Drosophila as a Genetic Model System to Study Organismal Energy Metabolism
Abstract
1. Metabolic Dysfunction and Obesity
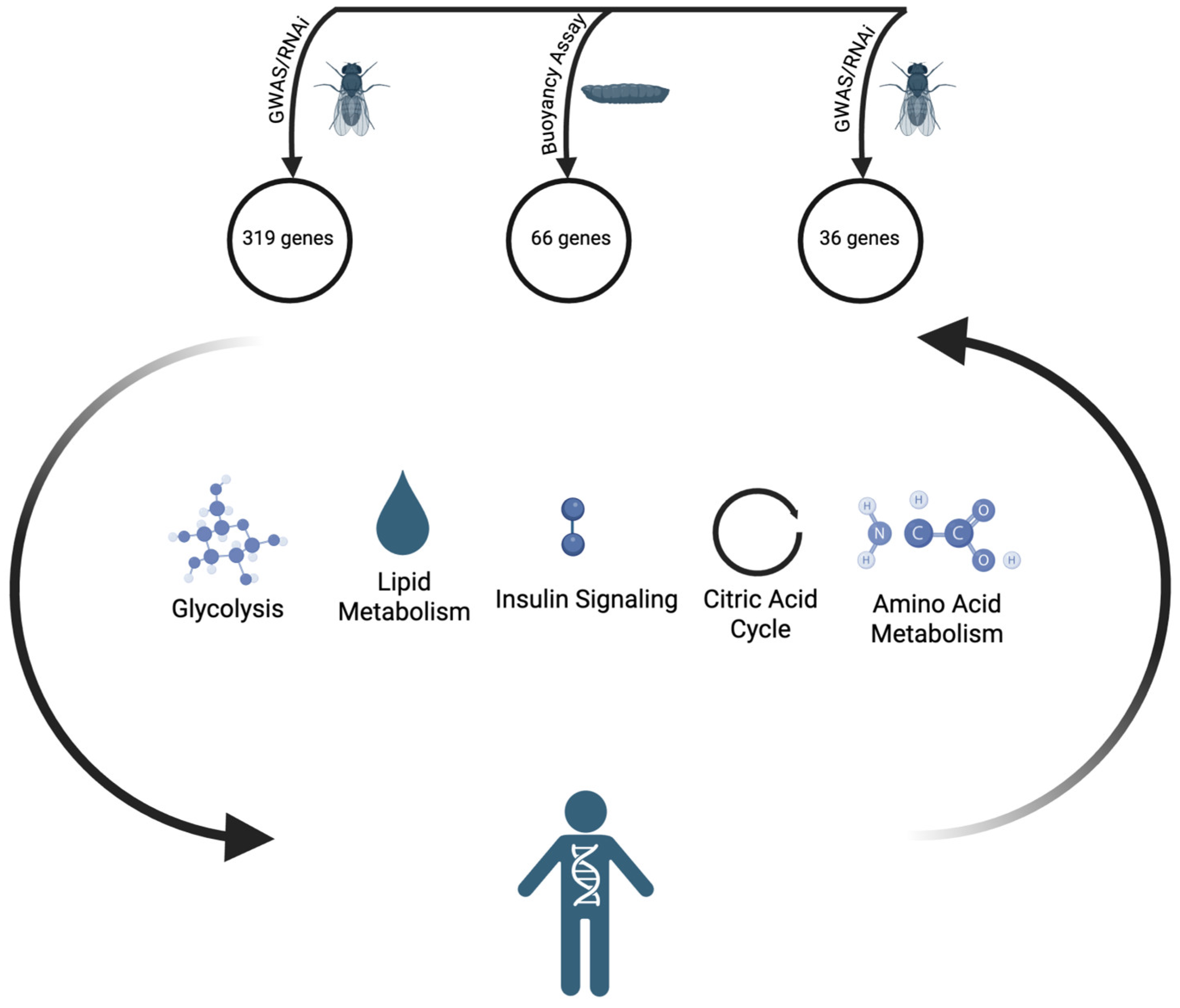
2. From Humans to Flies
| Fly Gene | Human Gene |
|---|---|
| Reis 2010 [31] (larval buoyancy, viable homozygous P-element insertions) | |
| clt | ACHE |
| boi | DCC, NEO1, ROBO2 |
| Gdi | GDI2 |
| lilli | AFF3 |
| trx | KMT2A |
| Fas2 | NCAM1, NCAM2 |
| NFAT | NFATC1, NFATC2, NFATC5 |
| Fur1 | PCSK6 |
| Eip75B | PPARA, PPARD, PPARG, RARB, THRA |
| shep | RBMS1, RBMS3 |
| Glut1 | SLC2A2 |
| tmod | TMOD1, LMOD1 |
| msn | TNIK, MINK1 |
| kibra | WWC1 |
| hdc | HECA |
| trn | LINGO1, LINGO2, RTN4RL1, ELFN1 |
| jim | ZNF257, ZNF713 |
| jing | AEBP2 |
| Pospisilik 2010 [30] (adult triglyceride levels, unbiased RNAi collection) | |
| CHES-1-like | FOXN3 |
| Ets96B | ETV5 |
| ttv | EXT1 |
| eya | EYA1, EYA2 |
| CG17026 | IMPA2 |
| CG34404 | MCC |
| DIP-alpha | NCAM1, NCAM2, CHL1, IGSF9B |
| RabX6 | RAB1A |
| RasGAP1 | RASA2 |
| CG8654 | SLC22A3, SLC22A2, SLC22A11 |
| org-1 | TBX15, MGA |
| CG6689 | ZNF229, ZNF268, ZNF585B |
| Ostgamma | TUSC3 |
| mim | MTSS1 |
| Vha100-5 | ATP6V0A2, (pro)renin receptor |
| Odc1 | ODC1 |
| Su(fu) | SUFU |
| kmr | PLEKHA5 |
| didum | MYO5C |
| Kif3C | KIF17 |
| foi | SLC39A8 |
| Zir | DOCK8 |
| CG30486 | CRISPLD2 |
| Baranski 2018 [28] (adult triglyceride levels, RNAi targeting human GWAS candidates) | |
| BCL7-like | BCL7A |
| emp | SCARB2 |
| fne | ELAVL4 |
| Egfr | ERBB3, ERBB4 |
| foxo | FOXO3 |
| trh | NPAS1 |
| park | PRKN |
| sv | PAX4, PAX5 |
| CLIP-190 | CLIP1 |
| Ets98B | SPI1 |
| pan | TCF7L2 |
| Nrx-1 | NRXN3, NRXN1, NRXN2 |
| TTLL4A | TTLL4 |
| CG1695 | SGSM2 |
| Aos1 | SAE1 |
| cert | CERT1 |
| CG42458 | RALY |
| Su(Tpl) | ELL2 |
| Cluap1 | CLUAP1 |
| su(sable) | ZC3H4 |
| spidey | HSD17B12 |
| stumps | BANK1 |
| CG4622 | ZCCHC8 |
| Nna1 | AGBL2 |
| mof | KAT8 |
| Sec16 | SEC16B |
| Pdi | PDILT |
| CG10465 | KCTD13 |
| CG4945 | SBK1 |
3. Fat Flies: A Well-Controlled Animal Model for the Study of Gene-Diet Interactions
4. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elks, C.E.; den Hoed, M.; Zhao, J.H.; Sharp, S.J.; Wareham, N.J.; Loos, R.J.; Ong, K.K. Variability in the heritability of body mass index: A systematic review and meta-regression. Front. Endocrinol. 2012, 3, 29. [Google Scholar] [CrossRef] [PubMed]
- Maes, H.H.; Neale, M.C.; Eaves, L.J. Genetic and environmental factors in relative body weight and human adiposity. Behav. Genet. 1997, 27, 325–351. [Google Scholar] [CrossRef]
- Zhang, Y.; Proenca, R.; Maffei, M.; Barone, M.; Leopold, L.; Friedman, J.M. Positional cloning of the mouse obese gene and its human homologue. Nature 1994, 372, 425–432. [Google Scholar] [CrossRef]
- Asgari, R.; Caceres-Valdiviezo, M.; Wu, S.; Hamel, L.; Humber, B.E.; Agarwal, S.M.; Fletcher, P.J.; Fulton, S.; Hahn, M.K.; Pereira, S. Regulation of energy balance by leptin as an adiposity signal and modulator of the reward system. Mol. Metab. 2025, 91, 102078. [Google Scholar] [CrossRef]
- Skoracka, K.; Hryhorowicz, S.; Schulz, P.; Zawada, A.; Ratajczak-Pawlowska, A.E.; Rychter, A.M.; Slomski, R.; Dobrowolska, A.; Krela-Kazmierczak, I. The role of leptin and ghrelin in the regulation of appetite in obesity. Peptides 2025, 186, 171367. [Google Scholar] [CrossRef]
- Montague, C.T.; Farooqi, I.S.; Whitehead, J.P.; Soos, M.A.; Rau, H.; Wareham, N.J.; Sewter, C.P.; Digby, J.E.; Mohammed, S.N.; Hurst, J.A.; et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature 1997, 387, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Clement, K.; Vaisse, C.; Lahlou, N.; Cabrol, S.; Pelloux, V.; Cassuto, D.; Gourmelen, M.; Dina, C.; Chambaz, J.; Lacorte, J.M.; et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature 1998, 392, 398–401. [Google Scholar] [CrossRef]
- Dosda, S.; Renard, E.; Meyre, D. Sequencing methods, functional characterization, prevalence, and penetrance of rare coding mutations in panels of monogenic obesity genes from the leptin-melanocortin pathway: A systematic review. Obes. Rev. 2024, 25, e13754. [Google Scholar] [CrossRef] [PubMed]
- Si, R.; Pan, D.; Wang, Z.; Chen, Y.; Cao, J. Regulation of the central melanocortin system on energy balance in mammals and birds. Neuropeptides 2022, 95, 102267. [Google Scholar] [CrossRef]
- Challis, B.G.; Coll, A.P.; Yeo, G.S.; Pinnock, S.B.; Dickson, S.L.; Thresher, R.R.; Dixon, J.; Zahn, D.; Rochford, J.J.; White, A.; et al. Mice lacking pro-opiomelanocortin are sensitive to high-fat feeding but respond normally to the acute anorectic effects of peptide-YY(3-36). Proc. Natl. Acad. Sci. USA 2004, 101, 4695–4700. [Google Scholar] [CrossRef]
- Fan, W.; Boston, B.A.; Kesterson, R.A.; Hruby, V.J.; Cone, R.D. Role of melanocortinergic neurons in feeding and the agouti obesity syndrome. Nature 1997, 385, 165–168. [Google Scholar] [CrossRef]
- Huszar, D.; Lynch, C.A.; Fairchild-Huntress, V.; Dunmore, J.H.; Fang, Q.; Berkemeier, L.R.; Gu, W.; Kesterson, R.A.; Boston, B.A.; Cone, R.D.; et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 1997, 88, 131–141. [Google Scholar] [CrossRef]
- Jackson, R.S.; Creemers, J.W.; Ohagi, S.; Raffin-Sanson, M.L.; Sanders, L.; Montague, C.T.; Hutton, J.C.; O’Rahilly, S. Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat. Genet. 1997, 16, 303–306. [Google Scholar] [CrossRef] [PubMed]
- Krude, H.; Biebermann, H.; Luck, W.; Horn, R.; Brabant, G.; Gruters, A. Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nat. Genet. 1998, 19, 155–157. [Google Scholar] [CrossRef]
- Lu, D.; Willard, D.; Patel, I.R.; Kadwell, S.; Overton, L.; Kost, T.; Luther, M.; Chen, W.; Woychik, R.P.; Wilkison, W.O.; et al. Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor. Nature 1994, 371, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Vaisse, C.; Clement, K.; Guy-Grand, B.; Froguel, P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat. Genet. 1998, 20, 113–114. [Google Scholar] [CrossRef] [PubMed]
- Yaswen, L.; Diehl, N.; Brennan, M.B.; Hochgeschwender, U. Obesity in the mouse model of pro-opiomelanocortin deficiency responds to peripheral melanocortin. Nat. Med. 1999, 5, 1066–1070. [Google Scholar] [CrossRef]
- Yeo, G.S.; Farooqi, I.S.; Aminian, S.; Halsall, D.J.; Stanhope, R.G.; O’Rahilly, S. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat. Genet. 1998, 20, 111–112. [Google Scholar] [CrossRef]
- Loos, R.J.F.; Yeo, G.S.H. The genetics of obesity: From discovery to biology. Nat. Rev. Genet. 2022, 23, 120–133. [Google Scholar] [CrossRef]
- Bernards, A.; Hariharan, I.K. Of flies and men--studying human disease in Drosophila. Curr. Opin. Genet. Dev. 2001, 11, 274–278. [Google Scholar] [CrossRef]
- Reiter, L.T.; Potocki, L.; Chien, S.; Gribskov, M.; Bier, E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001, 11, 1114–1125. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Perrimon, N. What fuels the fly: Energy metabolism in Drosophila and its application to the study of obesity and diabetes. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef]
- Musselman, L.P.; Kuhnlein, R.P. Drosophila as a model to study obesity and metabolic disease. J. Exp. Biol. 2018, 221. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Rulifson, E.J. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature 2004, 431, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Park, J.H. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics 2004, 167, 311–323. [Google Scholar] [CrossRef]
- Rulifson, E.J.; Kim, S.K.; Nusse, R. Ablation of insulin-producing neurons in flies: Growth and diabetic phenotypes. Science 2002, 296, 1118–1120. [Google Scholar] [CrossRef]
- Agrawal, N.; Lawler, K.; Davidson, C.M.; Keogh, J.M.; Legg, R.; Interval; Barroso, I.; Farooqi, I.S.; Brand, A.H. Predicting novel candidate human obesity genes and their site of action by systematic functional screening in Drosophila. PLoS Biol. 2021, 19, e3001255. [Google Scholar] [CrossRef]
- Baranski, T.J.; Kraja, A.T.; Fink, J.L.; Feitosa, M.; Lenzini, P.A.; Borecki, I.B.; Liu, C.T.; Cupples, L.A.; North, K.E.; Province, M.A. A high throughput, functional screen of human Body Mass Index GWAS loci using tissue-specific RNAi Drosophila melanogaster crosses. PLoS Genet. 2018, 14, e1007222. [Google Scholar] [CrossRef]
- Baumbach, J.; Hummel, P.; Bickmeyer, I.; Kowalczyk, K.M.; Frank, M.; Knorr, K.; Hildebrandt, A.; Riedel, D.; Jäckle, H.; Kühnlein, R.P. A Drosophila In Vivo Screen Identifies Store-Operated Calcium Entry as a Key Regulator of Adiposity. Cell Metab. 2014, 19, 331–343. [Google Scholar] [CrossRef]
- Pospisilik, J.A.; Schramek, D.; Schnidar, H.; Cronin, S.J.; Nehme, N.T.; Zhang, X.; Knauf, C.; Cani, P.D.; Aumayr, K.; Todoric, J.; et al. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell 2010, 140, 148–160. [Google Scholar] [CrossRef]
- Reis, T.; Van Gilst, M.R.; Hariharan, I.K. A buoyancy-based screen of Drosophila larvae for fat-storage mutants reveals a role for Sir2 in coupling fat storage to nutrient availability. PLoS Genet. 2010, 6, e1001206. [Google Scholar] [CrossRef] [PubMed]
- Trinh, I.; Gluscencova, O.B.; Boulianne, G.L. An in vivo screen for neuronal genes involved in obesity identifies Diacylglycerol kinase as a regulator of insulin secretion. Mol. Metab. 2019, 19, 13–23. [Google Scholar] [CrossRef]
- Suh, J.M.; Gao, X.; McKay, J.; McKay, R.; Salo, Z.; Graff, J.M. Hedgehog signaling plays a conserved role in inhibiting fat formation. Cell Metab. 2006, 3, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, Q.; Lee, J.H.; Shu, Y. Reduction in Tcf7l2 expression decreases diabetic susceptibility in mice. Int. J. Biol. Sci. 2012, 8, 791–801. [Google Scholar] [CrossRef]
- Akhtar, S.; Culver, S.A.; Siragy, H.M. Novel regulation of renal gluconeogenesis by Atp6ap2 in response to high fat diet via PGC1-alpha/AKT-1 pathway. Sci. Rep. 2021, 11, 11367. [Google Scholar] [CrossRef]
- Kanda, A.; Noda, K.; Ishida, S. ATP6AP2/(pro)renin receptor contributes to glucose metabolism via stabilizing the pyruvate dehydrogenase E1 beta subunit. J. Biol. Chem. 2015, 290, 9690–9700. [Google Scholar] [CrossRef]
- Lyne, R.; Smith, R.; Rutherford, K.; Wakeling, M.; Varley, A.; Guillier, F.; Janssens, H.; Ji, W.; McLaren, P.; North, P.; et al. FlyMine: An integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007, 8, R129. [Google Scholar] [CrossRef] [PubMed]
- Lyne, R.; Bazaga, A.; Butano, D.; Contrino, S.; Heimbach, J.; Hu, F.; Kalderimis, A.; Lyne, M.; Reierskog, K.; Stepan, R.; et al. HumanMine: Advanced data searching, analysis and cross-species comparison. Database 2022, 2022. [Google Scholar] [CrossRef]
- Al-Anzi, B.; Sapin, V.; Waters, C.; Zinn, K.; Wyman, R.J.; Benzer, S. Obesity-Blocking Neurons in Drosophila. Neuron 2009, 63, 329–341. [Google Scholar] [CrossRef]
- Mosher, J.; Zhang, W.; Blumhagen, R.Z.; D’Alessandro, A.; Nemkov, T.; Hansen, K.C.; Hesselberth, J.R.; Reis, T. Coordination between Drosophila Arc1 and a specific population of brain neurons regulates organismal fat. Dev. Biol. 2015, 405, 280–290. [Google Scholar] [CrossRef]
- Ashley, J.; Cordy, B.; Lucia, D.; Fradkin, L.G.; Budnik, V.; Thomson, T. Retrovirus-like Gag Protein Arc1 Binds RNA and Traffics across Synaptic Boutons. Cell 2018, 172, 262–274.e211. [Google Scholar] [CrossRef] [PubMed]
- Pastuzyn, E.D.; Day, C.E.; Kearns, R.B.; Kyrke-Smith, M.; Taibi, A.V.; McCormick, J.; Yoder, N.; Belnap, D.M.; Erlendsson, S.; Morado, D.R.; et al. The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein that Mediates Intercellular RNA Transfer. Cell 2018, 172, 275–288.e218. [Google Scholar] [CrossRef] [PubMed]
- Gillette, C.M.; Tennessen, J.M.; Reis, T. Balancing energy expenditure and storage with growth and biosynthesis during Drosophila development. Dev. Biol. 2021, 475, 234–244. [Google Scholar] [CrossRef]
- Tennessen, J.M.; Baker, K.D.; Lam, G.; Evans, J.; Thummel, C.S. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 2011, 13, 139–148. [Google Scholar] [CrossRef]
- Tennessen, J.M.; Bertagnolli, N.M.; Evans, J.; Sieber, M.H.; Cox, J.; Thummel, C.S. Coordinated metabolic transitions during Drosophila embryogenesis and the onset of aerobic glycolysis. G3 2014, 4, 839–850. [Google Scholar] [CrossRef]
- Drummond-Barbosa, D.; Spradling, A.C. Stem Cells and Their Progeny Respond to Nutritional Changes during Drosophila Oogenesis. Dev. Biol. 2001, 231, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Weaver, L.N.; Drummond-Barbosa, D. Maintenance of Proper Germline Stem Cell Number Requires Adipocyte Collagen in Adult Drosophila Females. Genetics 2018, 209, 1155–1166. [Google Scholar] [CrossRef]
- Armstrong, A.R.; Drummond-Barbosa, D. Insulin signaling acts in adult adipocytes via GSK-3β and independently of FOXO to control Drosophila female germline stem cell numbers. Dev. Biol. 2018, 440, 31–39. [Google Scholar] [CrossRef]
- Armstrong, A.R.; Laws, K.M.; Drummond-Barbosa, D. Adipocyte amino acid sensing controls adult germline stem cell number via the amino acid response pathway and independently of Target of Rapamycin signaling in Drosophila. Development 2014, 141, 4479–4488. [Google Scholar] [CrossRef]
- Bi, Y.; Chang, Y.; Liu, Q.; Mao, Y.; Zhai, K.; Zhou, Y.; Jiao, R.; Ji, G. ERp44/CG9911 promotes fat storage in Drosophila adipocytes by regulating ER Ca(2+) homeostasis. Aging 2021, 13, 15013–15031. [Google Scholar] [CrossRef]
- Johnstun, J.A.; Shankar, V.; Mokashi, S.S.; Sunkara, L.T.; Ihearahu, U.E.; Lyman, R.L.; Mackay, T.F.C.; Anholt, R.R.H. Functional Diversification, Redundancy, and Epistasis among Paralogs of the Drosophila melanogaster Obp50a-d Gene Cluster. Mol. Biol. Evol. 2021, 38, 2030–2044. [Google Scholar] [CrossRef]
- Losurdo, N.A.; Bibo, A.; Bedke, J.; Link, N. A novel adipose loss-of-function mutant in Drosophila. Fly 2024, 18, 2352938. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.W.; Tian, Y. The F-box gene Ppa promotes lipid storage in Drosophila. Yi Chuan 2021, 43, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Janssens, J.; De Waegeneer, M.; Kolluru, S.S.; Davie, K.; Gardeux, V.; Saelens, W.; David, F.P.A.; Brbic, M.; Spanier, K.; et al. Fly Cell Atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Science 2022, 375, eabk2432. [Google Scholar] [CrossRef]
- Deuschle, K.; Fehr, M.; Hilpert, M.; Lager, I.; Lalonde, S.; Looger, L.L.; Okumoto, S.; Persson, J.; Schmidt, A.; Frommer, W.B. Genetically encoded sensors for metabolites. Cytometry A 2005, 64, 3–9. [Google Scholar] [CrossRef]
- Hudry, B.; de Goeij, E.; Mineo, A.; Gaspar, P.; Hadjieconomou, D.; Studd, C.; Mokochinski, J.B.; Kramer, H.B.; Placais, P.Y.; Preat, T.; et al. Sex Differences in Intestinal Carbohydrate Metabolism Promote Food Intake and Sperm Maturation. Cell 2019, 178, 901–918.e916. [Google Scholar] [CrossRef]
- Takanaga, H.; Chaudhuri, B.; Frommer, W.B. GLUT1 and GLUT9 as major contributors to glucose influx in HepG2 cells identified by a high sensitivity intramolecular FRET glucose sensor. Biochim. Biophys. Acta 2008, 1778, 1091–1099. [Google Scholar] [CrossRef] [PubMed]
- Volkenhoff, A.; Hirrlinger, J.; Kappel, J.M.; Klambt, C.; Schirmeier, S. Live imaging using a FRET glucose sensor reveals glucose delivery to all cell types in the Drosophila brain. J. Insect Physiol. 2018, 106, 55–64. [Google Scholar] [CrossRef]
- Birse, R.T.; Choi, J.; Reardon, K.; Rodriguez, J.; Graham, S.; Diop, S.; Ocorr, K.; Bodmer, R.; Oldham, S. High-Fat-Diet-Induced Obesity and Heart Dysfunction Are Regulated by the TOR Pathway in Drosophila. Cell Metab. 2010, 12, 533–544. [Google Scholar] [CrossRef]
- Palanker Musselman, L.; Fink, J.L.; Narzinski, K.; Ramachandran, P.V.; Sukumar Hathiramani, S.; Cagan, R.L.; Baranski, T.J. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis. Models Mech. 2011, 4, 842–849. [Google Scholar] [CrossRef]
- Amitani, M.; Asakawa, A.; Amitani, H.; Inui, A. The role of leptin in the control of insulin-glucose axis. Front. Neurosci. 2013, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Beshel, J.; Dubnau, J.; Zhong, Y. A Leptin Analog Locally Produced in the Brain Acts via a Conserved Neural Circuit to Modulate Obesity-Linked Behaviors in Drosophila. Cell Metab. 2017, 25, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Rajan, A.; Perrimon, N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 2012, 151, 123–137. [Google Scholar] [CrossRef]
- Reed, L.K.; Williams, S.; Springston, M.; Brown, J.; Freeman, K.; DesRoches, C.E.; Sokolowski, M.B.; Gibson, G. Genotype-by-Diet Interactions Drive Metabolic Phenotype Variation in Drosophila melanogaster. Genetics 2010, 185, 1009–1019. [Google Scholar] [CrossRef]
- Francis, D.; Ghazanfar, S.; Havula, E.; Krycer, J.R.; Strbenac, D.; Senior, A.; Minard, A.Y.; Geddes, T.; Nelson, M.E.; Weiss, F.; et al. Genome-wide analysis in Drosophila reveals diet-by-gene interactions and uncovers diet-responsive genes. G3 2021, 11, jkab171. [Google Scholar] [CrossRef]
- Havula, E.; Ghazanfar, S.; Lamichane, N.; Francis, D.; Hasygar, K.; Liu, Y.; Alton, L.A.; Johnstone, J.; Needham, E.J.; Pulpitel, T.; et al. Genetic variation of macronutrient tolerance in Drosophila melanogaster. Nat. Commun. 2022, 13, 1637. [Google Scholar] [CrossRef] [PubMed]
- Aw, W.C.; Towarnicki, S.G.; Melvin, R.G.; Youngson, N.A.; Garvin, M.R.; Hu, Y.; Nielsen, S.; Thomas, T.; Pickford, R.; Bustamante, S.; et al. Genotype to phenotype: Diet-by-mitochondrial DNA haplotype interactions drive metabolic flexibility and organismal fitness. PLoS Genet. 2018, 14, e1007735. [Google Scholar] [CrossRef]
- Bousquet, F.; Chauvel, I.; Flaven-Pouchon, J.; Farine, J.P.; Ferveur, J.F. Dietary rescue of altered metabolism gene reveals unexpected Drosophila mating cues. J. Lipid Res. 2016, 57, 443–450. [Google Scholar] [CrossRef]
- Pasco, M.Y.; Leopold, P. High sugar-induced insulin resistance in Drosophila relies on the lipocalin Neural Lazarillo. PLoS ONE 2012, 7, e36583. [Google Scholar] [CrossRef]
- Hazegh, K.E.; Nemkov, T.; D’Alessandro, A.; Diller, J.D.; Monks, J.; McManaman, J.L.; Jones, K.L.; Hansen, K.C.; Reis, T. An autonomous metabolic role for Spen. PLoS Genet. 2017, 13, e1006859. [Google Scholar] [CrossRef]
- Gillette, C.M.; Hazegh, K.E.; Nemkov, T.; Stefanoni, D.; D’Alessandro, A.; Taliaferro, J.M.; Reis, T. Gene–Diet Interactions: Dietary Rescue of Metabolic Defects in spen-Depleted Drosophila melanogaster. Genetics 2020, 214, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Rai, M.; Buddika, K.; Sterrett, M.C.; Luhur, A.; Mahmoudzadeh, N.H.; Julick, C.R.; Pletcher, R.C.; Chawla, G.; Gosney, C.J.; et al. Lactate dehydrogenase and glycerol-3-phosphate dehydrogenase cooperatively regulate growth and carbohydrate metabolism during Drosophila melanogaster larval development. Development 2019, 146. [Google Scholar] [CrossRef]
- Cox, J.E.; Thummel, C.S.; Tennessen, J.M. Metabolomic Studies in Drosophila. Genetics 2017, 206, 1169–1185. [Google Scholar] [CrossRef]
- Yoshinari, Y.; Nishimura, T.; Yoshii, T.; Kondo, S.; Tanimoto, H.; Kobayashi, T.; Matsuyama, M.; Niwa, R. A high-protein diet-responsive gut hormone regulates behavioral and metabolic optimization in Drosophila melanogaster. Nat. Commun. 2024, 15, 10819. [Google Scholar] [CrossRef] [PubMed]
- Reis, T. Effects of Synthetic Diets Enriched in Specific Nutrients on Drosophila Development, Body Fat, and Lifespan. PLoS ONE 2016, 11, e0146758. [Google Scholar] [CrossRef]
- Sorge, S.; Girard, V.; Lampe, L.; Tixier, V.; Weaver, A.; Higgins, T.; Gould, A.P. A Drosophila holidic diet optimized for growth and development. Dev. Cell 2025. [Google Scholar] [CrossRef] [PubMed]
- Martelli, F.; Quig, A.; Mele, S.; Lin, J.; Fulton, T.L.; Wansbrough, M.; Barlow, C.K.; Schittenhelm, R.B.; Johnson, T.K.; Piper, M.D.W. A defined diet for pre-adult Drosophila melanogaster. Sci. Rep. 2024, 14, 6974. [Google Scholar] [CrossRef]
- Lüersen, K.; Röder, T.; Rimbach, G. Drosophila melanogaster in nutrition research-the importance of standardizing experimental diets. Genes. Nutr. 2019, 14, 3. [Google Scholar] [CrossRef]
- Piper, M.D. Using artificial diets to understand the nutritional physiology of Drosophila melanogaster. Curr. Opin. Insect Sci. 2017, 23, 104–111. [Google Scholar] [CrossRef]
- Piper, M.D.; Blanc, E.; Leitão-Gonçalves, R.; Yang, M.; He, X.; Linford, N.J.; Hoddinott, M.P.; Hopfen, C.; Soultoukis, G.A.; Niemeyer, C.; et al. A holidic medium for Drosophila melanogaster. Nat. Methods 2014, 11, 100–105. [Google Scholar] [CrossRef]
- Komarov, N.; Fritsch, C.; Maier, G.L.; Bues, J.; Biočanin, M.; Avalos, C.B.; Dodero, A.; Kwon, J.Y.; Deplancke, B.; Sprecher, S.G. Food hardness preference reveals multisensory contributions of fly larval gustatory organs in behaviour and physiology. PLoS Biol. 2025, 23, e3002730. [Google Scholar] [CrossRef] [PubMed]
- Mank, J.E.; Rideout, E.J. Developmental mechanisms of sex differences: From cells to organisms. Development 2021, 148. [Google Scholar] [CrossRef] [PubMed]
- Camara, N.; Whitworth, C.; Van Doren, M. The creation of sexual dimorphism in the Drosophila soma. Curr. Top. Dev. Biol. 2008, 83, 65–107. [Google Scholar] [CrossRef] [PubMed]
- Cortes, H.D.; Wevrick, R. Genetic analysis of very obese children with autism spectrum disorder. Mol. Genet. Genom. 2018, 293, 725–736. [Google Scholar] [CrossRef]
- Haussmann, I.U.; Bodi, Z.; Sanchez-Moran, E.; Mongan, N.P.; Archer, N.; Fray, R.G.; Soller, M. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 2016, 540, 301–304. [Google Scholar] [CrossRef]
- Kan, L.; Grozhik, A.V.; Vedanayagam, J.; Patil, D.P.; Pang, N.; Lim, K.S.; Huang, Y.C.; Joseph, B.; Lin, C.J.; Despic, V.; et al. The m(6)A pathway facilitates sex determination in Drosophila. Nat. Commun. 2017, 8, 15737. [Google Scholar] [CrossRef]
- Knuckles, P.; Lence, T.; Haussmann, I.U.; Jacob, D.; Kreim, N.; Carl, S.H.; Masiello, I.; Hares, T.; Villasenor, R.; Hess, D.; et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes. Dev. 2018, 32, 415–429. [Google Scholar] [CrossRef]
- Lence, T.; Akhtar, J.; Bayer, M.; Schmid, K.; Spindler, L.; Ho, C.H.; Kreim, N.; Andrade-Navarro, M.A.; Poeck, B.; Helm, M.; et al. m(6)A modulates neuronal functions and sex determination in Drosophila. Nature 2016, 540, 242–247. [Google Scholar] [CrossRef]
- Yan, D.; Perrimon, N. spenito is required for sex determination in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2015, 112, 11606–11611. [Google Scholar] [CrossRef]
- Diaz, A.V.; Stephenson, D.; Nemkov, T.; D’Alessandro, A.; Reis, T. Spenito-dependent metabolic sexual dimorphism intrinsic to fat storage cells. Genetics 2023, 225. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diaz, A.V.; Tekin, I.; Reis, T. Drosophila as a Genetic Model System to Study Organismal Energy Metabolism. Biomolecules 2025, 15, 652. https://doi.org/10.3390/biom15050652
Diaz AV, Tekin I, Reis T. Drosophila as a Genetic Model System to Study Organismal Energy Metabolism. Biomolecules. 2025; 15(5):652. https://doi.org/10.3390/biom15050652
Chicago/Turabian StyleDiaz, Arely V., Izel Tekin, and Tânia Reis. 2025. "Drosophila as a Genetic Model System to Study Organismal Energy Metabolism" Biomolecules 15, no. 5: 652. https://doi.org/10.3390/biom15050652
APA StyleDiaz, A. V., Tekin, I., & Reis, T. (2025). Drosophila as a Genetic Model System to Study Organismal Energy Metabolism. Biomolecules, 15(5), 652. https://doi.org/10.3390/biom15050652






