AFM for Studying the Functional Activity of Enzymes
Abstract
1. Introduction
2. The AFM Principle
3. AFM-Based Force Spectroscopy
4. AFM Imaging
5. Combining AFM with Infrared Spectroscopy
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saghatelian, A.; Cravatt, B. Assignment of protein function in the postgenomic era. Nat. Chem. Biol. 2005, 1, 130–142. [Google Scholar] [CrossRef] [PubMed]
- Qaradakhi, T.; Gadanec, L.K.; McSweeney, K.R.; Tacey, A.; Apostolopoulos, V.; Levinger, I.; Zulli, A. The potential actions of angiotensin-converting enzyme II (ACE2) activator diminazene aceturate (DIZE) in various diseases. Clin. Exp. Pharmacol. Physiol. 2020, 47, 751–758. [Google Scholar] [CrossRef]
- Leake, M.C. The Physics of Life: One Molecule at a Time. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120248. [Google Scholar] [CrossRef]
- Miller, H.; Zhou, Z.; Shepherd, J.; Wollman, A.J.M.; Leake, M.C. Single-Molecule Techniques in Biophysics: A Review of the Progress in Methods and Applications. Rep. Prog. Phys. 2018, 81, 024601. [Google Scholar] [CrossRef]
- Chen, P.; Zhou, X.; Andoy, N.M.; Han, K.S.; Choudhary, E.; Zou, N.; Shen, H. Spatiotemporal catalytic dynamics within single nanocatalysts revealed by single-molecule microscopy. Chem. Soc. Rev. 2014, 43, 1107–1117. [Google Scholar]
- Margolin, G.; Barkai, E. Single-molecule chemical reactions: Reexamination of the Kramers approach. Phys. Rev. E 2005, 72, 025101. [Google Scholar]
- English, B.P.; Min, W.; van Oijen, A.M.; Lee, K.T.; Luo, G.; Sun, H.; Cherayil, B.J.; Kou, S.C.; Xie, X.S. Ever-Fluctuating Single Enzyme Molecules: Michaelis-Menten Equation Revisited. Nat. Chem. Biol. 2006, 2, 87–94. [Google Scholar] [CrossRef]
- Harriman, O.; Leake, M. Single Molecule Experimentation in Biological Physics: Exploring the Living Component of Soft Condensed Matter One Molecule at a Time. J. Phys. Condens. Matter Inst. Phys. J. 2011, 23, 503101. [Google Scholar] [CrossRef]
- Lenn, T.; Leake, M.C. Experimental Approaches for Addressing Fundamental Biological Questions in Living, Functioning Cells with Single Molecule Precision. Open Biol. 2012, 2, 120090. [Google Scholar] [CrossRef]
- Leake, M.C. Analytical Tools for Single-Molecule Fluorescence Imaging in Cellulo. Phys. Chem. Chem. Phys. 2014, 16, 12635–12647. [Google Scholar] [CrossRef]
- Leake, M. Shining the Spotlight on Functional Molecular Complexes. Commun. Integr. Biol. 2010, 3, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Gomes, G.N.W.; Krzeminski, M.; Namini, A.; Martin, E.W.; Mittag, T.; Head-Gordon, T.; Gradinaru, C.C. Conformational ensembles of an intrinsically disordered protein consistent with NMR, SAXS, and single-molecule FRET. J. Am. Chem. Soc. 2020, 142, 15697–15710. [Google Scholar] [CrossRef] [PubMed]
- Akutsu, H. Strategies for Elucidation of the Structure and Function of the Large Membrane Protein Complex, FoF1-ATP Synthase, by Nuclear Magnetic Resonance. Biophys. Chem. 2023, 296, 106988. [Google Scholar] [CrossRef]
- Wagner, G.; Wüthrich, K. Dynamic model of globular protein conformations based on NMR studies in solution. Nature 1978, 275, 247–248. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, C.; Li, D. Visualizing Enzyme Catalytic Process Using Single-Molecule Techniques. TrAC Trends Anal. Chem. 2023, 163, 117083. [Google Scholar] [CrossRef]
- Lu, M.; Ma, X.; Castillo-Menendez, L.R.; Gorman, J.; Alsahafi, N.; Ermel, U.; Terry, D.S.; Chambers, M.; Peng, D.; Zhang, B.; et al. Associating HIV-1 Envelope Glycoprotein Structures with States on the Virus Observed by smFRET. Nature 2019, 568, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Schmid, S.; Hugel, T. Controlling Protein Function by Fine-Tuning Conformational Flexibility. eLife 2020, 9, e57180. [Google Scholar] [CrossRef] [PubMed]
- Maître, J.-L.; Heisenberg, C.-P. The Role of Adhesion Energy in Controlling Cell–Cell Contacts. Curr. Opin. Cell Biol. 2011, 23, 508–514. [Google Scholar] [CrossRef]
- Wruck, F.; Katranidis, A.; Nierhaus, K.H.; Büldt, G.; Hegner, M. Translation and Folding of Single Proteins in Real Time. Proc. Natl. Acad. Sci. USA 2017, 114, E4399–E4407. [Google Scholar] [CrossRef]
- Banerjee, S.; Chakraborty, S.; Sreepada, A.; Banerji, D.; Goyal, S.; Khurana, Y.; Haldar, S. Cutting-Edge Single-Molecule Technologies Unveil New Mechanics in Cellular Biochemistry. Annu. Rev. Biophys. 2021, 50, 419–445. [Google Scholar] [CrossRef]
- Bhatt, P.; Joshi, T.; Bhatt, K.; Zhang, W.; Huang, Y.; Chen, S. Binding Interaction of Glyphosate with Glyphosate Oxidoreductase and C–P Lyase: Molecular Docking and Molecular Dynamics Simulation Studies. J. Hazard. Mater. 2021, 409, 124927. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.-Q.; Zhu, X.; Song, C.-P. Single-Molecule Technique: A Revolutionary Approach to Exploring Fundamental Questions in Plant Science. New Phytol. 2019, 223, 508–510. [Google Scholar]
- Sheldon, R.A.; Woodley, J.M. Role of biocatalysis in sustainable chemistry. Chem. Rev. 2018, 118, 801–838. [Google Scholar] [CrossRef]
- López_Marzo, A.M. Techniques for Characterizing Biofunctionalized Surfaces for Bioanalysis Purposes. Biosens. Bioelectron. 2024, 263, 116599. [Google Scholar] [CrossRef] [PubMed]
- Josephs, E.A.; Ye, T. Nanoscale spatial distribution of thiolated DNA on model nucleic acid sensor surfaces. ACS Nano 2013, 7, 3653–3660. [Google Scholar] [CrossRef] [PubMed]
- Josephs, E.A.; Ye, T. A single-molecule view of conformational switching of DNA tethered to a gold electrode. J. Am. Chem. Soc. 2012, 134, 10021–10030. [Google Scholar]
- Branden, C.I.; Tooze, J. Introduction to Protein Structure, 2nd ed.; Garland Science: New York, NY, USA, 1998; ISBN 978-0-429-06209-4. [Google Scholar]
- Ukraintsev, A.A.; Kutuzov, M.M.; Lavrik, O.I. Studying Structure and Functions of Nucleosomes with Atomic Force Microscopy. Biochem. Mosc. 2024, 89, 674–687. [Google Scholar] [CrossRef]
- Mou, J.; Yang, J.; Shao, Z. Atomic Force Microscopy of Cholera Toxin B-Oligomers Bound to Bilayers of Biologically Relevant Lipids. J. Mol. Biol. 1995, 248, 507–512. [Google Scholar] [CrossRef]
- Müller, D.J.; Amrein, M.; Engel, A. Adsorption of Biological Molecules to a Solid Support for Scanning Probe Microscopy. J. Struct. Biol. 1997, 119, 172–188. [Google Scholar] [CrossRef]
- Czajkowsky, D.M.; Sheng, S.; Shao, Z. Staphylococcal α-Hemolysin Can Form Hexamers in Phospholipid Bilayers. J. Mol. Biol. 1998, 276, 325–330. [Google Scholar] [CrossRef]
- Müller, D.J.; Fotiadis, D.; Scheuring, S.; Müller, S.A.; Engel, A. Electrostatically Balanced Subnanometer Imaging of Biological Specimens by Atomic Force Microscope. Biophys. J. 1999, 76, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Hansma, P.K.; Cleveland, J.P.; Radmacher, M.; Walters, D.A.; Hillner, P.E.; Bezanilla, M.; Fritz, M.; Vie, D.; Hansma, H.G.; Prater, C.B.; et al. Tapping Mode Atomic Force Microscopy in Liquids. Appl. Phys. Lett. 1994, 64, 1738–1740. [Google Scholar] [CrossRef]
- Alsteens, D.; Newton, R.; Schubert, R.; Martinez-Martin, D.; Delguste, M.; Roska, B.; Müller, D.J. Nanomechanical Mapping of First Binding Steps of a Virus to Animal Cells. Nat. Nanotechnol. 2017, 12, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Shi, H.; Yang, X.; Wang, J.; Yang, W.; Zhang, H.; Liu, L. Recent Advances in AFM-Based Biological Characterization and Applications at Multiple Levels. Soft Matter 2020, 16, 8962–8984. [Google Scholar] [CrossRef]
- Alegre-Cebollada, J.; Perez-Jimenez, R.; Kosuri, P.; Fernandez, J.M. Single-Molecule Force Spectroscopy Approach to Enzyme Catalysis. J. Biol. Chem. 2010, 285, 18961–18966. [Google Scholar] [CrossRef]
- Arslan, B.; Colpan, M.; Ju, X.; Zhang, X.; Kostyukova, A.; Abu-Lail, N.I. The Effects of Noncellulosic Compounds on the Nanoscale Interaction Forces Measured between Carbohydrate-Binding Module and Lignocellulosic Biomass. Biomacromolecules 2016, 17, 1705–1715. [Google Scholar] [CrossRef]
- Qin, C.; Clarke, K.; Li, K. Interactive forces between lignin and cellulase as determined by atomic force microscopy. Biotechnol. Biofuels 2014, 7, 65. [Google Scholar] [CrossRef]
- Radmacher, M.; Fritz, M.; Hansma, H.; Hansma, P.K. Direct Observation of Enzyme Activity with the Atomic Force Microscope. Science 1994, 265, 1577–1579. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Bukharina, N.S.; Pleshakova, T.O.; Frantsuzov, P.A.; Krokhin, N.V.; Ziborov, V.S.; Archakov, A.I. Atomic Force Microscopy Visualization and Measurement of the Activity and Physicochemical Properties of Single Monomeric and Oligomeric Enzymes. Biophysics 2011, 56, 892–896. [Google Scholar] [CrossRef]
- Balashev, K.; Nielsen, L.K.; Callisen, T.; Svendsen, A. In Situ Studies of Single Enzymes and Enzyme Kinetics by Atomic Force Microscopy (AFM). Probe Microsc. 2001, 2, 177–185. [Google Scholar]
- Lambert, E.; Aguié-Béghin, V.; Dessaint, D.; Foulon, L.; Chabbert, B.; Paës, G.; Molinari, M. Real time and quantitative imaging of lignocellulosic films hydrolysis by atomic force microscopy reveals lignin recalcitrance at nanoscale. Biomacromolecules 2018, 20, 515–527. [Google Scholar] [PubMed]
- Uchihashi, T.; Watanabe, Y.; Nakazaki, Y.; Yamasaki, T.; Watanabe, H.; Maruno, T.; Ishii, K.; Uchiyama, S.; Song, C.; Murata, K.; et al. Dynamic Structural States of ClpB Involved in Its Disaggregation Function. Nat. Commun. 2018, 9, 2147. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.; Jang, J.; Kang, Y.; Watanabe, H.; Uchihashi, T.; Kim, S.J.; Kato, K.; Lee, J.Y.; Song, J.-J. Structural Basis of Nucleosome Assembly by the Abo1 AAA+ ATPase Histone Chaperone. Nat. Commun. 2019, 10, 5764. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, S.; Suzuki, K.; Imamura, M.; Sasaki, H.; Matsunami, H.; Mizutani, K.; Saito, Y.; Imai, F.L.; Ishizuka-Katsura, Y.; Kimura-Someya, T.; et al. Metastable Asymmetrical Structure of a Shaftless V1 Motor. Sci. Adv. 2019, 5, eaau8149. [Google Scholar] [CrossRef]
- Shibata, M.; Nishimasu, H.; Kodera, N.; Hirano, S.; Ando, T.; Uchihashi, T.; Nureki, O. Real-Space and Real-Time Dynamics of CRISPR-Cas9 Visualized by High-Speed Atomic Force Microscopy. Nat. Commun. 2017, 8, 1430. [Google Scholar] [CrossRef]
- Akter, L.; Flechsig, H.; Marchesi, A.; Franz, C.M. Observing Dynamic Conformational Changes within the Coiled-Coil Domain of Different Laminin Isoforms Using High-Speed Atomic Force Microscopy. Int. J. Mol. Sci. 2024, 25, 1951. [Google Scholar] [CrossRef]
- Best, R.B.; Brockwell, D.J.; Toca-Herrera, J.L.; Blake, A.W.; Smith, D.A.; Radford, S.E.; Clarke, J. Force Mode Atomic Force Microscopy as a Tool for Protein Folding Studies. Anal. Chim. Acta 2003, 479, 87–105. [Google Scholar] [CrossRef]
- Cappella, B.; Dietler, G. Force-Distance Curves by Atomic Force Microscopy. Surf. Sci. Rep. 1999, 34, 1–104. [Google Scholar] [CrossRef]
- Zhao, X.; Meng, X.; Ragauskas, A.J.; Lai, C.; Ling, Z.; Huang, C.; Yong, Q. Unlocking the Secret of Lignin-Enzyme Interactions: Recent Advances in Developing State-of-the-Art Analytical Techniques. Biotechnol. Adv. 2022, 54, 107830. [Google Scholar] [CrossRef]
- Beyer, M.K.; Clausen-Schaumann, H. Mechanochemistry: The mechanical activation of covalent bonds. Chem. Rev. 2005, 105, 2921–2948. [Google Scholar] [CrossRef]
- Schlierf, M.; Li, H.; Fernandez, J.M. The Unfolding Kinetics of Ubiquitin Captured with Single-Molecule Force-Clamp Techniques. Proc. Natl. Acad. Sci. USA 2004, 101, 7299–7304. [Google Scholar] [CrossRef] [PubMed]
- Oberhauser, A.F.; Hansma, P.K.; Carrion-Vazquez, M.; Fernandez, J.M. Stepwise Unfolding of Titin under Force-Clamp Atomic Force Microscopy. Proc. Natl. Acad. Sci. USA 2001, 98, 468–472. [Google Scholar] [CrossRef]
- Yoshida, T.; Nakamura, H.; Masutani, H.; Yodoi, J. The involvement of thioredoxin and thioredoxin binding protein-2 on cellular proliferation and aging process. Ann. New York Acad. Sci. 2005, 1055, 1–12. [Google Scholar] [CrossRef]
- Perez-Jimenez, R.; Wiita, A.P.; Rodriguez-Larrea, D.; Kosuri, P.; Gavira, J.A.; Sanchez-Ruiz, J.M.; Fernandez, J.M. Force-Clamp Spectroscopy Detects Residue Co-Evolution in Enzyme Catalysis. J. Biol. Chem. 2008, 283, 27121–27129. [Google Scholar] [CrossRef]
- Wiita, A.; Perez-Jimenez, R.; Walther, K.; Gräter, F.; Berne, B.J.; Holmgren, A.; Sanchez-Ruiz, J.M.; Fernandez, J.M. Probing the chemistry of thioredoxin catalysis with force. Nature 2007, 450, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Vermaas, J.V.; Petridis, L.; Qi, X.; Schulz, R.; Lindner, B.; Smith, J.C. Mechanism of Lignin Inhibition of Enzymatic Biomass Deconstruction. Biotechnol. Biofuels 2015, 8, 217. [Google Scholar] [CrossRef]
- McCammon, J.A.; Harvey, S.C. Dynamics of Proteins and Nucleic Acids; Cambridge University Press: Cambridge, UK, 1988. [Google Scholar]
- Hall, D.; Foster, A.S. Practical considerations for feature assignment in high-speed AFM of live cell membranes. Biophys. Physicobiology 2022, 19, e190016. [Google Scholar]
- Umeda, K.; McArthur, S.J.; Kodera, N. Spatiotemporal Resolution in High-Speed Atomic Force Microscopy for Studying Biological Macromolecules in Action. Microscopy 2023, 72, 151–161. [Google Scholar] [CrossRef]
- Krishnamurthy, K.; Rajendran, A.; Nakata, E.; Morii, T. Near Quantitative Ligation Results in Resistance of DNA Origami Against Nuclease and Cell Lysate. Small Methods 2024, 8, 2300999. [Google Scholar]
- Fukuda, S.; Ando, T. Technical Advances in High-Speed Atomic Force Microscopy. Biophys. Rev. 2023, 15, 2045–2058. [Google Scholar] [CrossRef]
- Endo, M.; Sugiyama, H. Single-Molecule Visualization of B–Z Transition in DNA Origami Using High-Speed AFM. In Z-DNA: Methods and Protocols; Kim, K.K., Subramani, V.K., Eds.; Springer: New York, NY, USA, 2023; pp. 241–250. ISBN 978-1-07-163084-6. [Google Scholar]
- Rajendran, A.; Endo, M.; Sugiyama, H. State-of-the-Art High-Speed Atomic Force Microscopy for Investigation of Single-Molecular Dynamics of Proteins. Chem. Rev. 2014, 114, 1493–1520. [Google Scholar] [CrossRef] [PubMed]
- Walters, D.A.; Cleveland, J.P.; Thomson, N.H.; Hansma, P.K.; Wendman, M.A.; Gurley, G.; Elings, V. Short cantilevers for atomic force microscopy. Rev. Sci. Instrum. 1996, 67, 3583–3590. [Google Scholar]
- Schitter, G.; Astrom, K.J.; DeMartini, B.E.; Thurner, P.J.; Turner, K.L.; Hansma, P.K. Design and modeling of a high-speed AFM-scanner. IEEE Trans. Control. Syst. Technol. 2007, 15, 906–915. [Google Scholar]
- Kodera, N.; Yamamoto, D.; Ishikawa, R.; Ando, T. Video Imaging of Walking Myosin V by High-Speed Atomic Force Microscopy. Nature 2010, 468, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Uchihashi, T.; Iino, R.; Ando, T.; Noji, H. High-Speed Atomic Force Microscopy Reveals Rotary Catalysis of Rotorless F1-ATPase. Science 2011, 333, 755–758. [Google Scholar] [CrossRef]
- Ando, T.; Kodera, N.; Uchihashi, T.; Miyagi, A.; Nakakita, R.; Yamashita, H.; Matada, K. High-Speed Atomic Force Microscopy for Capturing Dynamic Behavior of Protein Molecules at Work. E J. Surf. Sci. Nanotechnol. 2005, 3, 384–392. [Google Scholar] [CrossRef]
- Ando, T.; Uchihashi, T.; Fukuma, T. High-Speed Atomic Force Microscopy for Nano-Visualization of Dynamic Biomolecular Processes. Prog. Surf. Sci. 2008, 83, 337–437. [Google Scholar] [CrossRef]
- Okumura, M.; Noi, K.; Kanemura, S.; Kinoshita, M.; Saio, T.; Inoue, Y.; Hikima, T.; Akiyama, S.; Ogura, T.; Inaba, K. Dynamic Assembly of Protein Disulfide Isomerase in Catalysis of Oxidative Folding. Nat. Chem. Biol. 2019, 15, 499–509. [Google Scholar] [CrossRef]
- Maegawa, K.I.; Watanabe, S.; Noi, K.; Okumura, M.; Amagai, Y.; Inoue, M.; Inaba, K. The highly dynamic nature of ERdj5 is key to efficient elimination of aberrant protein oligomers through ER-associated degradation. Structure 2017, 25, 846–857. [Google Scholar]
- Okumura, M.; Noi, K.; Inaba, K. Visualization of Structural Dynamics of Protein Disulfide Isomerase Enzymes in Catalysis of Oxidative Folding and Reductive Unfolding. Curr. Opin. Struct. Biol. 2021, 66, 49–57. [Google Scholar] [CrossRef]
- Ivanova, I.A.; Ershova, M.O.; Pleshakova, T.O. Data Processing Algorithm for Determining Biomacromolecule Height Fluctuations in AFM Measurements; Institute of Biomedical Chemistry: Moscow, Russia, 2024; pp. 23–24. [Google Scholar]
- Ruggeri, F.S.; Habchi, J.; Cerreta, A.; Dietler, G. AFM-Based Single Molecule Techniques: Unraveling the Amyloid Pathogenic Species. Curr. Pharm. Des. 2016, 22, 3950–3970. [Google Scholar] [PubMed]
- Amenabar, I.; Poly, S.; Nuansing, W.; Hubrich, E.H.; Govyadinov, A.A.; Huth, F.; Krutokhvostov, R.; Zhang, L.; Knez, M.; Heberle, J.; et al. Structural Analysis and Mapping of Individual Protein Complexes by Infrared Nanospectroscopy. Nat. Commun. 2013, 4, 2890. [Google Scholar] [CrossRef] [PubMed]
- VD dos Santos, A.C.; Hondl, N.; Ramos-Garcia, V.; Kuligowski, J.; Lendl, B.; Ramer, G. AFM-IR for Nanoscale Chemical Characterization in Life Sciences: Recent Developments and Future Directions. ACS Meas. Sci. Au 2023, 3, 301–314. [Google Scholar] [CrossRef]
- Ramer, G.; Ruggeri, F.S.; Levin, A.; Knowles, T.P.J.; Centrone, A. Determination of Polypeptide Conformation with Nanoscale Resolution in Water. ACS Nano 2018, 12, 6612–6619. [Google Scholar] [CrossRef]
- O’Callahan, B.T.; Park, K.-D.; Novikova, I.V.; Jian, T.; Chen, C.-L.; Muller, E.A.; El-Khoury, P.Z.; Raschke, M.B.; Lea, A.S. In Liquid Infrared Scattering Scanning Near-Field Optical Microscopy for Chemical and Biological Nanoimaging. Nano Lett. 2020, 20, 4497–4504. [Google Scholar] [CrossRef]
- Virmani, D.; Bylinkin, A.; Dolado, I.; Janzen, E.; Edgar, J.H.; Hillenbrand, R. Amplitude- and Phase-Resolved Infrared Nanoimaging and Nanospectroscopy of Polaritons in a Liquid Environment. Nano Lett. 2021, 21, 1360–1367. [Google Scholar] [CrossRef]


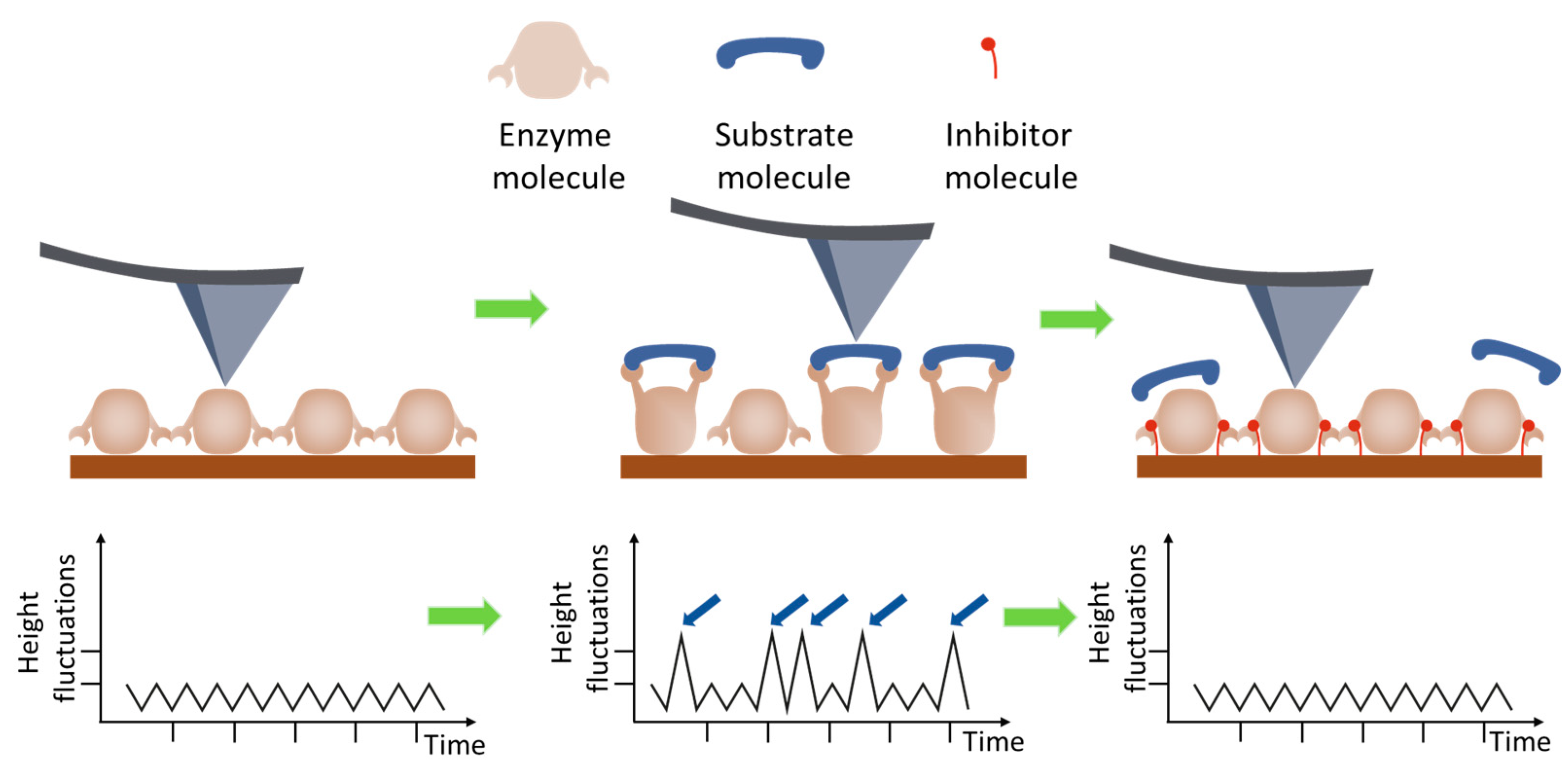



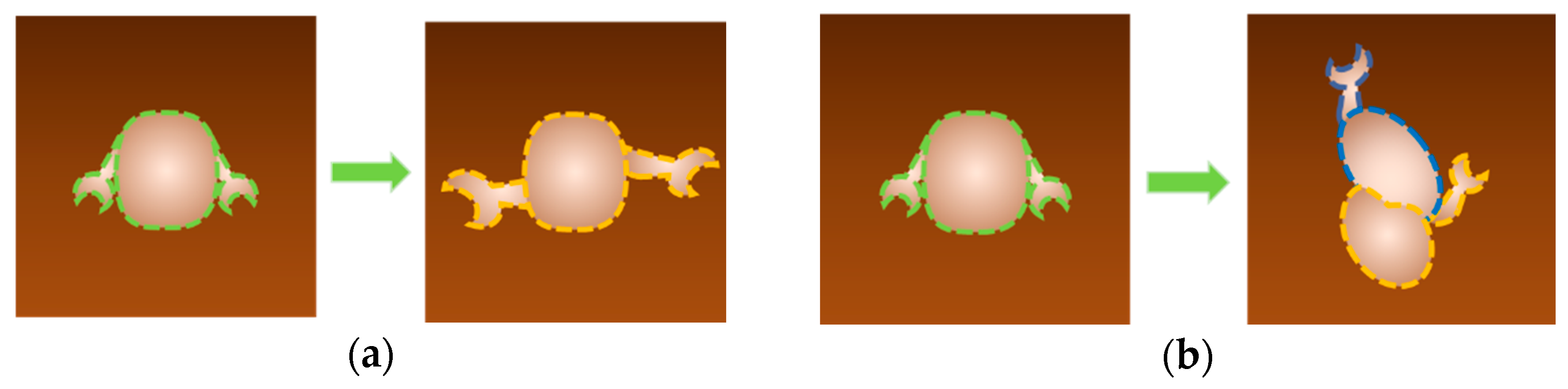
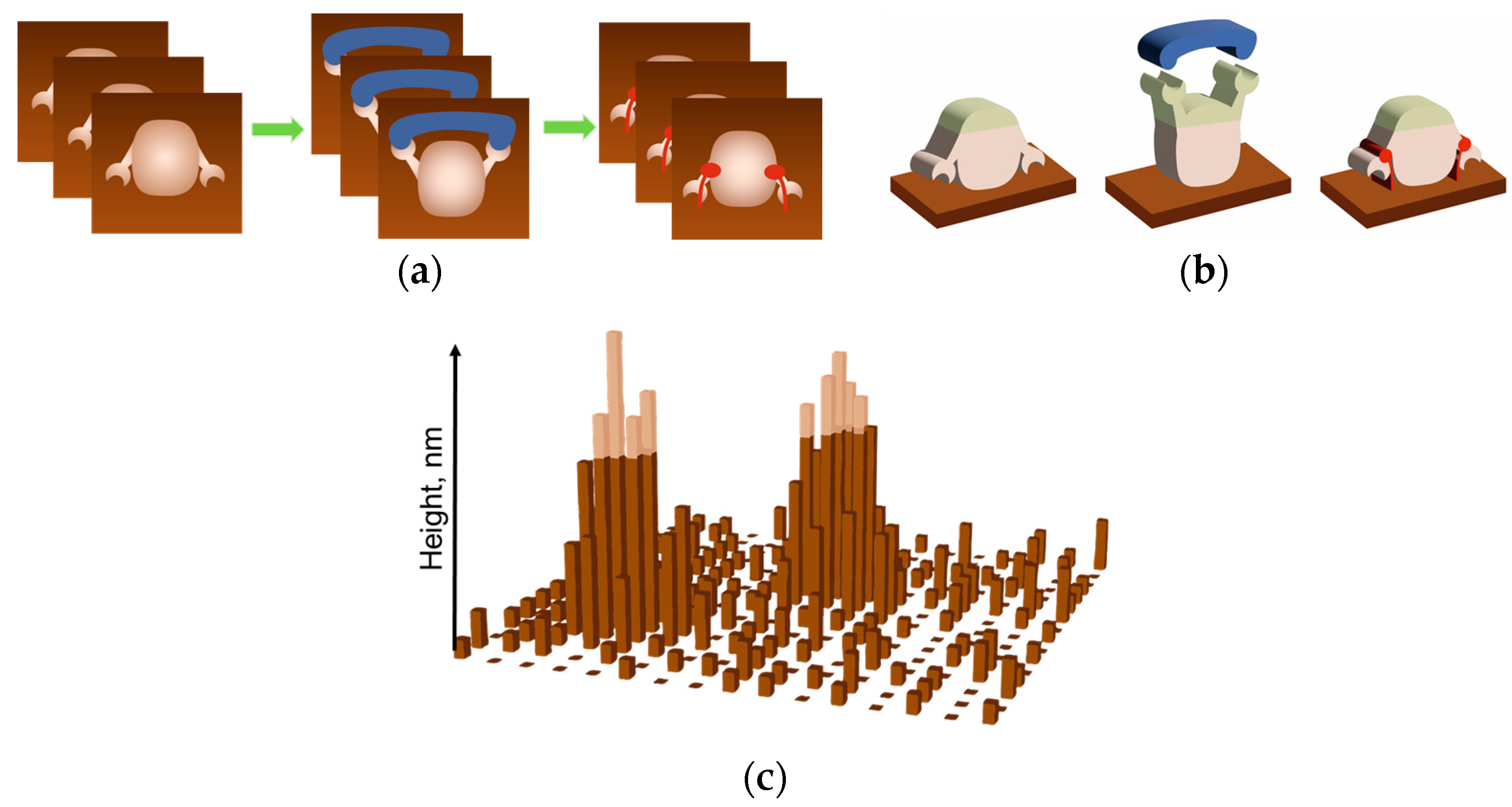
| AFM Mode | Enzyme System | Parameter | Reference |
|---|---|---|---|
| AFM-FS | Thioredoxin family | Mechanism of reduction in disulfide bonds | Alegre-Cebollada et al. [36] |
| Cellulase (CBM 1, CBH I, and Trichoderma reesei) | The bond strength of individual molecules | Arslan et al. [37] | |
| Cellulase | Comparison of the adhesion forces between cellulase and lignin with those between cellulase and cellulose and the examination of the moiety groups involved in cellulase binding to lignin | Qin et al. [38] | |
| AFM imaging | Lysozyme | Height fluctuations of lysozyme protein molecules | Radmacher et al. [39] |
| P450 CYP102A1 | Height fluctuations of protein molecules | Ivanov et al. [40] | |
| Lipase | Layers degradation induced by the lipase enzyme | Balashev et al. [41] | |
| Lignin | Layers degradation of lignocellulose films during hydrolysis | Lambert et al. [42] | |
| AFM imaging (HS-AFM) | Caseinolytic peptidase B protein homolog (ClpB) | The dynamics of changes in protein globule morphology and their relationship to catalytic activity | Uchihashi et al. [43] |
| ATPase histone chaperone Abo1 | Cho et al. [44] | ||
| V1-ATPase | Maruyama et al. [45] | ||
| Laminin-111 and laminin-332 | Akter et al. [46] | ||
| Cas9 nuclease | Shibata et al. [47] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanova, I.A.; Valueva, A.A.; Ershova, M.O.; Pleshakova, T.O. AFM for Studying the Functional Activity of Enzymes. Biomolecules 2025, 15, 574. https://doi.org/10.3390/biom15040574
Ivanova IA, Valueva AA, Ershova MO, Pleshakova TO. AFM for Studying the Functional Activity of Enzymes. Biomolecules. 2025; 15(4):574. https://doi.org/10.3390/biom15040574
Chicago/Turabian StyleIvanova, Irina A., Anastasia A. Valueva, Maria O. Ershova, and Tatiana O. Pleshakova. 2025. "AFM for Studying the Functional Activity of Enzymes" Biomolecules 15, no. 4: 574. https://doi.org/10.3390/biom15040574
APA StyleIvanova, I. A., Valueva, A. A., Ershova, M. O., & Pleshakova, T. O. (2025). AFM for Studying the Functional Activity of Enzymes. Biomolecules, 15(4), 574. https://doi.org/10.3390/biom15040574





