, 86. [Google Scholar] [CrossRef]
- Olaniyi, K.S.; Areloegbe, S.E. Butyrate Alleviates Adipose Mitochondrial Dysfunction and Inflammation in Experimental Model of Polycystic Ovarian Syndrome by Modulating SIRT1-Dependent Mechanism. Nutrire 2024, 49, 31. [Google Scholar] [CrossRef]
- Lu, N.; Li, M.; Lei, H.; Jiang, X.; Tu, W.; Lu, Y.; Xia, D. Butyric Acid Regulates Progesterone and Estradiol Secretion via cAMP Signaling Pathway in Porcine Granulosa Cells. J. Steroid Biochem. Mol. Biol. 2017, 172, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, J.L.; Bäckhed, F. Diet-Microbiota Interactions as Moderators of Human Metabolism. Nature 2016, 535, 56–64. [Google Scholar] [CrossRef]
- Zhenyukh, O.; Civantos, E.; Ruiz-Ortega, M.; Sánchez, M.S.; Vázquez, C.; Peiró, C.; Egido, J.; Mas, S. High Concentration of Branched-Chain Amino Acids Promotes Oxidative Stress, Inflammation and Migration of Human Peripheral Blood Mononuclear Cells via mTORC1 Activation. Free Radic. Biol. Med. 2017, 104, 165–177. [Google Scholar] [CrossRef]
- Zhou, M.; Yu, J.; Li, X.; Ruan, Z.; Yu, S. Role of the Gut Microbiota and Innate Immunity in Polycystic Ovary Syndrome: Current Updates and Future Prospects. J. Cell. Mol. Med. 2024, 28, e18258. [Google Scholar] [CrossRef]
- Hiippala, K.; Jouhten, H.; Ronkainen, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The Potential of Gut Commensals in Reinforcing Intestinal Barrier Function and Alleviating Inflammation. Nutrients 2018, 10, 988. [Google Scholar] [CrossRef] [PubMed]
- Duval, C.; Wyse, B.A.; Tsang, B.K.; Librach, C.L. Extracellular Vesicles and Their Content in the Context of Polycystic Ovarian Syndrome and Endometriosis: A Review. J. Ovarian Res. 2024, 17, 160. [Google Scholar] [CrossRef]
- Zhao, Y.; Tao, M.; Wei, M.; Du, S.; Wang, H.; Wang, X. Mesenchymal Stem Cells Derived Exosomal miR-323-3p Promotes Proliferation and Inhibits Apoptosis of Cumulus Cells in Polycystic Ovary Syndrome (PCOS). Artif. Cells Nanomed. Biotechnol. 2019, 47, 3804–3813. [Google Scholar] [CrossRef]
- Lima, P.D.A.; Nivet, A.-L.; Wang, Q.; Chen, Y.-A.; Leader, A.; Cheung, A.; Tzeng, C.-R.; Tsang, B.K. Polycystic Ovary Syndrome: Possible Involvement of Androgen-Induced, Chemerin-Mediated Ovarian Recruitment of Monocytes/Macrophages†. Biol. Reprod. 2018, 99, 838–852. [Google Scholar] [CrossRef]
- Salehi, R.; Asare-Werehene, M.; Wyse, B.A.; Abedini, A.; Pan, B.; Gutsol, A.; Jahangiri, S.; Szaraz, P.; Burns, K.D.; Vanderhyden, B. Granulosa Cell-Derived miR-379-5p Regulates Macrophage Polarization in Polycystic Ovarian Syndrome. Front. Immunol. 2023, 14, 1104550. [Google Scholar] [CrossRef]
- Cui, C.; Wang, J.; Han, X.; Wang, Q.; Zhang, S.; Liang, S.; Li, H.; Meng, L.; Zhang, C.; Chen, H. Identification of Small Extracellular Vesicle-Linked miRNA Specifically Derived from Intrafollicular Cells in Women with Polycystic Ovary Syndrome. Reprod. Biomed. Online 2021, 42, 870–880. [Google Scholar] [CrossRef]
- Cui, X.; Lei, X.; Huang, T.; Mao, X.; Shen, Z.; Yang, X.; Peng, W.; Yu, J.; Zhang, S.; Huo, P. Follicular Fluid-Derived Extracellular Vesicles miR-34a-5p Regulates Granulosa Cell Glycolysis in Polycystic Ovary Syndrome by Targeting LDHA. J. Ovarian Res. 2024, 17, 223. [Google Scholar] [CrossRef] [PubMed]
- Che, X.; Jian, F.; Chen, C.; Liu, C.; Liu, G.; Feng, W. PCOS Serum-Derived Exosomal miR-27a-5p Stimulates Endometrial Cancer Cells Migration and Invasion. J. Mol. Endocrinol. 2020, 64, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Deswal, R.; Dang, A.S. Dissecting the Role of Micro-RNAs as a Diagnostic Marker for Polycystic Ovary Syndrome: A Systematic Review and Meta-Analysis. Fertil. Steril. 2020, 113, 661–669.e2. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Huo, P.; Cui, K.; Wei, H.; Cao, J.; Wang, J.; Liu, Q.; Lei, X.; Zhang, S. Follicular Fluid-Derived Exosomal miR-143-3p/miR-155-5p Regulate Follicular Dysplasia by Modulating Glycolysis in Granulosa Cells in Polycystic Ovary Syndrome. Cell Commun. Signal. 2022, 20, 61. [Google Scholar] [CrossRef]
- Udesen, P.B.; Sørensen, A.E.; Svendsen, R.; Frisk, N.L.S.; Hess, A.L.; Aziz, M.; Wissing, M.L.M.; Englund, A.L.M.; Dalgaard, L.T. Circulating miRNAs in Women with Polycystic Ovary Syndrome: A Longitudinal Cohort Study. Cells 2023, 12, 983. [Google Scholar] [CrossRef]
- Wang, M.; Liu, M.; Sun, J.; Jia, L.; Ma, S.; Gao, J.; Xu, Y.; Zhang, H.; Tsang, S.Y.; Li, X. MicroRNA-27a-3p Affects Estradiol and Androgen Imbalance by Targeting Creb1 in the Granulosa Cells in Mouse Polycytic Ovary Syndrome Model. Reprod. Biol. 2017, 17, 295–304. [Google Scholar] [CrossRef]
- Wang, M.; Sun, J.; Xu, B.; Chrusciel, M.; Gao, J.; Bazert, M.; Stelmaszewska, J.; Xu, Y.; Zhang, H.; Pawelczyk, L.; et al. Functional Characterization of MicroRNA-27a-3p Expression in Human Polycystic Ovary Syndrome. Endocrinology 2018, 159, 297–309. [Google Scholar] [CrossRef]
- Jiang, X.; Li, J.; Zhang, B.; Hu, J.; Ma, J.; Cui, L.; Chen, Z.-J. Differential Expression Profile of Plasma Exosomal microRNAs in Women with Polycystic Ovary Syndrome. Fertil. Steril. 2021, 115, 782–792. [Google Scholar] [CrossRef]
- Hu, L.; Hong, G.; Li, J.; Chen, M.; Chang, C.-J.; Cheng, P.-J.; Zhang, Z.; Zhang, X.; Chen, H.; Zhuang, Y.; et al. Metformin Modifies Plasma Microbial-Derived Extracellular Vesicles in Polycystic Ovary Syndrome with Insulin Resistance. J. Ovarian Res. 2024, 17, 136. [Google Scholar] [CrossRef]
- Hotamisligil, G.S. Inflammation, Metaflammation and Immunometabolic Disorders. Nature 2017, 542, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Deeke, S.A.; Ning, Z.; Starr, A.E.; Butcher, J.; Li, J.; Mayne, J.; Cheng, K.; Liao, B.; Li, L.; et al. Metaproteomics Reveals Associations between Microbiome and Intestinal Extracellular Vesicle Proteins in Pediatric Inflammatory Bowel Disease. Nat. Commun. 2018, 9, 2873. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Shorakae, S.; Teede, H.; de Courten, B.; Lambert, G.; Boyle, J.; Moran, L. The Emerging Role of Chronic Low-Grade Inflammation in the Pathophysiology of Polycystic Ovary Syndrome. Semin. Reprod. Med. 2015, 33, 257–269. [Google Scholar] [CrossRef]
- Godbole, N.; Quinn, A.; Carrion, F.; Pelosi, E.; Salomon, C. Extracellular Vesicles as a Potential Delivery Platform for CRISPR-Cas Based Therapy in Epithelial Ovarian Cancer. Semin. Cancer Biol. 2023, 96, 64–81. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, I.A.; Kuehn, M.J. Offense and Defense: Microbial Membrane Vesicles Play Both Ways. Res. Microbiol. 2012, 163, 607–618. [Google Scholar] [CrossRef]
- Schooling, S.R.; Beveridge, T.J. Membrane Vesicles: An Overlooked Component of the Matrices of Biofilms. J. Bacteriol. 2006, 188, 5945–5957. [Google Scholar] [CrossRef]
- Sahu, G.; Karnati, M.; Rajput, A.S.; Chaudhary, M.; Maurya, R.; Dutta, M.K. Attention-Based Transfer Learning Approach Using Spatial Pyramid Pooling for Diagnosis of Polycystic Ovary Syndrome. In Proceedings of the 2023 9th International Conference on Signal Processing and Communication (ICSC), Noida, India, 21–23 December 2023; pp. 238–243. [Google Scholar]
- Walters, K.A.; Bertoldo, M.J.; Handelsman, D.J. Evidence from Animal Models on the Pathogenesis of PCOS. Best Pract. Res. Clin. Endocrinol. Metab. 2018, 32, 271–281. [Google Scholar] [CrossRef]
- Cao, M.; Zhao, Y.; Chen, T.; Zhao, Z.; Zhang, B.; Yuan, C.; Wang, X.; Chen, L.; Wang, N.; Li, C.; et al. Adipose Mesenchymal Stem Cell–Derived Exosomal microRNAs Ameliorate Polycystic Ovary Syndrome by Protecting against Metabolic Disturbances. Biomaterials 2022, 288, 121739. [Google Scholar] [CrossRef]
- Escobar-Morreale, H.F. Polycystic Ovary Syndrome: Definition, Aetiology, Diagnosis and Treatment. Nat. Rev. Endocrinol. 2018, 14, 270–284. [Google Scholar] [CrossRef]
- Hall, J.E.; Shekhar, S. AI and Machine Learning Can Successfully Diagnose Polycystic Ovary Syndrome. Available online: https://www.nih.gov/news-events/news-releases/ai-machine-learning-can-successfully-diagnose-polycystic-ovary-syndrome (accessed on 5 December 2024).
- Khanna, V.V.; Chadaga, K.; Sampathila, N.; Prabhu, S.; Bhandage, V.; Hegde, G.K. A Distinctive Explainable Machine Learning Framework for Detection of Polycystic Ovary Syndrome. Appl. Syst. Innov. 2023, 6, 32. [Google Scholar] [CrossRef]
- Kodipalli, A.; Devi, S. Prediction of PCOS and Mental Health Using Fuzzy Inference and SVM. Front Public Health 2021, 9, 789569. [Google Scholar] [CrossRef]
- Emara, H.M.; El-Shafai, W.; Soliman, N.F.; Algarni, A.D.; Alkanhel, R.; Abd El-Samie, F.E. A Stacked Learning Framework for Accurate Classification of Polycystic Ovary Syndrome with Advanced Data Balancing and Feature Selection Techniques. Front. Physiol. 2025, 16, 1435036. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.-N.; Wang, F.-F.; Zhou, J.; Liu, C.; Qu, F. Establishment and Analysis of a Combined Diagnostic Model of Polycystic Ovary Syndrome with Random Forest and Artificial Neural Network. Biomed. Res. Int. 2020, 2020, 2613091. [Google Scholar] [CrossRef]
- Mahesswari, G.U.; Maheswari, P.U. SmartScanPCOS: A Feature-Driven Approach to Cutting-Edge Prediction of Polycystic Ovary Syndrome Using Machine Learning and Explainable Artificial Intelligence. Heliyon 2024, 10. [Google Scholar] [CrossRef]
- Fu, J.; Zhang, Y.; Cai, X.; Huang, Y. Predicting Metformin Efficacy in Improving Insulin Sensitivity Among Women With Polycystic Ovary Syndrome and Insulin Resistance: A Machine Learning Study. Endocr. Pract. 2024, 30, 1023–1030. [Google Scholar] [CrossRef] [PubMed]
- Lecot-Connan, T.; Rives, M.; Bachelot, G.; Sow, C.; Fourati, S.; Tejedor, I.; Godon, T.; Lamaziere, A.; Bachelot, A. Combining Machine Learning and Metabolomics to Identify the Metabolic Signatures of Polycystic Ovary Syndrome Patients According to Body Mass Index. Endocr. Abstr. 2025, 110, P1045. [Google Scholar] [CrossRef]
- Chitra, P.; Srilatha, K.; Sumathi, M.; Jayasudha, F.V.; Bernatin, T.; Jagadeesh, M. Classification of Ultrasound PCOS Image Using Deep Learning Based Hybrid Models. In Proceedings of the 2023 Second International Conference on Electronics and Renewable Systems (ICEARS), Tuticorin, India, 2–4 March 2023; pp. 1389–1394. [Google Scholar]
- Sumathi, M.; Chitra, P.; Prabha, R.S.; Srilatha, K. Study and Detection of PCOS Related Diseases Using CNN. IOP Conf. Ser. Mater. Sci. Eng. 2021, 1070, 012062. [Google Scholar] [CrossRef]
- Gülhan, P.G.; Özmen, G.; Alptekin, H. CNN Based Determination of Polycystic Ovarian Syndrome Using Automatic Follicle Detection Methods. Politek. Derg. 2024, 27, 1589–1601. [Google Scholar] [CrossRef]
- Suha, S.A.; Islam, M.N. An Extended Machine Learning Technique for Polycystic Ovary Syndrome Detection Using Ovary Ultrasound Image. Sci Rep 2022, 12, 17123. [Google Scholar] [CrossRef]
- Paramasivam, G.B.; Ramasamy Rajammal, R. Modelling a Self-Defined CNN for Effectual Classification of PCOS from Ultrasound Images. Technol. Health Care 2024, 32, 2893–2909. [Google Scholar] [CrossRef] [PubMed]
- Karthik, Y.; Sruthi, R.; Sujithra, M. Polycystic Ovary Syndrome Prediction through CNN Based Image Analysis: A Deep Learning Based Approach. In Proceedings of the 2024 8th International Conference on I-SMAC (IoT in Social, Mobile, Analytics and Cloud) (I-SMAC), Kirtipur, Nepal, 3–5 October 2024; pp. 1547–1553. [Google Scholar]
- Moral, P.; Mustafi, D.; Mustafi, A.; Sahana, S.K. CystNet: An AI Driven Model for PCOS Detection Using Multilevel Thresholding of Ultrasound Images. Sci. Rep. 2024, 14, 25012. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Song, Y.; Fu, R.; Lin, X.; Su, Y.; Jin, X.; Yang, H.; Shan, X.; Du, W.; Huang, Q.; et al. Deep Learning Algorithm for Automated Detection of Polycystic Ovary Syndrome Using Scleral Images. Front. Endocrinol. 2022, 12, 789878. [Google Scholar] [CrossRef]
- Hosain, A.K.M.S.; Mehedi, M.H.K.; Kabir, I.E. PCONet: A Convolutional Neural Network Architecture to Detect Polycystic Ovary Syndrome (PCOS) from Ovarian Ultrasound Images. In Proceedings of the 2022 International Conference on Engineering and Emerging Technologies (ICEET), Kuala Lumpur, Malaysia, 27–28 October 2022; pp. 1–6. [Google Scholar]
- Alamoudi, A.; Khan, I.U.; Aslam, N.; Alqahtani, N.; Alsaif, H.S.; Al Dandan, O.; Al Gadeeb, M.; Al Bahrani, R. A Deep Learning Fusion Approach to Diagnosis the Polycystic Ovary Syndrome (PCOS). Appl. Comput. Intell. Soft Comput. 2023, 2023, 9686697. [Google Scholar] [CrossRef]
- Kermanshahchi, J.; Reddy, A.J.; Xu, J.; Mehrok, G.K.; Nausheen, F. Development of a Machine Learning-Based Model for Accurate Detection and Classification of Polycystic Ovary Syndrome on Pelvic Ultrasound. Cureus 2024, 16, e65134. [Google Scholar] [CrossRef]
- Garzia, E.; Galiano, V.; Marfia, G.; Navone, S.; Grossi, E.; Marconi, A.M. Hyperandrogenism and Menstrual Imbalance Are the Best Predictors of Metformin Response in PCOS Patients. Reprod Biol Endocrinol 2022, 20, 6. [Google Scholar] [CrossRef]
- Bhat, S.A. Detection of Polycystic Ovary Syndrome Using Machine Learning Algorithms. Ph.D. Thesis, National College of Ireland, Dublin, Ireland, 2021. [Google Scholar]
- Murri, M.; Insenser, M.; Fernández-Durán, E.; San-Millán, J.L.; Escobar-Morreale, H.F. Effects of Polycystic Ovary Syndrome (PCOS), Sex Hormones, and Obesity on Circulating miRNA-21, miRNA-27b, miRNA-103, and miRNA-155 Expression. J. Clin. Endocrinol. Metab. 2013, 98, E1835–E1844. [Google Scholar] [CrossRef]
- Murri, M.; Insenser, M.; Fernández-Durán, E.; San-Millán, J.L.; Luque-Ramírez, M.; Escobar-Morreale, H.F. Non-Targeted Profiling of Circulating microRNAs in Women with Polycystic Ovary Syndrome (PCOS): Effects of Obesity and Sex Hormones. Metab. Clin. Exp. 2018, 86, 49–60. [Google Scholar] [CrossRef]
- Butler, A.E.; Ramachandran, V.; Hayat, S.; Dargham, S.R.; Cunningham, T.K.; Benurwar, M.; Sathyapalan, T.; Najafi-Shoushtari, S.H.; Atkin, S.L. Expression of microRNA in Follicular Fluid in Women with and without PCOS. Sci. Rep. 2019, 9, 16306. [Google Scholar] [CrossRef]
- Topol, E.J. High-Performance Medicine: The Convergence of Human and Artificial Intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Zhang, F.; Li, S.-P.; Zhang, T.; Yu, B.; Zhang, J.; Ding, H.-G.; Ye, F.-J.; Yuan, H.; Ma, Y.-Y.; Pan, H.-T.; et al. High Throughput microRNAs Sequencing Profile of Serum Exosomes in Women with and without Polycystic Ovarian Syndrome. PeerJ 2021, 9, e10998. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hu, R.; Huang, Y.; Zhou, F.; Li, F.; Liu, Z.; Geng, Y.; Dong, H.; Ma, W.; Song, K.; et al. Present and Future: Crosstalks Between Polycystic Ovary Syndrome and Gut Metabolites Relating to Gut Microbiota. Front. Endocrinol. 2022, 13, 933110. [Google Scholar] [CrossRef]
- Rahman, M.M.; Islam, A.; Islam, F.; Zaman, M.; Islam, M.R.; Alam Sakib, M.S.; Hasan Babu, H.M. Empowering Early Detection: A Web-Based Machine Learning Approach for PCOS Prediction. Inform. Med. Unlocked 2024, 47, 101500. [Google Scholar] [CrossRef]
- Tiwari, S.; Kane, L.; Koundal, D.; Jain, A.; Alhudhaif, A.; Polat, K.; Zaguia, A.; Alenezi, F.; Althubiti, S.A. SPOSDS: A Smart Polycystic Ovary Syndrome Diagnostic System Using Machine Learning. Expert Syst. Appl. 2022, 203, 117592. [Google Scholar] [CrossRef]
- Song, H.; Qin, Q.; Yuan, C.; Li, H.; Zhang, F.; Fan, L. Metabolomic Profiling of Poor Ovarian Response Identifies Potential Predictive Biomarkers. Front. Endocrinol. 2021, 12, 774667. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Li, J.; Feng, X.; Feng, L.; Xia, Y.; Xiao, X.; Wang, Y.; Xu, Z. Machine Learning Classification of Polycystic Ovary Syndrome Based on Radial Pulse Wave Analysis. BMC Complement. Med. Ther. 2023, 23, 409. [Google Scholar] [CrossRef]
- Rashid, S.; Karnati, M.; Aggarwal, G.; Dutta, M.K.; Sikora, P.; Burget, R. Attention-Based Multiscale Deep Neural Network for Diagnosis of Polycystic Ovary Syndrome Using Ovarian Ultrasound Images. In Proceedings of the 2023 15th International Congress on Ultra Modern Telecommunications and Control Systems and Workshops (ICUMT), Ghent, Belgium, 30 October–1 November 2023; pp. 44–49. [Google Scholar]
- Sheikdavood, K.; Bala, M.P. Polycystic Ovary Cyst Segmentation Using Adaptive K-Means with Reptile Search Algorith. Inf. Technol. Control 2023, 52, 85–99. [Google Scholar] [CrossRef]
- Srinithi, V.; Rekha, R. Machine Learning for Diagnosis of Polycystic Ovarian Syndrome (PCOS/PCOD). In Proceedings of the 2023 International Conference on Intelligent Systems for Communication, IoT and Security (ICISCoIS), Coimbatore, India, 9–11 February 2023; pp. 19–24. [Google Scholar]
- Danaei Mehr, H.; Polat, H. Diagnosis of Polycystic Ovary Syndrome through Different Machine Learning and Feature Selection Techniques. Health Technol. 2022, 12, 137–150. [Google Scholar] [CrossRef]
- Swapnarekha, H.; Dash, P.B.; Nayak, J.; Routray, A.R. An Optimistic Bayesian Optimization Based Extreme Learning Machine for Polycystic Ovary Syndrome Diagnosis. In Nature-Inspired Optimization Methodologies in Biomedical and Healthcare; Nayak, J., Das, A.K., Naik, B., Meher, S.K., Brahnam, S., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 175–193. ISBN 978-3-031-17544-2. [Google Scholar]
- Barrea, L.; Marzullo, P.; Muscogiuri, G.; Di Somma, C.; Scacchi, M.; Orio, F.; Aimaretti, G.; Colao, A.; Savastano, S. Source and Amount of Carbohydrate in the Diet and Inflammation in Women with Polycystic Ovary Syndrome. Nutr. Res. Rev. 2018, 31, 291–301. [Google Scholar] [CrossRef]
- Shanmugavadivel, K.; M S, M.D.; T R, M.; Al-Shehari, T.; Alsadhan, N.A.; Yimer, T.E. Optimized Polycystic Ovarian Disease Prognosis and Classification Using AI Based Computational Approaches on Multi-Modality Data. BMC Med. Inf. Decis. Mak. 2024, 24, 281. [Google Scholar] [CrossRef]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best Practices for Analysing Microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Byts, N.; Makieieva, O.; Zhyvolozhnyi, A.; Bart, G.; Korvala, J.; Hekkala, J.; Salmi, S.; Samoylenko, A.; Reunanen, J. Purification of Bacterial-Enriched Extracellular Vesicle Samples from Feces by Density Gradient Ultracentrifugation. Methods Mol. Biol. 2023, 2668, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Dapas, M.; Lin, F.T.J.; Nadkarni, G.N.; Sisk, R.; Legro, R.S.; Urbanek, M.; Hayes, M.G.; Dunaif, A. Distinct Subtypes of Polycystic Ovary Syndrome with Novel Genetic Associations: An Unsupervised, Phenotypic Clustering Analysis. PLoS Med. 2020, 17, e1003132. [Google Scholar] [CrossRef]
- Lim, Y.; Kim, H.Y.; Han, D.; Choi, B.-K. Proteome and Immune Responses of Extracellular Vesicles Derived from Macrophages Infected with the Periodontal Pathogen Tannerella Forsythia. J. Extracell. Vesicles 2023, 12, 12381. [Google Scholar] [CrossRef] [PubMed]
- Zad, Z.; Jiang, V.S.; Wolf, A.T.; Wang, T.; Cheng, J.J.; Paschalidis, I.C.; Mahalingaiah, S. Predicting Polycystic Ovary Syndrome (PCOS) with Machine Learning Algorithms from Electronic Health Records. medRxiv 2023. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Li, S.; Fang, X.; Zhang, C.; Wang, Z.; Zheng, Y.; Deng, H.; Xu, S.; Mi, Y. Exploring Acetylation-Related Gene Markers in Polycystic Ovary Syndrome: Insights into Pathogenesis and Diagnostic Potential Using Machine Learning. Gynecol. Endocrinol. 2024, 40, 2427202. [Google Scholar] [CrossRef]
- Qi, X.; Yun, C.; Pang, Y.; Qiao, J. The Impact of the Gut Microbiota on the Reproductive and Metabolic Endocrine System. Gut Microbes 2021, 13, 1894070. [Google Scholar] [CrossRef]
- Callaway, E. Microbiome Privacy Risk. Nature 2015, 521, 136. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Powers, B.; Vogeli, C.; Mullainathan, S. Dissecting Racial Bias in an Algorithm Used to Manage the Health of Populations. Science 2019, 366, 447–453. [Google Scholar] [CrossRef]
- Warraich, H.J.; Tazbaz, T.; Califf, R.M. FDA Perspective on the Regulation of Artificial Intelligence in Health Care and Biomedicine. JAMA 2025, 333, 241–247. [Google Scholar] [CrossRef]
| Category | Description | Key Statistics/Features | References |
|---|---|---|---|
| Epidemiology | Global prevalence | 6–19% of reproductive-age women worldwide using NIH criteria; 8–13% using Rotterdam criteria | [1,3,4,8] |
| Ethnic variations | Higher rates in South Asian (8–22%) and Middle Eastern populations (12–20%); lower in East Asian populations (2.2–7.4%) | [1,5,9] | |
| Clinical Features | Reproductive manifestations | Hyperandrogenism, ovulatory dysfunction, polycystic ovarian morphology | [4,6] |
| Metabolic manifestations | Insulin resistance (65–70% of PCOS patients), obesity, increased risk of type 2 diabetes | [10,11,12] | |
| Other health risks | Cardiovascular disease, endometrial cancer | [6,11,13,14,15,16] | |
| Traditional Diagnostic Approaches | NIH criteria (1992) | (1) Hyperandrogenism, (2) oligo/anovulation, (3) exclusion of other disorders | [3] |
| Rotterdam criteria (2003) | Requires two of three features: (1) oligo/anovulation, (2) clinical/biochemical hyperandrogenism, (3) polycystic ovaries on ultrasound | [4] | |
| Androgen excess society criteria | (1) Hyperandrogenism, (2) ovarian dysfunction (oligo/anovulation and/or polycystic ovaries) | [17,18] |
| AI/ML Technique | Data Type and Sample Size | Validation Method | Feature Selection/Preprocessing | Performance | Key Findings | References |
|---|---|---|---|---|---|---|
| Clinical Data Analysis | ||||||
| Random forest ensemble (multi-stack) | Clinical parameters (hormonal profiles, ultrasound findings, metabolic markers), N = 541 | 5-fold cross-validation | Mutual Information (MI) feature selection, SMOTEENN balancing | Accuracy: 98%, precision: 97%, recall: 98%, F1-score: 98% | Best performing model with explainable AI integration using SHAP, LIME | [113,114,115] |
| Random forest with ANN | Gene expression data (GEO database), N = 133 (76 PCOS, 57 controls) | Two training sets, two validation sets | 12 key genes selected from 264 DEGs | AUC: 0.7273 (microarray), 0.6488 (RNA-seq) | Combined RF and neural network approach for gene biomarker identification | [116] |
| Hierarchical random forest ensemble | Clinical features with XAI, N = 541 | 8-fold cross-validation, 25 runs | TOMIM, TOPCA, OSSM feature selection methods | Accuracy: 99.31% (top 17 features), overall: 99.32% | Two-level ensemble with explainable AI using Shapash library | [117] |
| Support vector machines (SVMs) | Serum metabolomic profiles, metformin efficacy prediction, study-specific cohorts | Cross-validation | Metabolomic profiling | AUC-ROC: 0.935 (95% CI: 0.898–0.972) | Metabolomics-based prediction of treatment response | [118,119] |
| Fuzzy-TOPSIS + SVM | Clinical data with linguistic responses, study-specific | Not specified | Fuzzy logic preprocessing | Fuzzy-TOPSIS: 98.20%, SVM: 94.01% | Integration of fuzzy logic with traditional ML | [114] |
| Image analysis | ||||||
| CNN (ResNet-50) | Ultrasound images, study-specific | Standard train/test split | Image preprocessing, augmentation | Accuracy: 92.3% (95% CI: 90.1–94.5%), Sensitivity: 91.4% (95% CI: 88.7–94.1%), Specificity: 93.1% (95% CI: 90.6–95.6%) | ResNet-50 architecture for ultrasound analysis | [120,121] |
| CNN (VGG16+XGBoost stacking) | Ultrasound images, N = 594 ovary USG images | Train/validation/test split | Transfer learning with VGG16, feature extraction | Accuracy: 99.89%, execution time optimized | Hybrid approach combining CNN and ensemble learning | [122,123] |
| CNN (various architectures) | Ultrasound images, variable by study | Train/test splits | Preprocessing: contrast enhancement, noise reduction | Raw images: 65.81%, preprocessed: 77.81% | Importance of image preprocessing demonstrated | [121,122,124,125] |
| CNN (CystNet hybrid model) | Ultrasound images, Kaggle PCOS dataset | 5-fold cross-validation | InceptionV3 + convolutional autoencoder | Dense layer: 96.54% accuracy, RF classifier: 97.75% accuracy | Hybrid architecture with multiple classification approaches | [126] |
| Deep learning (U-Net + ResNet) | Non-invasive eye imaging (scleral images), N = 721 (388 PCOS patients) | Multi-instance learning validation | Sclera segmentation, attention mechanism | AUC: 0.979, accuracy: 92.9% | Novel non-invasive screening using eye imaging | [127] |
| CNN (PCONet + InceptionV3) | Ultrasound images, Kaggle dataset | Transfer learning validation | Fine-tuned pre-trained models | PCONet: 98.12%, InceptionV3: 96.56% | Custom CNN architecture vs. transfer learning comparison | [128] |
| Microbiome analysis | ||||||
| Random forest classifier | Stool microbiome profiles, study-specific cohorts | Cross-validation | 16S rRNA sequencing, taxonomic profiling | Accuracy: 87.5% (95% CI: 84.2–90.8%), sensitivity: 89.3% (95% CI: 85.6–93.0%), specificity: 85.7% (95% CI: 81.4–90.0%), AUC-ROC: 0.93 (95% CI: 0.90–0.96) | Microbiome-based classification showing promise for non-invasive diagnosis | [22] |
| Random forest | Gut microbiome and clinical data, multiple cohorts | 5-fold cross-validation | Feature selection, diversity metrics | Accuracy: 85% (95% CI: 81–89%), sensitivity: 87% (95% CI: 82–92%), specificity: 83% (95% CI: 78–88%) | Integration of microbiome and clinical parameters | [20] |
| Random forest | β-diversity with hormonal correlation, study cohorts | Statistical correlation analysis | Microbiome profiling, hormonal measurements | Significant correlation with hyperandrogenism (p = 0.0009) | Direct correlation between microbiome and PCOS phenotype | [19] |
| Multi-modal approaches | ||||||
| Machine learning (integrated) | Gut microbiome, BEV-associated miRNAs, clinical parameters, multi-source data integration | Cross-validation | Multi-omics data fusion | Accuracy: 92.0% (CI: 88.9–95.1%), sensitivity: 93.0%, specificity: 91.0%, AUC-ROC: 0.96 | Comprehensive multi-omics approach for enhanced accuracy | [33,43,44,47,68] |
| Deep learning with ensemble | Clinical features and ultrasound images, combined datasets | Cross-validation | Multi-modal feature fusion | SVM: 94.44%, VGG16: 98.29% validation accuracy | Multi-modal data integration approach | [51,129,130] |
| Specialized applications | ||||||
| Artificial neural networks (Auto-CM) | Clinical characteristics, study-specific | Not specified | Automated feature selection | Performance not specified | Automated clinical decision-making system | [131] |
| Gradient boosting | Clinical, hormonal, metabolomic data, study cohorts | Cross-validation | Multi-dimensional data integration | AUC-ROC: 0.83 | Integration of diverse clinical data types | [118,123,132] |
| ROC analysis (meta-analysis) | EV-associated miRNAs (miR-29a-5p, miR-320, miR-93), meta-analysis of multiple studies | Multi-study validation | Biomarker standardization | AUC = 0.95 for miR-29a-5p | Meta-analysis approach for biomarker validation | [133,134,135] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kushawaha, B.; Rem, T.T.; Pelosi, E. Harnessing Microbiome, Bacterial Extracellular Vesicle, and Artificial Intelligence for Polycystic Ovary Syndrome Diagnosis and Management. Biomolecules 2025, 15, 834. https://doi.org/10.3390/biom15060834
Kushawaha B, Rem TT, Pelosi E. Harnessing Microbiome, Bacterial Extracellular Vesicle, and Artificial Intelligence for Polycystic Ovary Syndrome Diagnosis and Management. Biomolecules. 2025; 15(6):834. https://doi.org/10.3390/biom15060834
Chicago/Turabian StyleKushawaha, Bhawna, Tial T. Rem, and Emanuele Pelosi. 2025. "Harnessing Microbiome, Bacterial Extracellular Vesicle, and Artificial Intelligence for Polycystic Ovary Syndrome Diagnosis and Management" Biomolecules 15, no. 6: 834. https://doi.org/10.3390/biom15060834
APA StyleKushawaha, B., Rem, T. T., & Pelosi, E. (2025). Harnessing Microbiome, Bacterial Extracellular Vesicle, and Artificial Intelligence for Polycystic Ovary Syndrome Diagnosis and Management. Biomolecules, 15(6), 834. https://doi.org/10.3390/biom15060834





