Liquid Biopsy-Derived Tumor Biomarkers for Clinical Applications in Glioblastoma
Abstract
1. Introduction
1.1. Glioblastoma Overview
1.2. Liquid Biopsy Overview
2. Materials and Methods
3. Selected Papers and the Clinical Applicability of Biomarkers Derived from Liquid Biopsy in Glioblastoma
3.1. Panorama of the Selected Papers
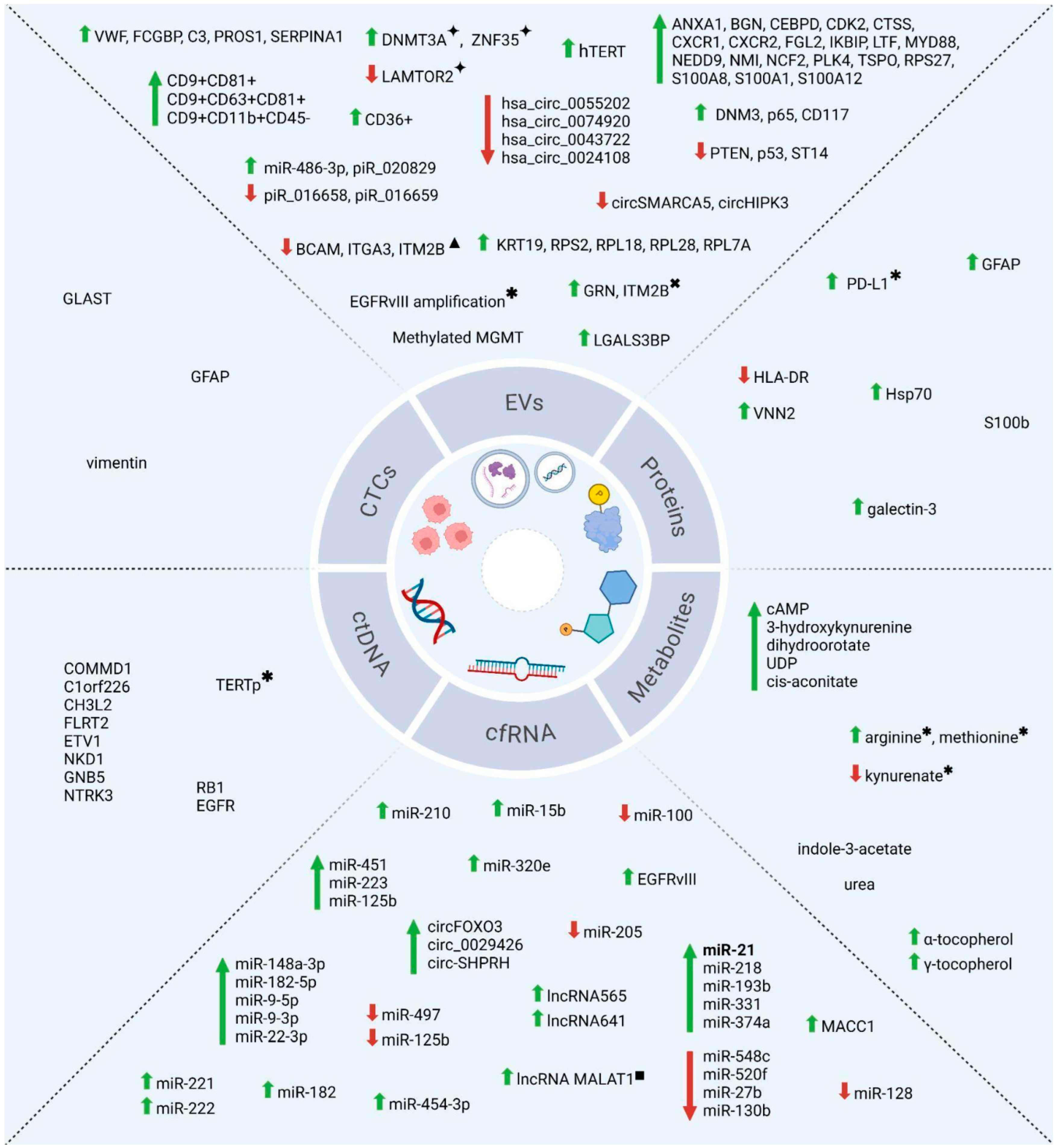
3.2. Circulating Tumor Cells (CTCs)
3.3. Extracellular Vesicles (EVs)
3.4. Circulating Tumor Nucleic Acids (ctNAs)
3.5. Circulating Proteins and Metabolites
4. Technical Challenges and Future Directions
5. Final Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GBM | Glioblastoma |
| IDH | Isocitrate dehydrogenase |
| TMZ | Temozolomide |
| BBB | Blood–brain barrier |
| CNS | Central nervous system |
| CSF | Cerebrospinal fluid |
| EVs | Extracellular vesicles |
| CTCs | Circulating tumor cells |
| miRNA | microRNA |
| ctNAs | Circulating tumor nucleic acids |
| cfDNA | Circulating free DNA |
| cfRNA | Circulating free RNA |
| lncRNA | Long non-coding RNA |
| circRNA | Circular ribonucleic acid |
| mRNA | Messenger ribonucleic acid |
| piRNA | PIWI-interacting RNA |
| sEVs | Small extracellular vesicles |
| MRI | Magnetic resonance imaging |
| MRgFUS | Magnetic resonance imaging-guided focused ultrasound |
| MR | Magnetic resonance |
| CT | Computerized tomography |
| PET | Positron emission tomography |
| GeLB | Epigenetic Glioma Liquid Biopsy |
| LB | Liquid biopsy |
| NGS | Next-generation sequencing |
| n | Sample size |
| PFS | Progression-free survival |
| PBMCs | Peripheral blood mononuclear cells |
| GSC-EV | Glioblastoma stemcell–extracellular vesicles |
| qPCR | Quantitative polymerase chain reaction |
| ddPCR | Droplet digital polymerase chain reaction |
| qRT-PCR | Quantitative reverse-transcribed PCR |
| TERT | Telomerase reverse transcriptase |
| hTERT | Human telomerase reverse transcriptase |
| TERTp | Telomerase reverse transcriptase promoter |
| EGFR | Epidermal growth factor receptor |
| MGMT | O6-methylguanine-DNA methyltransferase |
| PTEN | Phosphatase and tensin homolog |
| GFAP | Glial fibrillar acid protein |
| WT | Wild-type |
| IHC | Immunohistochemistry |
| FDA | Food and drugaAdministration |
| WHO | World health organization |
| CGGA | Chinese glioma genome atlas |
| FFPE | Formalin-fixed paraffin-embedded |
| ELISA | Enzyme-linked immunosorbent assay |
| LC-MS | Liquid chromatography–mass spectrometry |
| WB | Western blot |
| NA | Not available |
| OS | Overall survival |
| PFS | Progression-free survival |
References
- Oronsky, B.; Reid, T.R.; Oronsky, A.; Sandhu, N.; Knox, S.J. A Review of Newly Diagnosed Glioblastoma. Front. Oncol. 2021, 10, 574012. [Google Scholar] [CrossRef] [PubMed]
- Venkataramani, V.; Yang, Y.; Schubert, M.C.; Reyhan, E.; Tetzlaff, S.K.; Wißmann, N.; Botz, M.; Soyka, S.J.; Beretta, C.A.; Pramatarov, R.L.; et al. Glioblastoma Hijacks Neuronal Mechanisms for Brain Invasion. Cell 2022, 185, 2899–2917.e31. [Google Scholar] [CrossRef]
- Davis, M. Glioblastoma: Overview of Disease and Treatment. Clin. J. Oncol. Nurs. 2016, 20, S2–S8. [Google Scholar] [CrossRef]
- Young, R.M.; Jamshidi, A.; Davis, G.; Sherman, J.H. Current Trends in the Surgical Management and Treatment of Adult Glioblastoma. Ann. Transl. Med. 2015, 3, 121. [Google Scholar] [CrossRef] [PubMed]
- Lara-Velazquez, M.; Al-Kharboosh, R.; Jeanneret, S.; Vazquez-Ramos, C.; Mahato, D.; Tavanaiepour, D.; Rahmathulla, G.; Quinones-Hinojosa, A. Advances in Brain Tumor Surgery for Glioblastoma in Adults. Brain Sci. 2017, 7, 166. [Google Scholar] [CrossRef]
- Eibl, R.H.; Schneemann, M. Liquid Biopsy and Glioblastoma. Explor. Target. Anti-Tumor Ther. 2023, 4, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Iorgulescu, J.B.; Blewett, T.; Xiong, K.; Crnjac, A.; Liu, R.; Sridhar, S.; Braun, D.A.; Sellars, M.C.; Cheng, J.; Rhoades, J.; et al. Impact of Higher Cell-Free DNA Yields on Liquid Biopsy Testing in Glioblastoma Patients. Clin. Chem. 2025, 71, 215–225. [Google Scholar] [CrossRef]
- Wirsching, H.-G.; Galanis, E.; Weller, M. Glioblastoma. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 134, pp. 381–397. ISBN 978-0-12-802997-8. [Google Scholar]
- Lone, S.N.; Nisar, S.; Masoodi, T.; Singh, M.; Rizwan, A.; Hashem, S.; El-Rifai, W.; Bedognetti, D.; Batra, S.K.; Haris, M.; et al. Liquid Biopsy: A Step Closer to Transform Diagnosis, Prognosis and Future of Cancer Treatments. Mol. Cancer 2022, 21, 79. [Google Scholar] [CrossRef]
- Giordano, C.; Sabatino, G.; Romano, S.; Della Pepa, G.M.; Tufano, M.; D’Alessandris, Q.G.; Cottonaro, S.; Gessi, M.; Balducci, M.; Romano, M.F.; et al. Combining Magnetic Resonance Imaging with Systemic Monocyte Evaluation for the Implementation of GBM Management. Int. J. Mol. Sci. 2021, 22, 3797. [Google Scholar] [CrossRef]
- Bagley, S.J.; Nabavizadeh, S.A.; Mays, J.J.; Till, J.E.; Ware, J.B.; Levy, S.; Sarchiapone, W.; Hussain, J.; Prior, T.; Guiry, S.; et al. Clinical Utility of Plasma Cell-Free DNA in Adult Patients with Newly Diagnosed Glioblastoma: A Pilot Prospective Study. Clin. Cancer Res. 2020, 26, 397–407. [Google Scholar] [CrossRef]
- Senhaji, N.; Squalli Houssaini, A.; Lamrabet, S.; Louati, S.; Bennis, S. Molecular and Circulating Biomarkers in Patients with Glioblastoma. Int. J. Mol. Sci. 2022, 23, 7474. [Google Scholar] [CrossRef] [PubMed]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid Biopsy: Current Technology and Clinical Applications. J. Hematol. Oncol. 2022, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zhu, Q.; Yang, J.; Zheng, Y.; Du, S.; Song, M.; Peng, Q.; Yang, R.; Liu, Y.; Qi, L. Imaging and Liquid Biopsy for Distinguishing True Progression From Pseudoprogression in Gliomas, Current Advances and Challenges. Acad. Radiol. 2024, 31, 3366–3383. [Google Scholar] [CrossRef] [PubMed]
- Balana, C.; Castañer, S.; Carrato, C.; Moran, T.; Lopez-Paradís, A.; Domenech, M.; Hernandez, A.; Puig, J. Preoperative Diagnosis and Molecular Characterization of Gliomas With Liquid Biopsy and Radiogenomics. Front. Neurol. 2022, 13, 865171. [Google Scholar] [CrossRef]
- Gao, F.; Cui, Y.; Jiang, H.; Sui, D.; Wang, Y.; Jiang, Z.; Zhao, J.; Lin, S. Circulating Tumor Cell Is a Common Property of Brain Glioma and Promotes the Monitoring System. Oncotarget 2016, 7, 71330–71340. [Google Scholar] [CrossRef]
- Lynch, D.; Powter, B.; Po, J.W.; Cooper, A.; Garrett, C.; Koh, E.-S.; Sheridan, M.; Van Gelder, J.; Darwish, B.; Mckechnie, S.; et al. Isolation of Circulating Tumor Cells from Glioblastoma Patients by Direct Immunomagnetic Targeting. Appl. Sci. 2020, 10, 3338. [Google Scholar] [CrossRef]
- Müller Bark, J.; Kulasinghe, A.; Hartel, G.; Leo, P.; Warkiani, M.E.; Jeffree, R.L.; Chua, B.; Day, B.W.; Punyadeera, C. Isolation of Circulating Tumour Cells in Patients With Glioblastoma Using Spiral Microfluidic Technology–A Pilot Study. Front. Oncol. 2021, 11, 681130. [Google Scholar] [CrossRef]
- Kolostova, K.; Pospisilova, E.; Pavlickova, V.; Bartos, R.; Sames, M.; Pawlak, I.; Bobek, V. Next Generation Sequencing of Glioblastoma Circulating Tumor Cells: Non-Invasive Solution for Disease Monitoring. Am. J. Transl. Res. 2021, 13, 4489–4499. [Google Scholar]
- Yang, J.; Song, J.; Huo, H.; Zhao, Y.; Zhang, G.; Zhao, Z.; Sun, G.; Jiao, B. DNM3, P65 and P53 from Exosomes Represent Potential Clinical Diagnosis Markers for Glioblastoma Multiforme. Ther. Adv. Med. Oncol. 2017, 9, 741–754. [Google Scholar] [CrossRef]
- Hallal, S.; Azimi, A.; Wei, H.; Ho, N.; Lee, M.Y.T.; Sim, H.-W.; Sy, J.; Shivalingam, B.; Buckland, M.E.; Alexander-Kaufman, K.L. A Comprehensive Proteomic SWATH-MS Workflow for Profiling Blood Extracellular Vesicles: A New Avenue for Glioma Tumour Surveillance. Int. J. Mol. Sci. 2020, 21, 4754. [Google Scholar] [CrossRef]
- Dobra, G.; Bukva, M.; Szabo, Z.; Bruszel, B.; Harmati, M.; Gyukity-Sebestyen, E.; Jenei, A.; Szucs, M.; Horvath, P.; Biro, T.; et al. Small Extracellular Vesicles Isolated from Serum May Serve as Signal-Enhancers for the Monitoring of CNS Tumors. Int. J. Mol. Sci. 2020, 21, 5359. [Google Scholar] [CrossRef] [PubMed]
- Cilibrasi, C.; Simon, T.; Vintu, M.; Tolias, C.; Samuels, M.; Mazarakis, N.K.; Eravci, M.; Stewart, N.; Critchley, G.; Giamas, G. Definition of an Inflammatory Biomarker Signature in Plasma-Derived Extracellular Vesicles of Glioblastoma Patients. Biomedicines 2022, 10, 125. [Google Scholar] [CrossRef]
- Hallal, S.M.; Tűzesi, Á.; Sida, L.A.; Xian, E.; Madani, D.; Muralidharan, K.; Shivalingam, B.; Buckland, M.E.; Satgunaseelan, L.; Alexander, K.L. Glioblastoma Biomarkers in Urinary Extracellular Vesicles Reveal the Potential for a ‘Liquid Gold’ Biopsy. Br. J. Cancer 2024, 130, 836–851. [Google Scholar] [CrossRef]
- Brahmer, A.; Geiß, C.; Lygeraki, A.; Neuberger, E.; Tzaridis, T.; Nguyen, T.T.; Luessi, F.; Régnier-Vigouroux, A.; Hartmann, G.; Simon, P.; et al. Assessment of Technical and Clinical Utility of a Bead-Based Flow Cytometry Platform for Multiparametric Phenotyping of CNS-Derived Extracellular Vesicles. Cell Commun. Signal. 2023, 21, 276. [Google Scholar] [CrossRef]
- Dufrusine, B.; Capone, E.; Ponziani, S.; Lattanzio, R.; Lanuti, P.; Giansanti, F.; De Laurenzi, V.; Iacobelli, S.; Ippoliti, R.; Mangiola, A.; et al. Extracellular LGALS3BP: A Potential Disease Marker and Actionable Target for Antibody–Drug Conjugate Therapy in Glioblastoma. Mol. Oncol. 2023, 17, 1460–1473. [Google Scholar] [CrossRef] [PubMed]
- Lennartz, P.; Thölke, D.; Bashiri Dezfouli, A.; Pilz, M.; Lobinger, D.; Messner, V.; Zanth, H.; Ainslie, K.; Kafshgari, M.H.; Rammes, G.; et al. Biomarkers in Adult-Type Diffuse Gliomas: Elevated Levels of Circulating Vesicular Heat Shock Protein 70 Serve as a Biomarker in Grade 4 Glioblastoma and Increase NK Cell Frequencies in Grade 3 Glioma. Biomedicines 2023, 11, 3235. [Google Scholar] [CrossRef]
- Werner, C.; Stangl, S.; Salvermoser, L.; Schwab, M.; Shevtsov, M.; Xanthopoulos, A.; Wang, F.; Dezfouli, A.B.; Thölke, D.; Ostheimer, C.; et al. Hsp70 in Liquid Biopsies-A Tumor-Specific Biomarker for Detection and Response Monitoring in Cancer. Cancers 2021, 13, 3706. [Google Scholar] [CrossRef] [PubMed]
- Aibaidula, A.; Fain, C.E.; Garcia, L.C.; Wier, A.; Bouchal, S.M.; Bauman, M.M.; Jung, M.-Y.; Sarkaria, J.N.; Johnson, A.J.; Parney, I.F. Spectral Flow Cytometry Identifies Distinct Nonneoplastic Plasma Extracellular Vesicle Phenotype in Glioblastoma Patients. Neuro-Oncol. Adv. 2023, 5, vdad082. [Google Scholar] [CrossRef]
- Ricklefs, F.L.; Wollmann, K.; Salviano-Silva, A.; Drexler, R.; Maire, C.L.; Kaul, M.G.; Reimer, R.; Schüller, U.; Heinemann, S.; Kolbe, K.; et al. Circulating Extracellular Vesicles as Biomarker for Diagnosis, Prognosis, and Monitoring in Glioblastoma Patients. Neuro-Oncol. 2024, 26, 1280–1291. [Google Scholar] [CrossRef]
- Figueroa, J.M.; Skog, J.; Akers, J.; Li, H.; Komotar, R.; Jensen, R.; Ringel, F.; Yang, I.; Kalkanis, S.; Thompson, R.; et al. Detection of Wild-Type EGFR Amplification and EGFRvIII Mutation in CSF-Derived Extracellular Vesicles of Glioblastoma Patients. Neuro-Oncology 2017, 19, 1494–1502. [Google Scholar] [CrossRef]
- Hallal, S.; Ebrahim Khani, S.; Wei, H.; Lee, M.Y.T.; Sim, H.-W.; Sy, J.; Shivalingam, B.; Buckland, M.E.; Alexander-Kaufman, K.L. Deep Sequencing of Small RNAs from Neurosurgical Extracellular Vesicles Substantiates miR-486-3p as a Circulating Biomarker That Distinguishes Glioblastoma from Lower-Grade Astrocytoma Patients. Int. J. Mol. Sci. 2020, 21, 4954. [Google Scholar] [CrossRef] [PubMed]
- Yekula, A.; Hsia, T.; Kitchen, R.R.; Chakrabortty, S.K.; Yu, W.; Batool, S.M.; Lewis, B.; Szeglowski, A.J.; Weissleder, R.; Lee, H.; et al. Longitudinal Analysis of Serum-Derived Extracellular Vesicle RNA to Monitor Dacomitinib Treatment Response in EGFR-Amplified Recurrent Glioblastoma Patients. Neuro-Oncol. Adv. 2023, 5, vdad104. [Google Scholar] [CrossRef]
- Uziel, O.; Kanner, A.A.; Beery, E.; Lev, S.; Lahav, M.; Horn-Fichman, S.; Nof, S.H.; Laviv, Y.; Yust-Katz, S.; Amiel, A.; et al. Is Serum-derived Exosomal hTERT Transcript a Marker of Oncogenic Activity in Primary Brain Tumors? An Exploratory Study. Cancer Med. 2024, 13, e6784. [Google Scholar] [CrossRef] [PubMed]
- Manda, S.V.; Kataria, Y.; Tatireddy, B.R.; Ramakrishnan, B.; Ratnam, B.G.; Lath, R.; Ranjan, A.; Ray, A. Exosomes as a Biomarker Platform for Detecting Epidermal Growth Factor Receptor–Positive High-Grade Gliomas. J. Neurosurg. 2018, 128, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Hua, W.; Li, H.; Ramakrishnan, V.; Yang, Z.; Quan, K.; Zhu, W.; Li, J.; Figueroa, J.; Hirshman, B.R.; et al. A Cerebrospinal Fluid microRNA Signature as Biomarker for Glioblastoma. Oncotarget 2017, 8, 68769–68779. [Google Scholar] [CrossRef]
- Shi, R.; Wang, P.-Y.; Li, X.-Y.; Chen, J.-X.; Li, Y.; Zhang, X.-Z.; Zhang, C.-G.; Jiang, T.; Li, W.-B.; Ding, W.; et al. Exosomal Levels of miRNA-21 from Cerebrospinal Fluids Associated with Poor Prognosis and Tumor Recurrence of Glioma Patients. Oncotarget 2015, 6, 26971–26981. [Google Scholar] [CrossRef]
- Rosas-Alonso, R.; Colmenarejo-Fernández, J.; Pernía, O.; Burdiel, M.; Rodríguez-Antolín, C.; Losantos-García, I.; Rubio, T.; Moreno-Velasco, R.; Esteban-Rodríguez, I.; Martínez-Marín, V.; et al. Evaluation of the Clinical Use of MGMT Methylation in Extracellular Vesicle-Based Liquid Biopsy as a Tool for Glioblastoma Patient Management. Sci. Rep. 2024, 14, 11398. [Google Scholar] [CrossRef]
- Wang, Y.; Springer, S.; Zhang, M.; McMahon, K.W.; Kinde, I.; Dobbyn, L.; Ptak, J.; Brem, H.; Chaichana, K.; Gallia, G.L.; et al. Detection of Tumor-Derived DNA in Cerebrospinal Fluid of Patients with Primary Tumors of the Brain and Spinal Cord. Proc. Natl. Acad. Sci. USA 2015, 112, 9704–9709. [Google Scholar] [CrossRef]
- Miller, A.M.; Shah, R.H.; Pentsova, E.I.; Pourmaleki, M.; Briggs, S.; Distefano, N.; Zheng, Y.; Skakodub, A.; Mehta, S.A.; Campos, C.; et al. Tracking Tumour Evolution in Glioma through Liquid Biopsies of Cerebrospinal Fluid. Nature 2019, 565, 654–658. [Google Scholar] [CrossRef]
- Juratli, T.A.; Stasik, S.; Zolal, A.; Schuster, C.; Richter, S.; Daubner, D.; Juratli, M.A.; Thowe, R.; Hennig, S.; Makina, M.; et al. TERT Promoter Mutation Detection in Cell-Free Tumor-Derived DNA in Patients with IDH Wild-Type Glioblastomas: A Pilot Prospective Study. Clin. Cancer Res. 2018, 24, 5282–5291. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, C.; Li, M.; Shen, Y.; Feng, S.; Liu, J.; Li, F.; Hou, L.; Chen, Z.; Jiang, J.; et al. Applications of Cerebrospinal Fluid Circulating Tumor DNA in the Diagnosis of Gliomas. Jpn. J. Clin. Oncol. 2020, 50, 325–332. [Google Scholar] [CrossRef]
- Wang, Q.; Liang, Q.; Wei, W.; Niu, W.; Liang, C.; Wang, X.; Wang, X.; Pan, H. Concordance Analysis of Cerebrospinal Fluid with the Tumor Tissue for Integrated Diagnosis in Gliomas Based on Next-Generation Sequencing. Pathol. Oncol. Res. 2023, 29, 1611391. [Google Scholar] [CrossRef] [PubMed]
- Piccioni, D.E.; Achrol, A.S.; Kiedrowski, L.A.; Banks, K.C.; Boucher, N.; Barkhoudarian, G.; Kelly, D.F.; Juarez, T.; Lanman, R.B.; Raymond, V.M.; et al. Analysis of Cell-Free Circulating Tumor DNA in 419 Patients with Glioblastoma and Other Primary Brain Tumors. CNS Oncol. 2019, 8, CNS34. [Google Scholar] [CrossRef]
- De Mattos-Arruda, L.; Mayor, R.; Ng, C.K.Y.; Weigelt, B.; Martínez-Ricarte, F.; Torrejon, D.; Oliveira, M.; Arias, A.; Raventos, C.; Tang, J.; et al. Cerebrospinal Fluid-Derived Circulating Tumour DNA Better Represents the Genomic Alterations of Brain Tumours than Plasma. Nat. Commun. 2015, 6, 8839. [Google Scholar] [CrossRef] [PubMed]
- Bagley, S.J.; Till, J.; Abdalla, A.; Sangha, H.K.; Yee, S.S.; Freedman, J.; Black, T.A.; Hussain, J.; Binder, Z.A.; Brem, S.; et al. Association of Plasma Cell-Free DNA with Survival in Patients with IDH Wild-Type Glioblastoma. Neuro-Oncol. Adv. 2021, 3, vdab011. [Google Scholar] [CrossRef]
- Meng, Y.; Pople, C.B.; Suppiah, S.; Llinas, M.; Huang, Y.; Sahgal, A.; Perry, J.; Keith, J.; Davidson, B.; Hamani, C.; et al. MR-Guided Focused Ultrasound Liquid Biopsy Enriches Circulating Biomarkers in Patients with Brain Tumors. Neuro-Oncology 2021, 23, 1789–1797. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Liu, Z.; Zhu, Y.; Ma, L. Genome-Wide Methylation Analysis of Circulating Tumor DNA: A New Biomarker for Recurrent Glioblastom. Heliyon 2023, 9, e14339. [Google Scholar] [CrossRef]
- Lucero, R.; Zappulli, V.; Sammarco, A.; Murillo, O.D.; Cheah, P.S.; Srinivasan, S.; Tai, E.; Ting, D.T.; Wei, Z.; Roth, M.E.; et al. Glioma-Derived miRNA-Containing Extracellular Vesicles Induce Angiogenesis by Reprogramming Brain Endothelial Cells. Cell Rep. 2020, 30, 2065–2074.e4. [Google Scholar] [CrossRef]
- Drusco, A.; Bottoni, A.; Laganà, A.; Acunzo, M.; Fassan, M.; Cascione, L.; Antenucci, A.; Kumchala, P.; Vicentini, C.; Gardiman, M.P.; et al. A Differentially Expressed Set of microRNAs in Cerebro-Spinal Fluid (CSF) Can Diagnose CNS Malignancies. Oncotarget 2015, 6, 20829–20839. [Google Scholar] [CrossRef]
- Qu, K.; Lin, T.; Pang, Q.; Liu, T.; Wang, Z.; Tai, M.; Meng, F.; Zhang, J.; Wan, Y.; Mao, P.; et al. Extracellular miRNA-21 as a Novel Biomarker in Glioma: Evidence from Meta-Analysis, Clinical Validation and Experimental Investigations. Oncotarget 2016, 7, 33994–34010. [Google Scholar] [CrossRef]
- Ivo D’Urso, P.; Fernando D’Urso, O.; Damiano Gianfreda, C.; Mezzolla, V.; Storelli, C.; Marsigliante, S. miR-15b and miR-21 as Circulating Biomarkers for Diagnosis of Glioma. Curr. Genom. 2015, 16, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Liao, K.; Wu, X.; Huang, J.; Zhang, S.; Lu, X. Serum microRNA-128 as a Biomarker for Diagnosis of Glioma. Int. J. Clin. Exp. Med. 2015, 8, 456–463. [Google Scholar] [PubMed]
- Regazzo, G.; Terrenato, I.; Spagnuolo, M.; Carosi, M.; Cognetti, G.; Cicchillitti, L.; Sperati, F.; Villani, V.; Carapella, C.; Piaggio, G.; et al. A Restricted Signature of Serum miRNAs Distinguishes Glioblastoma from Lower Grade Gliomas. J. Exp. Clin. Cancer Res. 2016, 35, 124. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, L.; Song, Z.; Guo, C.; Zhu, J.; Li, Z.; Zhu, S. Potential Diagnostic and Prognostic Value of Plasma Circulating MicroRNA-182 in Human Glioma. Med. Sci. Monit. 2016, 22, 855–862. [Google Scholar] [CrossRef]
- Lai, N.; Wu, D.; Fang, X.; Lin, Y.; Chen, S.; Li, Z.; Xu, S. Serum microRNA-210 as a Potential Noninvasive Biomarker for the Diagnosis and Prognosis of Glioma. Br. J. Cancer 2015, 112, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, J.; Wang, Z.; Ruan, C.; Wang, L.; Guo, H. Serum miR-100 Is a Potential Biomarker for Detection and Outcome Prediction of Glioblastoma Patients. Cancer Biomark. 2019, 24, 43–49. [Google Scholar] [CrossRef]
- Morokoff, A.; Jones, J.; Nguyen, H.; Ma, C.; Lasocki, A.; Gaillard, F.; Bennett, I.; Luwor, R.; Stylli, S.; Paradiso, L.; et al. Serum microRNA Is a Biomarker for Post-Operative Monitoring in Glioma. J. Neurooncol. 2020, 149, 391–400. [Google Scholar] [CrossRef]
- Yue, X.; Lan, F.; Hu, M.; Pan, Q.; Wang, Q.; Wang, J. Downregulation of Serum microRNA-205 as a Potential Diagnostic and Prognostic Biomarker for Human Glioma. J. Neurosurg. 2016, 124, 122–128. [Google Scholar] [CrossRef]
- Swellam, M.; Ezz El Arab, L.; Al-Posttany, A.S.; Said, S.B. Clinical Impact of Circulating Oncogenic MiRNA-221 and MiRNA-222 in Glioblastoma Multiform. J. Neurooncol. 2019, 144, 545–551. [Google Scholar] [CrossRef]
- Shao, N.; Wang, L.; Xue, L.; Wang, R.; Lan, Q. Plasma miR-454-3p as a Potential Prognostic Indicator in Human Glioma. Neurol. Sci. 2015, 36, 309–313. [Google Scholar] [CrossRef]
- Stella, M.; Falzone, L.; Caponnetto, A.; Gattuso, G.; Barbagallo, C.; Battaglia, R.; Mirabella, F.; Broggi, G.; Altieri, R.; Certo, F.; et al. Serum Extracellular Vesicle-Derived circHIPK3 and circSMARCA5 Are Two Novel Diagnostic Biomarkers for Glioblastoma Multiforme. Pharmaceuticals 2021, 14, 618. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Zhong, L.; Ju, K.; Lu, T.; Lv, J.; Cao, H. Plasmatic circRNA Predicting the Occurrence of Human Glioblastoma. Cancer Manag. Res. 2020, 12, 2917–2923. [Google Scholar] [CrossRef]
- Amer, R.G.; Ezz El Arab, L.R.; Abd El Ghany, D.; Saad, A.S.; Bahie-Eldin, N.; Swellam, M. Prognostic Utility of lncRNAs (LINC00565 and LINC00641) as Molecular Markers in Glioblastoma Multiforme (GBM). J. Neurooncol. 2022, 158, 435–444. [Google Scholar] [CrossRef]
- Xia, D.; Gu, X. Plasmatic Exosome-Derived circRNAs Panel Act as Fingerprint for Glioblastoma. Aging 2021, 13, 19575–19586. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Xu, X.-K.; Li, J.-L.; Kong, K.-K.; Li, H.; Chen, C.; He, J.; Wang, F.; Li, P.; Ge, X.-S.; et al. MALAT1 Is a Prognostic Factor in Glioblastoma Multiforme and Induces Chemoresistance to Temozolomide through Suppressing miR-203 and Promoting Thymidylate Synthase Expression. Oncotarget 2017, 8, 22783–22799. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, C.; Neuhaus, N.; Dahlmann, M.; Kessler, A.; Kobelt, D.; Herrmann, P.; Eyrich, M.; Freitag, B.; Linsenmann, T.; Monoranu, C.; et al. Circulating MACC1 Transcripts in Glioblastoma Patients Predict Prognosis and Treatment Response. Cancers 2019, 11, 825. [Google Scholar] [CrossRef]
- Masood, A.B.; Batool, S.; Bhatti, S.N.; Ali, A.; Valko, M.; Jomova, K.; Kuca, K. Plasma PD-L1 as a Biomarker in the Clinical Management of Glioblastoma Multiforme—A Retrospective Cohort Study. Front. Immunol. 2023, 14, 1202098. [Google Scholar] [CrossRef]
- Tsvetkov, P.O.; Eyraud, R.; Ayache, S.; Bougaev, A.A.; Malesinski, S.; Benazha, H.; Gorokhova, S.; Buffat, C.; Dehais, C.; Sanson, M.; et al. An AI-Powered Blood Test to Detect Cancer Using NanoDSF. Cancers 2021, 13, 1294. [Google Scholar] [CrossRef]
- Soler, D.C.; Young, A.B.; Cooper, K.D.; Kerstetter-Fogle, A.; Barnholtz-Sloan, J.S.; Gittleman, H.; McCormick, T.S.; Sloan, A.E. The Ratio of HLA-DR and VNN2+ Expression on CD14+ Myeloid Derived Suppressor Cells Can Distinguish Glioblastoma from Radiation Necrosis Patients. J. Neurooncol. 2017, 134, 189–196. [Google Scholar] [CrossRef]
- Ghorbani, A.; Chatanaka, M.K.; Avery, L.M.; Wang, M.; Brown, J.; Cohen, R.; Gorham, T.; Misaghian, S.; Padmanabhan, N.; Romero, D.; et al. Glial Fibrillary Acidic Protein, Neurofilament Light, Matrix Metalloprotease 3 and Fatty Acid Binding Protein 4 as Non-Invasive Brain Tumor Biomarkers. Clin. Proteom. 2024, 21, 41. [Google Scholar] [CrossRef]
- Björkblom, B.; Wibom, C.; Jonsson, P.; Mörén, L.; Andersson, U.; Johannesen, T.B.; Langseth, H.; Antti, H.; Melin, B. Metabolomic Screening of Pre-Diagnostic Serum Samples Identifies Association between α- and γ-Tocopherols and Glioblastoma Risk. Oncotarget 2016, 7, 37043–37053. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Chen, Y.; Meng, Z.; Chu, Z. The Causal Relationship between CSF Metabolites and GBM: A Two-Sample Mendelian Randomization Analysis. BMC Cancer 2024, 24, 1119. [Google Scholar] [CrossRef]
- Zhao, H.; Heimberger, A.B.; Lu, Z.; Wu, X.; Hodges, T.R.; Song, R.; Shen, J. Metabolomics Profiling in Plasma Samples from Glioma Patients Correlates with Tumor Phenotypes. Oncotarget 2016, 7, 20486–20495. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Song, R.; Hodges, T.R.; Heimberger, A.B.; Zhao, H. Identification of Metabolites in Plasma for Predicting Survival in Glioblastoma. Mol. Carcinog. 2018, 57, 1078–1084. [Google Scholar] [CrossRef]
- Liu, Y.A.; Aboud, O.; Dahabiyeh, L.A.; Bloch, O.; Fiehn, O. Metabolomic Characterization of Human Glioblastomas and Patient Plasma: A Pilot Study. F1000Research 2024, 13, 98. [Google Scholar] [CrossRef]
- Muller Bark, J.; Karpe, A.V.; Doecke, J.D.; Leo, P.; Jeffree, R.L.; Chua, B.; Day, B.W.; Beale, D.J.; Punyadeera, C. A Pilot Study: Metabolic Profiling of Plasma and Saliva Samples from Newly Diagnosed Glioblastoma Patients. Cancer Med. 2023, 12, 11427–11437. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Ji, N.; Wang, G.; Zhang, Y.; Song, H.; Yuan, Y.; Yang, C.; Jin, Y.; Zhang, Z.; Zhang, L.; et al. Metabolic Detection of Malignant Brain Gliomas through Plasma Lipidomic Analysis and Support Vector Machine-Based Machine Learning. EBioMedicine 2022, 81, 104097. [Google Scholar] [CrossRef]
- Soylemez, B.; Bulut, Z.; Şahin-Bölükbaşı, S. Investigating the Potential of Lipids for Use as Biomarkers for Glioblastoma via an Untargeted Lipidomics Approach. J. Korean Neurosurg. Soc. 2023, 66, 133–143. [Google Scholar] [CrossRef]
- Sareen, H.; Garrett, C.; Lynch, D.; Powter, B.; Brungs, D.; Cooper, A.; Po, J.; Koh, E.-S.; Vessey, J.Y.; McKechnie, S.; et al. The Role of Liquid Biopsies in Detecting Molecular Tumor Biomarkers in Brain Cancer Patients. Cancers 2020, 12, 1831. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.; Lin, H.; Zhu, Y.; Huang, D.; Lai, M.; Xi, X.; Huang, J.; Zhang, W.; Zhong, T. Research Progress of CTC, ctDNA, and EVs in Cancer Liquid Biopsy. Front. Oncol. 2024, 14, 1303335. [Google Scholar] [CrossRef]
- Śledzińska, P.; Bebyn, M.G.; Furtak, J.; Kowalewski, J.; Lewandowska, M.A. Prognostic and Predictive Biomarkers in Gliomas. Int. J. Mol. Sci. 2021, 22, 10373. [Google Scholar] [CrossRef]
- Müller Bark, J.; Kulasinghe, A.; Chua, B.; Day, B.W.; Punyadeera, C. Circulating Biomarkers in Patients with Glioblastoma. Br. J. Cancer 2020, 122, 295–305. [Google Scholar] [CrossRef]
- Hassan, S. Applications of RNA from Circulating Tumor Cells. Front. Biosci. 2020, 25, 874–892. [Google Scholar] [CrossRef] [PubMed]
- Lessi, F.; Morelli, M.; Franceschi, S.; Aretini, P.; Menicagli, M.; Marranci, A.; Pasqualetti, F.; Gambacciani, C.; Pieri, F.; Grimod, G.; et al. Innovative Approach to Isolate and Characterize Glioblastoma Circulating Tumor Cells and Correlation with Tumor Mutational Status. Int. J. Mol. Sci. 2023, 24, 10147. [Google Scholar] [CrossRef]
- Adamczyk, L.A.; Williams, H.; Frankow, A.; Ellis, H.P.; Haynes, H.R.; Perks, C.; Holly, J.M.P.; Kurian, K.M. Current Understanding of Circulating Tumor Cells–Potential Value in Malignancies of the Central Nervous System. Front. Neurol. 2015, 6, 174. [Google Scholar] [CrossRef]
- Indira Chandran, V.; Gopala, S.; Venkat, E.H.; Kjolby, M.; Nejsum, P. Extracellular Vesicles in Glioblastoma: A Challenge and an Opportunity. Npj Precis. Oncol. 2024, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Del Bene, M.; Osti, D.; Faletti, S.; Beznoussenko, G.V.; DiMeco, F.; Pelicci, G. Extracellular Vesicles: The Key for Precision Medicine in Glioblastoma. Neuro-Oncology 2022, 24, 184–196. [Google Scholar] [CrossRef]
- Hu, C.; Jiang, W.; Lv, M.; Fan, S.; Lu, Y.; Wu, Q.; Pi, J. Potentiality of Exosomal Proteins as Novel Cancer Biomarkers for Liquid Biopsy. Front. Immunol. 2022, 13, 792046. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Li, Y.; Wang, M.; Gu, J.; Xu, W.; Cai, H.; Fang, X.; Zhang, X. Exosomes as a New Frontier of Cancer Liquid Biopsy. Mol. Cancer 2022, 21, 56. [Google Scholar] [CrossRef]
- Yin, K.; Liu, X. CircMMP1 Promotes the Progression of Glioma through miR-433/HMGB3 Axis in Vitro and in Vivo. IUBMB Life 2020, 72, 2508–2524. [Google Scholar] [CrossRef]
- Khristov, V.; Lin, A.; Freedman, Z.; Staub, J.; Shenoy, G.; Mrowczynski, O.; Rizk, E.; Zacharia, B.; Connor, J. Tumor-Derived Biomarkers in Liquid Biopsy of Glioblastoma. World Neurosurg. 2023, 170, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Mafi, A.; Rahmati, A.; Babaei Aghdam, Z.; Salami, R.; Salami, M.; Vakili, O.; Aghadavod, E. Recent Insights into the microRNA-Dependent Modulation of Gliomas from Pathogenesis to Diagnosis and Treatment. Cell. Mol. Biol. Lett. 2022, 27, 65. [Google Scholar] [CrossRef]
- Ali, H.; Harting, R.; De Vries, R.; Ali, M.; Wurdinger, T.; Best, M.G. Blood-Based Biomarkers for Glioma in the Context of Gliomagenesis: A Systematic Review. Front. Oncol. 2021, 11, 665235. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Vishnu, V.Y. Lumbar Puncture: Indications, Challenges and Recent Advances. Neurology 2021, 17, 23. [Google Scholar] [CrossRef]
- Qiu, S.; Cai, Y.; Yao, H.; Lin, C.; Xie, Y.; Tang, S.; Zhang, A. Small Molecule Metabolites: Discovery of Biomarkers and Therapeutic Targets. Signal Transduct. Target. Ther. 2023, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Adhit, K.K.; Wanjari, A.; Menon, S.; Siddhaarth, K. Liquid Biopsy: An Evolving Paradigm for Non-Invasive Disease Diagnosis and Monitoring in Medicine. Cureus 2023, 15, e50176. [Google Scholar] [CrossRef]
- Skouras, P.; Markouli, M.; Kalamatianos, T.; Stranjalis, G.; Korkolopoulou, P.; Piperi, C. Advances on Liquid Biopsy Analysis for Glioma Diagnosis. Biomedicines 2023, 11, 2371. [Google Scholar] [CrossRef]
- Donnelly, D.; Aung, P.P.; Jour, G. The “-OMICS” Facet of Melanoma: Heterogeneity of Genomic, Proteomic and Metabolomic Biomarkers. Semin. Cancer Biol. 2019, 59, 165–174. [Google Scholar] [CrossRef]
- Yaghoubi Naei, V.; Bordhan, P.; Mirakhorli, F.; Khorrami, M.; Shrestha, J.; Nazari, H.; Kulasinghe, A.; Ebrahimi Warkiani, M. Advances in Novel Strategies for Isolation, Characterization, and Analysis of CTCs and ctDNA. Ther. Adv. Med. Oncol. 2023, 15, 17588359231192401. [Google Scholar] [CrossRef]
- Fujita, Y.; Nunez-Rubiano, L.; Dono, A.; Bellman, A.; Shah, M.; Rodriguez, J.C.; Putluri, V.; Kamal, A.H.M.; Putluri, N.; Riascos, R.F.; et al. IDH1 p.R132H ctDNA and D-2-Hydroxyglutarate as CSF Biomarkers in Patients with IDH-Mutant Gliomas. J. Neurooncol. 2022, 159, 261–270. [Google Scholar] [CrossRef]
- Saenz-Antoñanzas, A.; Auzmendi-Iriarte, J.; Carrasco-Garcia, E.; Moreno-Cugnon, L.; Ruiz, I.; Villanua, J.; Egaña, L.; Otaegui, D.; Samprón, N.; Matheu, A. Liquid Biopsy in Glioblastoma: Opportunities, Applications and Challenges. Cancers 2019, 11, 950. [Google Scholar] [CrossRef] [PubMed]
- Sievänen, H.; Kari, J.; Eskola, V.; Huurre, A.; Soukka, H.; Palmu, S. Incidence of Traumatic Lumbar Punctures in Adults: The Impact of a Patient’s First Procedure. Clin. Med. 2023, 23, 31–37. [Google Scholar] [CrossRef]
- Kim, K.T. Lumbar Puncture: Considerations, Procedure, and Complications. Encephalitis 2022, 2, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.W. Complications of Lumbar Puncture. Neurol. Clin. 1998, 16, 83–105. [Google Scholar] [CrossRef]
- Katzendobler, S.; Do, A.; Weller, J.; Dorostkar, M.M.; Albert, N.L.; Forbrig, R.; Niyazi, M.; Egensperger, R.; Thon, N.; Tonn, J.C.; et al. Diagnostic Yield and Complication Rate of Stereotactic Biopsies in Precision Medicine of Gliomas. Front. Neurol. 2022, 13, 822362. [Google Scholar] [CrossRef] [PubMed]
- Sabedot, T.S.; Malta, T.M.; Snyder, J.; Nelson, K.; Wells, M.; deCarvalho, A.C.; Mukherjee, A.; Chitale, D.A.; Mosella, M.S.; Sokolov, A.; et al. A Serum-Based DNA Methylation Assay Provides Accurate Detection of Glioma. Neuro-Oncology 2021, 23, 1494–1508. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, X.; Xie, P.; Zhang, W. Liquid Biopsy: An Arsenal for Tumour Screening and Early Diagnosis. Cancer Treat. Rev. 2024, 129, 102774. [Google Scholar] [CrossRef]
- Gatto, L.; Franceschi, E.; Di Nunno, V.; Tosoni, A.; Lodi, R.; Brandes, A.A. Liquid Biopsy in Glioblastoma Management: From Current Research to Future Perspectives. Oncologist 2021, 26, 865–878. [Google Scholar] [CrossRef]
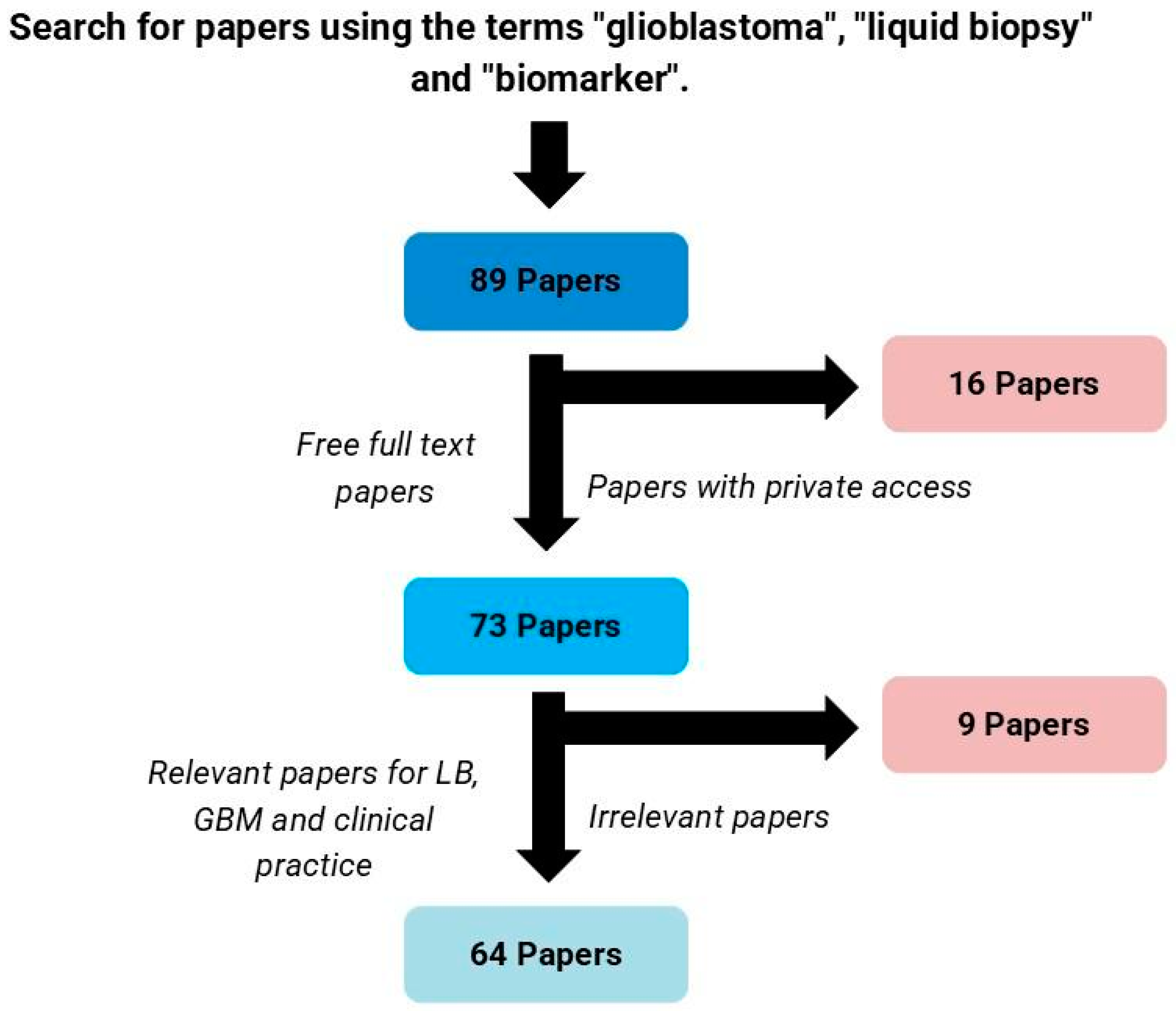
| Authors | Biological Matrix | GBM Sample Size (n) | Biomarker | Method (Platform of Analysis/Assay) | Ref. |
|---|---|---|---|---|---|
| Gao et al., 2016 | Blood | 11 ▲ | CTC (enumeration) | SE-iFISH (Olympus BX-53) | [16] |
| Lynch et al., 2020 | Blood and cell line | 13 ▲ | CTC (enumeration) | Immunocytostaining (CellCelector), flow cytometry (BD FACS Canto II) | [17] |
| Bark et al., 2021 | Blood | 20 (18 GBM IDH-WT) | CTC (enumeration) | Immunofluorescence (Zeiss Axio Imager Z2) | [18] |
| Kolostova et al., 2021 | Blood | 18 ▲ | CTC (DNA) | NGS (GeneReader) | [19] |
| Yang et al., 2017 | Plasma and tumor tissue | 4 ▲ | EV (RNA, protein), tissue RNA | Microaaray (NA), WB (NA) | [20] |
| Hallal et al., 2020a | Plasma | 41 (24 GBM IDH-WT) | EV (protein) | SWATH-MS (TripleTOF®6600/EskpertTM NanoLC 425) | [21] |
| Dobra et al., 2020 | Serum | 24 ▲ | EV (protein) | LC-MS (Q Exactive Plus) | [22] |
| Cilibrasi et al., 2022 | Plasma | 15 * | EV (protein) | Mass spectrometry (Q Exactive/Dionex Ultimate 3000 RSLCnano) | [23] |
| Hallal et al., 2024 | Urine | 24 * | EV (protein) | Liquid chromatography (Ultimate 3000) and mass spectrometry (Q-Exactive HFX3) | [24] |
| Brahmer et al., 2023 | Serum and cell line | 9 ▲ | EV (protein) | Multiplex bead-based flow cytometry (MACSPlex Neuro EV in Attune NxT) | [25] |
| Dufrusine et al., 2023 | Plasma, serum, tumor tissue, and cell line | 17 ▲ | EV (protein), protein | WB (NA), ELISA (KE00155), IHC (NA) | [26] |
| Lennartz et al., 2023 | Plasma, serum, and tumor cell | 99 * | EV (protein), protein | ELISA (Hsp70-exo, R&D Systems DuoSet), Multiparameter Flow Cytometry (BD FACSCalibur) | [27] |
| Werner et al., 2021 | Serum and cell line | 34 ▲ | EVs, protein | ELISA (R&D Systems DuoSet), WB (NA), Flow Cytometry (BD FACSCalibur™) | [28] |
| Aibaidula et al., 2023 | Plasma | 20 * | EV (surface protein) | Spectral flow cytometry (Cytek Aurora) | [29] |
| Ricklefs et al., 2024 | Plasma | 101 ▲ | EV (quantification, surface protein) | Nanoparticle tracking analysis (NanoSight LM14), and imaging flow cytometry (IFCM) (ImageStreamX Mark II Imaging) | [30] |
| Figueroa et al., 2017 | CSF and tumor tissue | 55 ▲ | EV (RNA) | qRT-PCR (ABI Prism 7500) | [31] |
| Hallal et al., 2020b | Surgical aspirate and plasma | 17 (12 GBM IDH-WT) | EV (RNA) | NGS (NextSeq 500) | [32] |
| Yekula et al., 2023 | Serum | 14 ▲ | EV (RNA) | NGS—RNA-seq (NextSeq 500) | [33] |
| Uziel et al., 2024 | Serum | 61 (60 GBM IDH-WT) | EV (RNA) | qRT-PCR (Step One) | [34] |
| Manda et al., 2018 | Serum and tumor tissue | 73 ▲ | EV (RNA), tissue RNA | End-point RT-PCR (Master Cycler Pro S) | [35] |
| Akers et al., 2017 | Tumor tissue andCSF (EV and total) | 111 ▲ | EV (miRNA), miRNA | TaqMan OpenArray® Human MicroRNA Panel (Taqman OpenArray), qRT-PCR (CFX96) | [36] |
| Shi et al., 2015 | Serum and CSF | 45 ▲ | EVs (miRNA) | qRT-PCR (NA) | [37] |
| Rosas-Alonso et al., 2024 | Plasma and FFPE tumor tissue | 50 * | EV (DNA), tissue DNA | Quantitative methylation-specific PCR (qMSP) (NA) | [38] |
| Wang et al., 2015 | CSF | 35 (9 GBM IDH-WT) | ctDNA | whole-exome sequencing (WES), SafeSeqS Pipeline | [39] |
| Miller et al., 2019 | CSF, plasma, and tumor tissue | 46 (44 GBM IDH-WT) | ctDNA | NGS (MSK-IMPACT) | [40] |
| Juratli et al., 2018 | Tumor tissue, CSF, and Plasma | 38 * | ctDNA | NGS (Ion Torrent PGM), ddPCR (QX200) | [41] |
| Zhao et al., 2020 | Tumor tissue and CSF | 4 * | ctDNA | NGS (Ion Proton) | [42] |
| Wang, Q. et al., 2023 | CSF and tumor tissue | 27 (12 GBM IDH-WT) | cfDNA | NGS (NovaSeq 6000 system), IHC (NA) | [43] |
| Piccioni et al., 2019 | Plasma | 222 ▲ | ctDNA | NGS (Guardant360) | [44] |
| Mattos-Arruda et al., 2015 | CSF, plasma, and tumor tissue | 4 ▲ | ctDNA, tissue DNA | NGS (HiSeq 2000), ddPCR (QX200) | [45] |
| Bagley et al., 2021 | Plasma | 62 * | cfDNA (quantification) | qPCR (ViiA 7) | [46] |
| Meng et al., 2021 | Plasma | 9 (8 GBM IDH-WT) | cfDNA (quantification and methylation profiling), EV, protein (quantification) | MethylationEPIC 850k array, ddPCR (QX200), ELISA (EZHS100B-33K) | [47] |
| Dai et al., 2023 | CSF and tumor tissue | 4 ▲ + 109 in sillico (CGGA) | RNA | RNA-seq (NovaSeq and NovaSeq 6000) | [48] |
| Lucero et al., 2020 | Human brain endothelial cells (HBMVECs) | Not applicable | miRNA | DNA methylation profiling (Human 450K Infinium Methylation BeadChip) and RNA-seq (NextSeq 500 and HiSeq 4000), histoepigenetic analyses (NA), | [49] |
| Drusco et al., 2015 | CSF | 4 ▲ | miRNA | Microarray (nCounter NanoString) | [50] |
| Qu et al., 2016 | CSF and tumor tissue | 35 ▲ | miRNA | qRT-PCR (NA) | [51] |
| D’Urso et al., 2015 | Serum and plasma | 16 ▲ | miRNA | qRT-PCR (7500), microarray (Affymetrix 428) | [52] |
| Sun et al., 2015 | Serum | 61 ▲ | miRNA | qRT-PCR (NA) | [53] |
| Regazzo et al., 2016 | Serum | 10 ▲ | miRNA | qRT-PCR (ABI PRISM 7900) | [54] |
| Xiao et al., 2016 | Plasma | 39 ▲ | miRNA | qRT-PCR (ABI PRISM 7300) | [55] |
| Lai et al., 2015 | Serum | 32 ▲ | miRNA | qRT-PCR (DNA Engine Opticon 2) | [56] |
| Zhang et al., 2019 | Serum | 95 (67 GBM IDH-WT) | miRNA | qRT-PCR (ABI PRISM 7900) | [57] |
| Morokoff et al., 2020 | Serum | 44 (29 GBM IDH-WT) | miRNA | Micro array (nCounter NanoString), ddPCR (NA) | [58] |
| Yue et al., 2016 | Serum | 27 ▲ | miRNA | qRT-PCR (ABI PRISM 7900) | [59] |
| Swellam et al., 2019 | Serum | 20 ▲ | miRNA | qRT-PCR (Max3005P) | [60] |
| Shao et al., 2015 | Plasma | 22 ▲ | miRNA | qRT-PCR (ABI PRISM 7500) | [61] |
| Stella et al., 2021 | Serum and tumor tissue | 23 ▲ | circRNA | ddPCR (QX200), qRT-PCR (ABI PRISM 7900 HT) | [62] |
| Chen et al., 2020 | Plasma | 100 ▲ | circRNA | qRT-PCR (NA) | [63] |
| Xia et al., 2021 | Plasma | 120 ▲ | circRNAs | qRT-PCR, circRNA microarray analysis | [65] |
| Amer et al., 2022 | Serum | 35 ▲ | lncRNA | qRT-PCR (Max 3005P) | [64] |
| Chen et al., 2017 | Serum | 140 ▲ | lncRNA | qRT-PCR (ABI PRISM 7500) | [66] |
| Hagemann et al., 2019 | Plasma | 45 (36 GBM IDH-WT) | mRNA | qRT-PCR (LightCycler 480) | [67] |
| Masood et al., 2023 | Plasma | 64 ▲ | mRNA, protein | qRT-PCR (NA), ELISA (Human PD-L1 Platinum) | [68] |
| Tsvetkov et al., 2021 | Plasma | 84 (19 IDH-WT) | Protein | nanoDSF Prometheus NT.Plex instrument (Nanotemper) | [69] |
| Soler et al., 2017 | Blood | 18 ▲ | PBMC (surface protein) | Flow cytometry (BD C6) | [70] |
| Ghorbani et al., 2024 | Plasma | 67 * | Protein | MSD® Electroluminescence multiplexed immunoassays | [71] |
| Björkblom et al., 2016 | Serum | 110 ▲ | Metabolite | Mass spectrometry/chromatography (Leco Pegasus 4D TOFMS/Agilent 6890) | [72] |
| Bao et al., 2024 | CSF | 91 | Metabolite | Mendelian randomization (GWAS-based) | [73] |
| Zhao et al., 2016 | Plasma | 18 ▲ | Metabolite | LC-QQQ-MS (Sciex 5500 QTRAP/Agilent 1200) | [74] |
| Shen et al., 2018 | Plasma | 159 (105 GBM IDH-WT) | Metabolite | Mass spectrometry (NA) | [75] |
| Liu et al., 2024 | Plasma and tumor tissue | 15 * | Metabolite | Mass spectrometry (1290 UHPLC/Sciex TripleTOF 6600 and 1290 UHPLC/Agilent 6530 QTOF and 6550 QTOF mass spectrometer) | [76] |
| Bark et al., 2023 | Plasma and saliva | 21 (20 GBM IDH-WT) | Metabolite, lipid | LC-QqQ-MS (Agilent 6470/Infinity II Flex UHPLC), LC-QTOF-MS (Agilent 6546/Infinity II Flex UHPLC) | [77] |
| Zhou et al., 2022 | Serum | 377 ▲ (139 in the validation cohort) | Lipid | Liquid chromatography/mass spectrometry (Ultimate 3000/Q-Exactive MS) | [78] |
| Soylemez et al., 2023 | Blood | 14 ▲ | Lipid | Liquid chromatography/mass spectrometry (6530 Accurate-Mass Q-TOF LC/MS) | [79] |
| Biomarker | n | Source | Sensitivity | Specificity | Notes | Ref. |
|---|---|---|---|---|---|---|
| EV (RNA) | 55 (23 GBM) | CSF | 61% | 98% | Compared to EGFRvIII status in tumor tissue. | [31] |
| EV (RNA) | 73 | Serum | 81.6% | 79.3% | Compared to EGFRvIII status in tumor tissue. | [35] |
| EV (miRNA) | 111 | CSF | 67% (cisternal); 28% (lumbar) | 80% (cisternal); 95% (lumbar) | 9-miRNA panel compared to tumor presence. | [36] |
| EV (DNA) | 50 | Plasma | 63.2% | 92.6% | Compared to tumor tissue MGMT methylation. | [38] |
| ctDNA | 35 (9 GBM) | CSF | 95% | Not reported | Concordance of mutations between CSF and matched tumor tissue. | [39] |
| ctDNA | 46 | CSF | 49% | Not reported | Compared to matched tumor tissue. | [40] |
| ctDNA | 38 | CSF and plasma | 92,1% (CSF); 7,9% (plasma) | 100% (CSF) | Compared to tumor tissue TERT promoter status. | [41] |
| ctDNA | 4 | CSF | 82% | Not reported | Compared to tumor tissue; reflected key molecular alterations. | [42] |
| ctDNA | 419 (222 GBM) | Plasma | 55% | Not reported | Compared plasma ctDNA to matched tumor sequencing. | [44] |
| ctDNA | 7 (4 GBM) | CSF and plasma | 58% (CSF); 0% (plasma) | Not reported | Compared to tumor tissue. CSF better reflects mutations in CNS-restricted disease than plasma. | [45] |
| Authors | Biomarker | Main Findings | Potential Clinical Application | Ref. |
|---|---|---|---|---|
| Gao et al., 2016 | CTC | CTCs helped monitor treatment response and differentiate radionecrosis from glioma recurrence. | Monitoring | [16] |
| Lynch et al., 2020 | CTC | The GLAST survey can complement GFAP probing to improve GBM-CTC identification. | Diagnosis | [17] |
| Bark et al., 2021 | CTC | Characterization for GFAP, vimentin protein expression and EGFR amplification. | Diagnosis | [18] |
| Kolostova et al., 2021 | CTC | CTCs showed high concordance with primary tumor samples and more mutations were detected. | Diagnosis and monitoring | [19] |
| Yang et al., 2017 | Exosomes | Detection of differentially expressed genes in blood exosomes of primary and recurrent GBM; increased expression of DNM3, p65 and CD117 and decreased expression of PTEN and p53 in primary tumors; increased expression of DNM3 and p65 in recurrent tumors. | Diagnosis and prognosis | [20] |
| Brahmer et al., 2023 | EV | EVs marker profiles were significantly increased in GBM compared to healthy controls. | Diagnosis | [25] |
| Dobra et al., 2020 | EV, protein | sEVs were more effective in discriminating between patient groups than whole serum. | Diagnosis | [22] |
| Aibaidula et al., 2023 | EV | Identification of a distinct phenotype (CD9+CD81+ and CD9+CD63+CD81+) and increased CD9+CD11b+CD45 phenotype of extracellular vesicles originating from nonneoplastic cells in the plasma of patients with GBM. | Diagnosis | [29] |
| Stella et al., 2021 | EV | circSMARCA5 and circHIPK3 were significantly decreased in the sera EVs of GBM patients compared with healthy controls. | Diagnosis | [62] |
| Cilibrasi et al., 2022 | EV | Inflammatory biomarker signature composed of several proteins (VWF, FCGBP, C3, PROS1, SERPINA1) present in EVs from GBM patients. | Diagnosis and monitoring | [23] |
| Hallal et al., 2020a | EV | Identification of 4054 proteins in plasma EVs. Protein profiles of EVs grouped according to glioma subtype and histological grade. | Diagnosis and monitoring | [21] |
| Hallal et al., 2020b | EV | miR-486-3p as well as piR_016658, 016659, and 020829 piRNAs were differentially expressed in GBM surgical aspirate and serum EVs. | Diagnosis and monitoring | [32] |
| Hallal et al., 2024 | EV | GBM-specific proteomic signatures were determined, and putative urinary EV biomarkers corresponding to diagnosis (KRT19, RPS2, RPL18, RPL28, RPL7A), tumor burden (BCAM, ITGA3, ITM2B), and recurrence (GRN, ITM2B) were identified. | Diagnosis and monitoring | [24] |
| Manda et al., 2018 | EV | EGFRvIII amplification in tumor tissues and exosomes correlated with poor survival. | Diagnosis and prognosis | [35] |
| Shi et al., 2015 | EV | Exosomal miR-21 levels were increased in the CSF of glioma patients compared to non-glioma (brain-trauma) patients. Higher levels of tissue miR-21 are indicative of poor prognosis in the CGGA cohort. | Diagnosis and prognosis | [37] |
| Ricklefs et al., 2024 | EV | Plasma EV concentration is increased in glioblastoma patients, and high EV levels are an independent negative prognostic parameter. | Diagnosis, prognosis and monitoring | [30] |
| Uziel et al., 2024 | EV | hTERT mRNA transcript levels from EVs can be measured in serum from GBM patients. Preoperative measurements correlated with tumor volume and disease course. | Monitoring | [34] |
| Yekula et al., 2023 | EV | Stratification of patients with recurrent GBM/EGFR-amplified after dacomitinib treatment; detection of a unique responder signature in the serum EV transcriptome through the biomarkers DNMT3A, ZNF35, and LAMTOR2. | Monitoring, patient stratification | [33] |
| Rosas-Alonso et al., 2024 | EV | Detection of methylated MGMT in sEV-DNA with sensitivity and specificity of 87.5% and 90%, respectively. | Prognosis and monitoring | [38] |
| Lennartz et al., 2023 | EV, protein | Increased Hsp70 protein levels in GBM patients associated with overall survival in different glioma subtypes. | Diagnosis and prognosis | [27] |
| Werner et al., 2021 | EVs, protein | Elevated levels of Hsp70 showed in serum from GBM patients, suggesting its use as a tumor-specific biomarker. | Diagnosis and treatment monitoring | [28] |
| Zhao et al., 2020 | ctDNA | Mutations in CSF ctDNA showed high concordance with tumor DNA, highlighting mutations in RB1 and EGFR. | Diagnosis | [42] |
| Wang et al., 2015 | ctDNA | Identified ctDNA in CSF from 9 GBM patients, highlighting the use of cfDNA for molecular characterization and tumor progression monitoring. | Diagnosis | [39] |
| Xia et al., 2021 | circRNAs | Identified circRNAs (hsa_circ_0055202, hsa_circ_0074920, hsa_circ_0043722) expressed in an elevated state in GBM, validated as potential diagnostic biomarkers with high specificity. | Diagnosis | [65] |
| Dai et al., 2023 | ctDNA | Identification of 8 differentially expressed and methylated hub genes (COMMD1, C1orf226, CH3L2, FLRT2, ETV1, NKD1, GNB5, and NTRK3) to build diagnostic and prognostic models of recurrent GBM with high accuracy. | Diagnosis and prognosis | [48] |
| Juratli et al., 2018 | ctDNA | The TERTp mutation was detected in CSF ctDNA with high sensitivity and specificity and associated with patient survival. | Diagnosis and prognosis | [41] |
| Wang Q. et al., 2023 | cfDNA | High concordance between cfDNA in CSF and tumor tissue DNA in GBM, correlating ctDNA levels with Ki67 expression. | Diagnosis and prognosis | [43] |
| Mattos-Arruda et al., 2015 | ctDNA | CSF-derived ctDNA was more representative of brain tumor genomic alterations than plasma ctDNA, allowing identification of somatic mutations and reflecting changes in tumor burden over time. | Diagnosis and monitoring | [45] |
| Miller et al., 2019 | ctDNA | Genomic characterization in CSF reflected tumor biopsies, allowing monitoring of glioma genome evolution. CtDNA positivity was correlated to shorter OS. | Diagnosis, monitoring and prognosis | [40] |
| Piccioni et al., 2019 | ctDNA | Alterations in plasma cfDNA were detected in 55% of GBM patients. | Diagnosis, monitoring and stratification | [44] |
| Bagley et al., 2021 | cfDNA | High preoperative cfDNA concentration associated with shorter progression-free survival and overall survival in patients with GBM. | Prognosis | [46] |
| Meng et al., 2021 | cfDNA, EV, protein | MRgFUS enriches the signal of brain-derived circulating biomarkers including cfDNA, EVs, and S100b after sonication. | Diagnosis | [47] |
| Lucero et al., 2020 | miRNA | Identification of 8 candidate miRNAs (miR-16-2-3p, miR-148a-3p, miR-182-5p, miR-9-5p, miR-9-3p, miR-22-3p, miR-186-5p, miR-378e) related to angiogenesis. | Prognosis | [49] |
| Akers et al., 2017 | miRNA | Profiling of miRNAs derived from EVs from tumor tissue and CSF of patients with GBM, identifying a tumor signature composed of 9 miRNAs (miR-21, -218, -193b, -331, -374a, miR-548c, -520f, 27b, and 130b). Sensitivity and specificity for GBM detection were 67% and 80% for cisternal CSF, and 28% and 95% for lumbar CSF. | Diagnosis | [36] |
| Drusco et al., 2015 | miRNA | Identification of miRNA signatures (miR-451, -711, -935, -223 and -125b) in CSF capable of distinguishing different types of brain tumors. | Diagnosis | [50] |
| D’Urso et al., 2015 | miRNA | Increased levels of miR-21 and miR-15b in serum EVs of glioma patients, while downregulation of miR-16 distinguished GBM from low grade and anaplastic gliomas. | Diagnosis | [52] |
| Regazzo et al., 2016 | miRNA | Decreased levels of miR-497 and miR-125b in the serum of glioma patients, with GBM having significant lower levels (AUC = 0.87 for miR-497 and 0.75 for miR-125b). | Diagnosis | [54] |
| Sun et al., 2015 | miRNA | Significant decrease of serum miR-128 in glioma patients compared to meningioma patients and healthy controls. | Diagnosis | [53] |
| Lai et al., 2015 | miRNA | Significantly increased miRNA-210 expression in GBM patients compared with healthy controls correlated with tumor grade and worse patient outcome. | Diagnosis and prognosis | [56] |
| Qu et al., 2016 | miRNA | Significant increase of miR-21 in the CSF and tumor tissue of glioma patients compared to healthy controls. MiR-21 from CSF outperformed its tumor tissue counterpart as a prognostic marker. | Diagnosis and prognosis | [51] |
| Shao et al., 2015 | miRNA | The levels of miR-454-3p in plasma in glioma patients were significantly higher than that from healthy controls (AUC= 0.9063). Increased miR-454-3p levels also correlated with higher tumor grades and poorer prognosis. | Diagnosis and prognosis | [61] |
| Swellam et al., 2019 | miRNA | Significant increase in the levels of miR-221 and miR-222 observed in the serum of GBM patients compared to healthy controls. Higher levels of miR-221 and miR-222 were indicative of poor OS. | Diagnosis and prognosis | [60] |
| Xiao et al., 2016 | miRNA | Significant higher levels of miR-182 in glioma patients compared to healthy controls (AUC = 0.778), specially in higher grade gliomas (AUC = 0.815). Higher miR-182 levels also correlated with shorter OS and PFS. | Diagnosis and prognosis | [55] |
| Yue et al., 2016 | miRNA | Significant decrease of serum miR-205 in glioma patients compared to healthy individuals and other brain tumors. MiR-205 levels were inversely proportional to the tumor grade (higher grade have lower miR-205). Low miR-205 levels were an independent factor associated to poor OS. | Diagnosis and prognosis | [59] |
| Zhang et al., 2019 | miRNA | Significantly decreased serum levels of miR-100 in GBM patients compared with healthy controls, with an increase after treatment. Low miR-100 expression was associated with unfavorable clinicopathological features and shorter survival. | Diagnosis and prognosis | [57] |
| Morokoff et al., 2020 | miRNA | Highly accurate 9-serum miRNA signature (miR320e, miR-223, miR-16-5p, miR-484, miR520a, miR-532, miR-630, miR651, miR-761) identification in gliomas. Observed dynamic changes in specific miRNAs correlating with tumor volume over long-term follow-up. | Monitoring | [58] |
| Chen et al., 2020 | cirRNA | Significant increase in the levels of circFOXO3, circ_0029426, and circ-SHPRH were found in the serum of GBM patients compared to healthy controls. | Diagnosis | [63] |
| Amer et al., 2022 | lncRNA | Significant increase in lncRNA565 and lncRNA641 expression in GBM patients compared with healthy controls, correlated with clinicopathological data and unfavorable survival pattern. | Diagnosis and prognosis | [64] |
| Chen et al., 2017 | lncRNA | High levels of serum lncRNA MALAT1 are predictive of resistance to TMZ in GBM patients. High expression of this lncRNA correlated with shorter OS and recurrence. | Prognosis and stratification | [66] |
| Hagemann et al., 2019 | mRNA | Increased circulating MACC1 gene transcripts in the blood of GBM patients. Elevated MACC1 levels were associated with a worse prognosis. | Diagnosis and prognosis | [67] |
| Figueroa et al., 2017 | mRNA, EV | Detection of DNA copy number amplification of wild-type EGFR and EGFRvIII variants. Sensitivity of 61% and specificity of 98% for detection of EGFRvIII-positive GBM. RNAs from EVs reflected the molecular genetic status of GBM, facilitating the guidance of specific therapies. | Diagnosis, monitoring and therapeutic decisions | [31] |
| Ghorbani et al., 2024 | Protein | High plasma GFAP concentration was associated with GBM. | Diagnosis | [71] |
| Soler et al., 2017 | Protein | The HLA-DR-/low/VNN2⁺ ratio in CD14+ PBMCs distinguishes patients with recurrent GBM from those with radiation necrosis. | Diagnosis | [70] |
| Tsvetkov et al., 2021 | Proteins | Identified protein denaturation profiles that differentiate gliomas from healthy controls, including 19 cases of IDH wild-type GBM. | Diagnosis | [69] |
| Dufrusine et al., 2023 | Protein, EV | Overexpression of LGALS3BP protein in EVs derived from plasma of GBM patients. Targeting extracellular LGALS3BP in xenografted mice increases survival. | Diagnosis and stratification | [26] |
| Masood et al., 2023 | Protein, mRNA | Elevated PD-L1 levels have been associated with poor overall survival, being a potential prognostic marker and selection tool for blockade therapy. | Prognosis and stratification | [68] |
| Björkblom et al., 2016 | Metabolite | Elevated levels of serum vitamin E (α- and γ-tocopherol) predict future GBM development years before onset. | Prediction | [72] |
| Bao et al., 2024 | Metabolite | 14 CSF metabolites were causally associated with GBM risk: 11 (α-tocopherol, butyrate, uracil, valine) linked to increased risk, and 3 (N1-methylinosine, succinylcarnitine) linked to decreased risk. | Prediction | [73] |
| Shen et al., 2018 | Metabolite | Decreased arginine and methionine added to increased kynurenate plasma levels were associated with poor OS and PFS in newly diagnosed GBM patients. | Prognosis | [75] |
| Zhao et al., 2016 | Metabolite | Eighteen metabolites (arginine and ornithine being the most relevant) distinguish high- from low-grade gliomas with 91.1% accuracy while six metabolites differed in quantities according to IDH mutational status. | Diagnosis, prognosis and stratification | [74] |
| Liu et al., 2024 | Metabolite | Distinct metabolic profiles were observed in GBM tissue and patient plasma at recurrence, including N-alpha-methylhistamine, glycerol-3-phosphate, phosphocholine, and succinic acid in tissue, and indole-3-acetate and urea in plasma. | Monitoring | [76] |
| Bark et al., 2023 | Metabolite, lipid | Detection of 151 metabolites and 197 lipids, highlighting an increase in specific metabolites in patients with unfavorable outcomes. The lipid profile showed greater heterogeneity in patients with unfavorable outcomes. | Prognosis | [77] |
| Zhou et al., 2022 | Lipid | A panel of 11 plasma lipids was identified as serum biomarkers to distinguish malignant gliomas from healthy controls with 0.9641 accuracy. These included several phosphatidylcholines, lysophosphatidylcholines and triglycerides. | Diagnosis | [78] |
| Soylemez et al., 2022 | Lipid | Differentially regulated lipids were identified in patients with GBM, including fatty acid, glycerolipid, glycerophospholipid, saccharolipid, sphingolipid, and sterol lipid. | Diagnosis | [79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Lima, B.P.; Ferraz, L.S.; Devalle, S.; Borges, H.L. Liquid Biopsy-Derived Tumor Biomarkers for Clinical Applications in Glioblastoma. Biomolecules 2025, 15, 658. https://doi.org/10.3390/biom15050658
de Lima BP, Ferraz LS, Devalle S, Borges HL. Liquid Biopsy-Derived Tumor Biomarkers for Clinical Applications in Glioblastoma. Biomolecules. 2025; 15(5):658. https://doi.org/10.3390/biom15050658
Chicago/Turabian Stylede Lima, Bruna Pereira, Leticia Silva Ferraz, Sylvie Devalle, and Helena Lobo Borges. 2025. "Liquid Biopsy-Derived Tumor Biomarkers for Clinical Applications in Glioblastoma" Biomolecules 15, no. 5: 658. https://doi.org/10.3390/biom15050658
APA Stylede Lima, B. P., Ferraz, L. S., Devalle, S., & Borges, H. L. (2025). Liquid Biopsy-Derived Tumor Biomarkers for Clinical Applications in Glioblastoma. Biomolecules, 15(5), 658. https://doi.org/10.3390/biom15050658







