Interactions Between Non-Coding RNAs and HIF-1alpha in the Context of Colorectal Cancer
Abstract
1. Introduction
1.1. Non-Coding RNAs
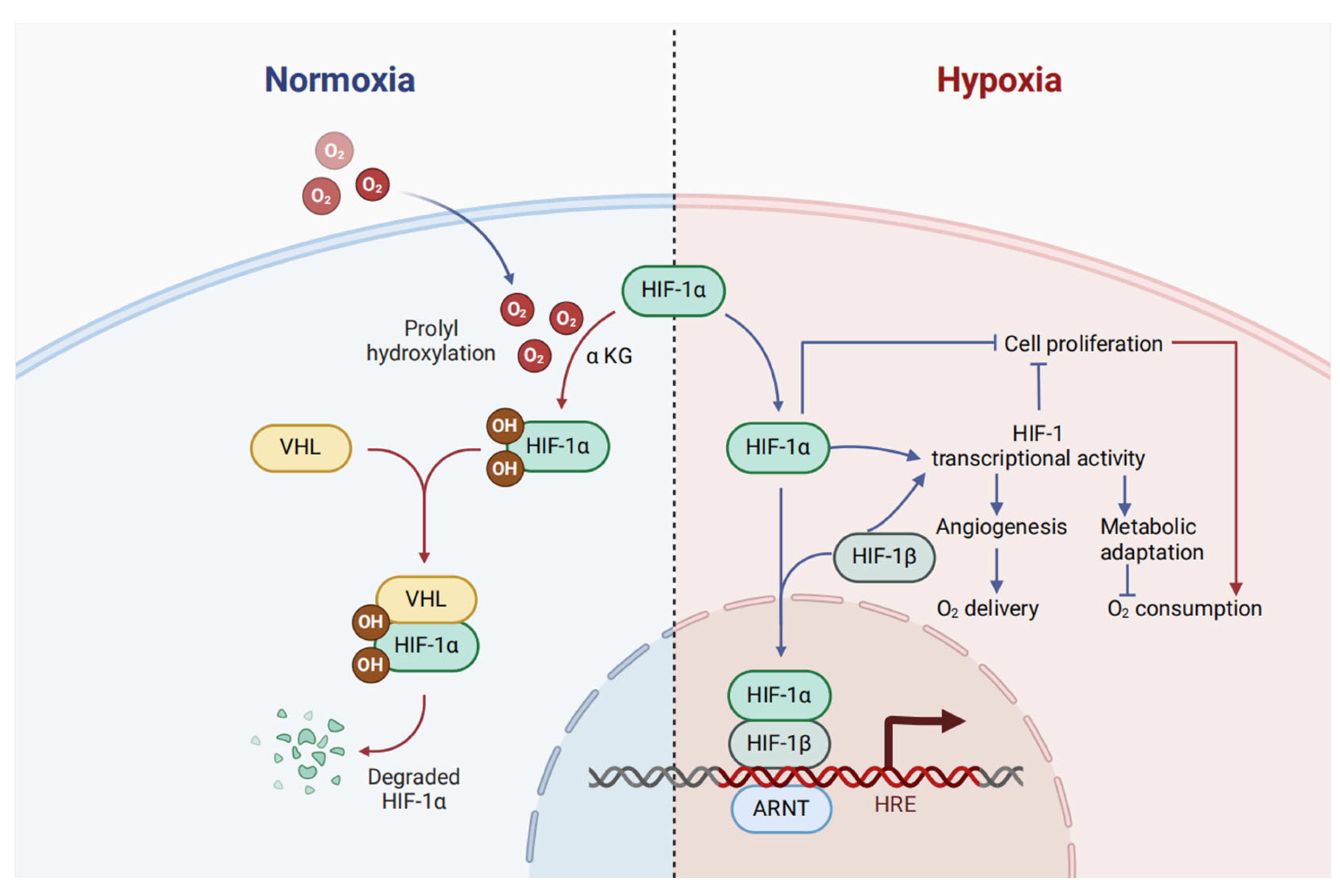
1.2. HIF-1α
1.3. Colorectal Cancer
2. HIF-1α Interacting ncRNAs in Colorectal Cancer
2.1. Long ncRNA
2.2. MicroRNAs
3. New Areas of ncRNA Research in Colorectal Cancer
3.1. RNA Vaccines Based on ncRNA
3.2. Non-Coding RNA and Disulfidptosis
3.3. Single Cell and Spatial Transcriptomics
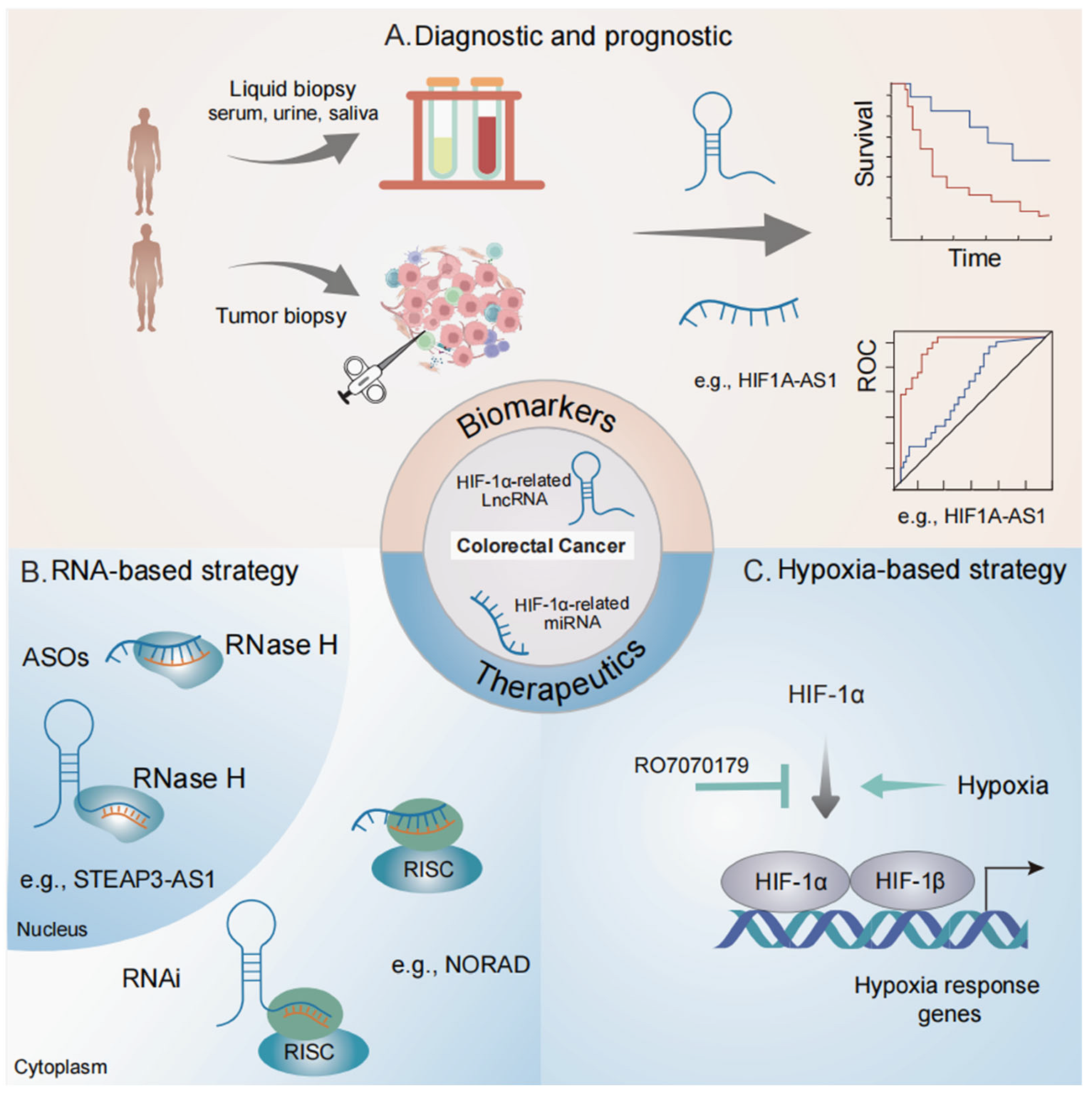
4. Clinical Relevance of HIF-1α-Related ncRNAs in Colorectal Cancer
4.1. Biomarker Potentials
4.2. Therapeutic Potential in Targeting HIF Signaling
4.3. Therapeutic Potential in Targeting ncRNAs
5. Concluding Remarks and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Nemeth, K.; Bayraktar, R.; Ferracin, M.; Calin, G.A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. 2024, 25, 211–232. [Google Scholar] [CrossRef]
- Bartel, D. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Ponting, C.; Oliver, P.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Mercer, T.; Dinger, M.; Mattick, J. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.; Gregersen, L.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Hansen, T.; Jensen, T.; Clausen, B.; Bramsen, J.; Finsen, B.; Damgaard, C.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Imaoka, H.; Toiyama, Y.; Fujikawa, H.; Hiro, J.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Mori, T.; Kato, T.; et al. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2016, 27, 1879–1886. [Google Scholar] [CrossRef]
- Yaman Agaoglu, F.; Kovancilar, M.; Dizdar, Y.; Darendeliler, E.; Holdenrieder, S.; Dalay, N.; Gezer, U. Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumour Biol. 2011, 32, 583–588. [Google Scholar] [CrossRef]
- Levin, A.A. Treating Disease at the RNA Level with Oligonucleotides. N. Engl. J. Med. 2019, 380, 57–70. [Google Scholar] [CrossRef]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef]
- Zhou, X.; Natino, D.; Qin, Z.; Wang, D.; Tian, Z.; Cai, X.; Wang, B.; He, X. Identification and functional characterization of circRNA-0008717 as an oncogene in osteosarcoma through sponging miR-203. Oncotarget 2018, 9, 22288–22300. [Google Scholar] [CrossRef]
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016, 166, 1055–1056. [Google Scholar] [CrossRef]
- Yang, Q.; Du, W.W.; Wu, N.; Yang, W.; Awan, F.M.; Fang, L.; Ma, J.; Li, X.; Zeng, Y.; Yang, Z.; et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017, 24, 1609–1620. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Li, X.; Awan, F.M.; Yang, Z.; Fang, L.; Lyu, J.; Li, F.; Peng, C.; Krylov, S.N.; et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 2018, 37, 5829–5842. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, Y.; Hao, W.; Zhao, W.; Yang, G.; Jing, C. Hypoxia-induced Circular RNA hsa_circ_0006508 Promotes the Warburg Effect in Colorectal Cancer Cells. Balk. Med. J. 2023, 40, 21–27. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Y.; Liu, Z.; Lin, C.; Meng, F.; Xu, L.; Zhang, X.; Zhang, C.; Zhang, P.; Gong, S.; et al. CircMYH9 drives colorectal cancer growth by regulating serine metabolism and redox homeostasis in a p53-dependent manner. Mol. Cancer 2021, 20, 114. [Google Scholar] [CrossRef]
- Chen, L.Y.; Wang, L.; Ren, Y.X.; Pang, Z.; Liu, Y.; Sun, X.D.; Tu, J.; Zhi, Z.; Qin, Y.; Sun, L.N.; et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation. Mol. Cancer 2020, 19, 164. [Google Scholar] [CrossRef]
- Zhou, P.; Xie, W.; Huang, H.L.; Huang, R.Q.; Tian, C.; Zhu, H.B.; Dai, Y.H.; Li, Z.Y. circRNA_100859 functions as an oncogene in colon cancer by sponging the miR-217-HIF-1α pathway. Aging 2020, 12, 13338–13353. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Yang, F.; Miao, S.; Liu, W.; Wang, C.; Shu, Y.; Shen, H. Role of hypoxia-induced exosomes in tumor biology. Mol. Cancer 2018, 17, 120. [Google Scholar] [CrossRef] [PubMed]
- Cowman, S.J.; Koh, M.Y. Revisiting the HIF switch in the tumor and its immune microenvironment. Trends Cancer 2022, 8, 28–42. [Google Scholar] [CrossRef]
- Wang, G.L.; Semenza, G.L. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 1995, 270, 1230–1237. [Google Scholar] [CrossRef]
- Wang, G.L.; Jiang, B.H.; Rue, E.A.; Semenza, G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 1995, 92, 5510–5514. [Google Scholar] [CrossRef]
- Semenza, G.L.; Jiang, B.H.; Leung, S.W.; Passantino, R.; Concordet, J.P.; Maire, P.; Giallongo, A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996, 271, 32529–32537. [Google Scholar] [CrossRef]
- Kaelin, W.G., Jr.; Ratcliffe, P.J. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 2008, 30, 393–402. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia-inducible factors in physiology and medicine. Cell 2012, 148, 399–408. [Google Scholar] [CrossRef]
- Semenza, G.L. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Investig. 2013, 123, 3664–3671. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Li, T.; Li, X.; Zhang, L.; Sun, L.; He, X.; Zhong, X.; Jia, D.; Song, L.; Semenza, G.L.; et al. HIF-1-mediated suppression of acyl-CoA dehydrogenases and fatty acid oxidation is critical for cancer progression. Cell Rep. 2014, 8, 1930–1942. [Google Scholar] [CrossRef] [PubMed]
- Hicklin, D.J.; Ellis, L.M. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2005, 23, 1011–1027. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef]
- Wen, X.; Liu, X.; Mao, Y.P.; Yang, X.J.; Wang, Y.Q.; Zhang, P.P.; Lei, Y.; Hong, X.H.; He, Q.M.; Ma, J.; et al. Long non-coding RNA DANCR stabilizes HIF-1α and promotes metastasis by interacting with NF90/NF45 complex in nasopharyngeal carcinoma. Theranostics 2018, 8, 5676–5689. [Google Scholar] [CrossRef]
- Zeng, Z.; Xu, F.Y.; Zheng, H.; Cheng, P.; Chen, Q.Y.; Ye, Z.; Zhong, J.X.; Deng, S.J.; Liu, M.L.; Huang, K.; et al. LncRNA-MTA2TR functions as a promoter in pancreatic cancer via driving deacetylation-dependent accumulation of HIF-1α. Theranostics 2019, 9, 5298–5314. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fedewa, S.A.; Ahnen, D.J.; Meester, R.G.S.; Barzi, A.; Jemal, A. Colorectal cancer statistics, 2017. CA A Cancer J. Clin. 2017, 67, 177–193. [Google Scholar] [CrossRef]
- Ma, B.; Wang, S.; Wu, W.; Shan, P.; Chen, Y.; Meng, J.; Xing, L.; Yun, J.; Hao, L.; Wang, X.; et al. Mechanisms of circRNA/lncRNA-miRNA interactions and applications in disease and drug research. Biomed. Pharmacother. 2023, 162, 114672. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Hussen, B.M.; Shoorei, H.; Abak, A.; Poornajaf, Y.; Taheri, M.; Samadian, M. Interactions between non-coding RNAs and HIF-1α in the context of cancer. Eur. J. Pharmacol. 2023, 943, 175535. [Google Scholar] [CrossRef]
- Lan, J.; Sun, L.; Xu, F.; Liu, L.; Hu, F.; Song, D.; Hou, Z.; Wu, W.; Luo, X.; Wang, J.; et al. M2 Macrophage-Derived Exosomes Promote Cell Migration and Invasion in Colon Cancer. Cancer Res. 2019, 79, 146–158. [Google Scholar] [CrossRef]
- Yin, J.; Qi, W.; Ji, C.G.; Zhang, D.X.; Xie, X.L.; Ding, Q.; Jiang, X.Y.; Han, J.; Jiang, H.Q. Small RNA sequencing revealed aberrant piRNA expression profiles in colorectal cancer. Oncol. Rep. 2019, 42, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhang, Y.; Jiang, B.; Li, A. Prognostic value of circulating long non-coding RNAs in colorectal cancer patients: A meta-analysis. Expert. Rev. Anticancer. Ther. 2024, 24, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhou, T.; Wu, J.; Lv, H.; Zhou, H.; Du, M.; Zhang, X.; Wu, N.; Gong, S.; Ren, Z.; et al. Plasma exosome-derived circGAPVD1 as a potential diagnostic marker for colorectal cancer. Transl. Oncol. 2023, 31, 101652. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Luo, X.; Li, C.; Wang, G. LncRNA LINC00525 activates HIF-1α through miR-338-3p/UBE2Q1/β-catenin axis to regulate the Warburg effect in colorectal cancer. Bioengineered 2022, 13, 2554–2567. [Google Scholar] [CrossRef]
- Li, X.; Wang, J.; Lin, W.; Yuan, Q.; Lu, Y.; Wang, H.; Chen, Y.; Chen, L.; Dai, P.; Long, H.; et al. circEXOC6B interacting with RRAGB, an mTORC1 activator, inhibits the progression of colorectal cancer by antagonizing the HIF1A-RRAGB-mTORC1 positive feedback loop. Mol. Cancer 2022, 21, 135. [Google Scholar] [CrossRef]
- Du, Z.; Fei, T.; Verhaak, R.G.; Su, Z.; Zhang, Y.; Brown, M.; Chen, Y.; Liu, X.S. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 2013, 20, 908–913. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.; Fu, C.; Yang, L.; Li, G.; Wu, Y.; Tong, H.; Tian, G.; Wang, K.; Wang, J.; et al. Exploration and validation of ceRNA regulatory networks in colorectal cancer based on associations whole transcriptome sequencing. Sci. Rep. 2024, 14, 20446. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Kim, J.; Yang, X.; Martindale, J.L.; Tominaga-Yamanaka, K.; White, E.J.; Orjalo, A.V.; Rinn, J.L.; Kreft, S.G.; et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 2013, 4, 2939. [Google Scholar] [CrossRef]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef]
- Zhang, W.; Yuan, W.; Song, J.; Wang, S.; Gu, X. LncRNA CPS1-IT1 suppresses EMT and metastasis of colorectal cancer by inhibiting hypoxia-induced autophagy through inactivation of HIF-1α. Biochimie 2018, 144, 21–27. [Google Scholar] [CrossRef]
- Qin, M.; Liu, Q.; Yang, W.; Wang, Q.; Xiang, Z. IGFL2-AS1-induced suppression of HIF-1α degradation promotes cell proliferation and invasion in colorectal cancer by upregulating CA9. Cancer Med. 2023, 12, 8415–8432. [Google Scholar] [CrossRef]
- Yang, X.; Luo, Y.; Li, M.; Jin, Z.; Chen, G.; Gan, C. Long non-coding RNA NBR2 suppresses the progression of colorectal cancer by downregulating miR-19a to regulate M2 macrophage polarization. Chin. J. Physiol. 2023, 66, 546–557. [Google Scholar] [CrossRef]
- Zhao, L.; Jiang, L.; Zhang, M.; Zhang, Q.; Guan, Q.; Li, Y.; He, M.; Zhang, J.; Wei, M. NF-κB-activated SPRY4-IT1 promotes cancer cell metastasis by downregulating TCEB1 mRNA via Staufen1-mediated mRNA decay. Oncogene 2021, 40, 4919–4929. [Google Scholar] [CrossRef]
- Xu, L.; Huan, L.; Guo, T.; Wu, Y.; Liu, Y.; Wang, Q.; Huang, S.; Xu, Y.; Liang, L.; He, X. LncRNA SNHG11 facilitates tumor metastasis by interacting with and stabilizing HIF-1α. Oncogene 2020, 39, 7005–7018. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, J.; Huang, Z.; Jin, P.; Peng, L.; Luo, M.; Zhang, Z.; Chen, Y.; Xie, N.; Gao, W.; et al. Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing m(6)A-mediated degradation of STEAP3 mRNA. Mol. Cancer 2022, 21, 168. [Google Scholar] [CrossRef]
- Li, Q.; Sun, H.; Luo, D.; Gan, L.; Mo, S.; Dai, W.; Liang, L.; Yang, Y.; Xu, M.; Li, J.; et al. Lnc-RP11-536 K7.3/SOX2/HIF-1α signaling axis regulates oxaliplatin resistance in patient-derived colorectal cancer organoids. J. Exp. Clin. Cancer Res. 2021, 40, 348. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, H.; Zhang, Y.; Xiao, X.; Chu, F.; Zhang, L. Induction of lncRNA NORAD accounts for hypoxia-induced chemoresistance and vasculogenic mimicry in colorectal cancer by sponging the miR-495-3p/hypoxia-inducible factor-1α (HIF-1α). Bioengineered 2022, 13, 950–962. [Google Scholar] [CrossRef]
- Sun, S.; Xia, C.; Xu, Y. HIF-1α induced lncRNA LINC00511 accelerates the colorectal cancer proliferation through positive feedback loop. Biomed. Pharmacother. 2020, 125, 110014. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Li, L.; Xue, X.; Xie, H.; Shi, H.; Hu, Y. A lncRNA coordinates with Ezh2 to inhibit HIF-1α transcription and suppress cancer cell adaption to hypoxia. Oncogene 2020, 39, 1860–1874. [Google Scholar] [CrossRef]
- Gong, W.; Tian, M.; Qiu, H.; Yang, Z. Elevated serum level of lncRNA-HIF1A-AS1 as a novel diagnostic predictor for worse prognosis in colorectal carcinoma. Cancer Biomark. 2017, 20, 417–424. [Google Scholar] [CrossRef]
- Wang, X.; Li, L.; Zhao, K.; Lin, Q.; Li, H.; Xue, X.; Ge, W.; He, H.; Liu, D.; Xie, H.; et al. A novel LncRNA HITT forms a regulatory loop with HIF-1α to modulate angiogenesis and tumor growth. Cell Death Differ. 2020, 27, 1431–1446. [Google Scholar] [CrossRef]
- Wu, L.; Xue, M.; Lai, S.; Chen, J.; Lin, Y.; Ding, N.; Zhong, J.; Chen, S.; Wang, L. Hypoxia derived exosomes promote the proliferation and metastasis of colorectal cancer through the regulation of HIF-1α/miR-4299/ZBTB4. Life Sci. 2023, 329, 121872. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Wang, D.; Huang, Y.; Zeng, Z.; Xu, P.; Xiong, H.; Ke, Z.; Zhang, Y.; Hu, Y.; Wang, F.; et al. A reciprocal feedback between colon cancer cells and Schwann cells promotes the proliferation and metastasis of colon cancer. J. Exp. Clin. Cancer Res. 2022, 41, 348. [Google Scholar] [CrossRef]
- Islam, S.U.; Ahmed, M.B.; Sonn, J.K.; Jin, E.J.; Lee, Y.S. PRP4 Induces Epithelial-Mesenchymal Transition and Drug Resistance in Colon Cancer Cells via Activation of p53. Int. J. Mol. Sci. 2022, 23, 3092. [Google Scholar] [CrossRef]
- Tsai, H.L.; Tsai, Y.C.; Chen, Y.C.; Huang, C.W.; Chen, P.J.; Li, C.C.; Su, W.C.; Chang, T.K.; Yeh, Y.S.; Yin, T.C.; et al. MicroRNA-148a induces apoptosis and prevents angiogenesis with bevacizumab in colon cancer through direct inhibition of ROCK1/c-Met via HIF-1α under hypoxia. Aging 2022, 14, 6668–6688. [Google Scholar] [CrossRef]
- Li, H.; Liang, J.; Wang, J.; Han, J.; Li, S.; Huang, K.; Liu, C. Mex3a promotes oncogenesis through the RAP1/MAPK signaling pathway in colorectal cancer and is inhibited by hsa-miR-6887-3p. Cancer Commun. 2021, 41, 472–491. [Google Scholar] [CrossRef]
- Jin, F.; Yang, R.; Wei, Y.; Wang, D.; Zhu, Y.; Wang, X.; Lu, Y.; Wang, Y.; Zen, K.; Li, L. HIF-1α-induced miR-23a∼27a∼24 cluster promotes colorectal cancer progression via reprogramming metabolism. Cancer Lett. 2019, 440–441, 211–222. [Google Scholar] [CrossRef]
- Pranzini, E.; Leo, A.; Rapizzi, E.; Ramazzotti, M.; Magherini, F.; Giovannelli, L.; Caselli, A.; Cirri, P.; Taddei, M.L.; Paoli, P. miR-210-3p mediates metabolic adaptation and sustains DNA damage repair of resistant colon cancer cells to treatment with 5-fluorouracil. Mol. Carcinog. 2019, 58, 2181–2192. [Google Scholar] [CrossRef]
- Tsai, H.L.; Miao, Z.F.; Chen, Y.T.; Huang, C.W.; Yeh, Y.S.; Yang, I.P.; Wang, J.Y. miR-148a inhibits early relapsed colorectal cancers and the secretion of VEGF by indirectly targeting HIF-1α under non-hypoxia/hypoxia conditions. J. Cell Mol. Med. 2019, 23, 3572–3582. [Google Scholar] [CrossRef]
- Xu, K.; Zhan, Y.; Yuan, Z.; Qiu, Y.; Wang, H.; Fan, G.; Wang, J.; Li, W.; Cao, Y.; Shen, X.; et al. Hypoxia Induces Drug Resistance in Colorectal Cancer through the HIF-1α/miR-338-5p/IL-6 Feedback Loop. Mol. Ther. 2019, 27, 1810–1824. [Google Scholar] [CrossRef]
- Xu, Z.; Zhu, C.; Chen, C.; Zong, Y.; Feng, H.; Liu, D.; Feng, W.; Zhao, J.; Lu, A. CCL19 suppresses angiogenesis through promoting miR-206 and inhibiting Met/ERK/Elk-1/HIF-1α/VEGF-A pathway in colorectal cancer. Cell Death Dis. 2018, 9, 974. [Google Scholar] [CrossRef]
- Shang, Y.; Chen, H.; Ye, J.; Wei, X.; Liu, S.; Wang, R. HIF-1α/Ascl2/miR-200b regulatory feedback circuit modulated the epithelial-mesenchymal transition (EMT) in colorectal cancer cells. Exp. Cell Res. 2017, 360, 243–256. [Google Scholar] [CrossRef]
- Sun, Y.; Xing, X.; Liu, Q.; Wang, Z.; Xin, Y.; Zhang, P.; Hu, C.; Liu, Y. Hypoxia-induced autophagy reduces radiosensitivity by the HIF-1α/miR-210/Bcl-2 pathway in colon cancer cells. Int. J. Oncol. 2015, 46, 750–756. [Google Scholar] [CrossRef]
- Ye, H.; Pang, L.; Wu, Q.; Zhu, Y.; Guo, C.; Deng, Y.; Zheng, X. A critical role of mir-199a in the cell biological behaviors of colorectal cancer. Diagn. Pathol. 2015, 10, 65. [Google Scholar] [CrossRef]
- Yamakuchi, M.; Yagi, S.; Ito, T.; Lowenstein, C.J. MicroRNA-22 regulates hypoxia signaling in colon cancer cells. PLoS ONE 2011, 6, e20291. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Zanutto, S.; Ciniselli, C.M.; Belfiore, A.; Lecchi, M.; Masci, E.; Delconte, G.; Primignani, M.; Tosetti, G.; Dal Fante, M.; Fazzini, L.; et al. Plasma miRNA-based signatures in CRC screening programs. Int. J. Cancer 2020, 146, 1164–1173. [Google Scholar] [CrossRef]
- Che, J.; Wang, W.; Huang, Y.; Zhang, L.; Zhao, J.; Zhang, P.; Yuan, X. miR-20a inhibits hypoxia-induced autophagy by targeting ATG5/FIP200 in colorectal cancer. Mol. Carcinog. 2019, 58, 1234–1247. [Google Scholar] [CrossRef]
- Li, J.; Yang, P.; Chen, F.; Tan, Y.; Huang, C.; Shen, H.; Peng, C.; Feng, Y.; Sun, Y. Hypoxic colorectal cancer-derived extracellular vesicles deliver microRNA-361-3p to facilitate cell proliferation by targeting TRAF3 via the noncanonical NF-κB pathways. Clin. Transl. Med. 2021, 11, e349. [Google Scholar] [CrossRef]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, Y.; Wu, P.; Zhang, S.; Gong, Z.; Liao, Q.; Guo, C.; Wang, F.; Li, Y.; Zeng, Z.; et al. Application prospect of circular RNA-based neoantigen vaccine in tumor immunotherapy. Cancer Lett. 2023, 563, 216190. [Google Scholar] [CrossRef]
- Chen, R.; Wang, S.K.; Belk, J.A.; Amaya, L.; Li, Z.; Cardenas, A.; Abe, B.T.; Chen, C.K.; Wender, P.A.; Chang, H.Y. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 2023, 41, 262–272. [Google Scholar] [CrossRef]
- Seyhan, A.A. Trials and Tribulations of MicroRNA Therapeutics. Int. J. Mol. Sci. 2024, 25, 1469. [Google Scholar] [CrossRef]
- Khorkova, O.; Stahl, J.; Joji, A.; Volmar, C.H.; Zeier, Z.; Wahlestedt, C. Long non-coding RNA-targeting therapeutics: Discovery and development update. Expert. Opin. Drug Discov. 2023, 18, 1011–1029. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Dai, Z.; Cheng, Q.; Xu, L.; Huang, L.; Liu, Z.; Li, X.; Wang, N.; Wang, G.; Wang, L.; et al. LncRNA-targeting bio-scaffold mediates triple immune effects for postoperative colorectal cancer immunotherapy. Biomaterials 2022, 284, 121485. [Google Scholar] [CrossRef]
- Zheng, T.; Liu, Q.; Xing, F.; Zeng, C.; Wang, W. Disulfidptosis: A new form of programmed cell death. J. Exp. Clin. Cancer Res. 2023, 42, 137. [Google Scholar] [CrossRef]
- Zheng, P.; Zhou, C.; Ding, Y.; Duan, S. Disulfidptosis: A new target for metabolic cancer therapy. J. Exp. Clin. Cancer Res. 2023, 42, 103. [Google Scholar] [CrossRef]
- Dong, X.; Liao, P.; Liu, X.; Yang, Z.; Wang, Y.; Zhong, W.; Wang, B. Construction and Validation of a Reliable Disulfidptosis-Related LncRNAs Signature of the Subtype, Prognostic, and Immune Landscape in Colon Cancer. Int. J. Mol. Sci. 2023, 24, 12915. [Google Scholar] [CrossRef]
- Jung, G.; Hernández-Illán, E.; Moreira, L.; Balaguer, F.; Goel, A. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 111–130. [Google Scholar] [CrossRef]
- Liu, S.J.; Nowakowski, T.J.; Pollen, A.A.; Lui, J.H.; Horlbeck, M.A.; Attenello, F.J.; He, D.; Weissman, J.S.; Kriegstein, A.R.; Diaz, A.A.; et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016, 17, 67. [Google Scholar] [CrossRef]
- Connor, A.A.; Gallinger, S. Pancreatic cancer evolution and heterogeneity: Integrating omics and clinical data. Nat. Rev. Cancer 2022, 22, 131–142. [Google Scholar] [CrossRef]
- Liu, C.J.; Hu, F.F.; Xie, G.Y.; Miao, Y.R.; Li, X.W.; Zeng, Y.; Guo, A.Y. GSCA: An integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Brief. Bioinform. 2023, 24, bbac558. [Google Scholar] [CrossRef]
- Jiménez-Santos, M.J.; García-Martín, S.; Fustero-Torre, C.; Di Domenico, T.; Gómez-López, G.; Al-Shahrour, F. Bioinformatics roadmap for therapy selection in cancer genomics. Mol. Oncol. 2022, 16, 3881–3908. [Google Scholar] [CrossRef]
- Tang, Q.; Chen, J.; Di, Z.; Yuan, W.; Zhou, Z.; Liu, Z.; Han, S.; Liu, Y.; Ying, G.; Shu, X.; et al. TM4SF1 promotes EMT and cancer stemness via the Wnt/β-catenin/SOX2 pathway in colorectal cancer. J. Exp. Clin. Cancer Res. 2020, 39, 232. [Google Scholar] [CrossRef]
- Shigeyasu, K.; Toden, S.; Ozawa, T.; Matsuyama, T.; Nagasaka, T.; Ishikawa, T.; Sahoo, D.; Ghosh, P.; Uetake, H.; Fujiwara, T.; et al. The PVT1 lncRNA is a novel epigenetic enhancer of MYC, and a promising risk-stratification biomarker in colorectal cancer. Mol. Cancer 2020, 19, 155. [Google Scholar] [CrossRef]
- Schito, L.; Semenza, G.L. Hypoxia-Inducible Factors: Master Regulators of Cancer Progression. Trends Cancer 2016, 2, 758–770. [Google Scholar] [CrossRef]
- Ivan, M.; Haberberger, T.; Gervasi, D.C.; Michelson, K.S.; Günzler, V.; Kondo, K.; Yang, H.; Sorokina, I.; Conaway, R.C.; Conaway, J.W.; et al. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc. Natl. Acad. Sci. USA 2002, 99, 13459–13464. [Google Scholar] [CrossRef]
- Terzuoli, E.; Puppo, M.; Rapisarda, A.; Uranchimeg, B.; Cao, L.; Burger, A.M.; Ziche, M.; Melillo, G. Aminoflavone, a ligand of the aryl hydrocarbon receptor, inhibits HIF-1alpha expression in an AhR-independent fashion. Cancer Res. 2010, 70, 6837–6848. [Google Scholar] [CrossRef]
- Mohlin, S.; Hamidian, A.; von Stedingk, K.; Bridges, E.; Wigerup, C.; Bexell, D.; Påhlman, S. PI3K-mTORC2 but not PI3K-mTORC1 regulates transcription of HIF2A/EPAS1 and vascularization in neuroblastoma. Cancer Res. 2015, 75, 4617–4628. [Google Scholar] [CrossRef]
- Lee, S.H.; Jee, J.G.; Bae, J.S.; Liu, K.H.; Lee, Y.M. A group of novel HIF-1α inhibitors, glyceollins, blocks HIF-1α synthesis and decreases its stability via inhibition of the PI3K/AKT/mTOR pathway and Hsp90 binding. J. Cell Physiol. 2015, 230, 853–862. [Google Scholar] [CrossRef]
- Kummar, S.; Chen, A.; Ji, J.; Zhang, Y.; Reid, J.M.; Ames, M.; Jia, L.; Weil, M.; Speranza, G.; Murgo, A.J.; et al. Phase I study of PARP inhibitor ABT-888 in combination with topotecan in adults with refractory solid tumors and lymphomas. Cancer Res. 2011, 71, 5626–5634. [Google Scholar] [CrossRef]
- Norris, R.E.; Shusterman, S.; Gore, L.; Muscal, J.A.; Macy, M.E.; Fox, E.; Berkowitz, N.; Buchbinder, A.; Bagatell, R. Phase 1 evaluation of EZN-2208, a polyethylene glycol conjugate of SN38, in children adolescents and young adults with relapsed or refractory solid tumors. Pediatr. Blood Cancer 2014, 61, 1792–1797. [Google Scholar] [CrossRef]
- Welsh, S.J.; Williams, R.R.; Birmingham, A.; Newman, D.J.; Kirkpatrick, D.L.; Powis, G. The thioredoxin redox inhibitors 1-methylpropyl 2-imidazolyl disulfide and pleurotin inhibit hypoxia-induced factor 1alpha and vascular endothelial growth factor formation. Mol. Cancer Ther. 2003, 2, 235–243. [Google Scholar]
- Isaacs, J.S.; Jung, Y.J.; Mimnaugh, E.G.; Martinez, A.; Cuttitta, F.; Neckers, L.M. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J. Biol. Chem. 2002, 277, 29936–29944. [Google Scholar] [CrossRef]
- Hutt, D.M.; Roth, D.M.; Vignaud, H.; Cullin, C.; Bouchecareilh, M. The histone deacetylase inhibitor, Vorinostat, represses hypoxia inducible factor 1 alpha expression through translational inhibition. PLoS ONE 2014, 9, e106224. [Google Scholar] [CrossRef]
- Park, J.E.; Kong, D.; Fisher, R.; Cardellina, J.; Shoemaker, R.H.; Melillo, G. Targeting the PAS-A domain of HIF-1α for development of small molecule inhibitors of HIF-1. Cell Cycle 2006, 5, 1847–1853. [Google Scholar] [PubMed]
- Scheuermann, T.H.; Li, Q.; Ma, H.W.; Key, J.; Zhang, L.; Chen, R.; Garcia, J.A.; Naidoo, J.; Longgood, J.; Frantz, D.E.; et al. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat. Chem. Biol. 2013, 9, 271–276. [Google Scholar] [CrossRef]
- Kong, D.; Park, E.J.; Stephen, A.G.; Calvani, M.; Cardellina, J.H.; Monks, A.; Fisher, R.J.; Shoemaker, R.H.; Melillo, G. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Cancer Res. 2005, 65, 9047–9055. [Google Scholar] [CrossRef]
- Olenyuk, B.Z.; Zhang, G.J.; Klco, J.M.; Nickols, N.G.; Kaelin, W.G., Jr.; Dervan, P.B. Inhibition of vascular endothelial growth factor with a sequence-specific hypoxia response element antagonist. Proc. Natl. Acad. Sci. USA 2004, 101, 16768–16773. [Google Scholar] [CrossRef]
- Reece, K.M.; Richardson, E.D.; Cook, K.M.; Campbell, T.J.; Pisle, S.T.; Holly, A.J.; Venzon, D.J.; Liewehr, D.J.; Chau, C.H.; Price, D.K.; et al. Epidithiodiketopiperazines (ETPs) exhibit in vitro antiangiogenic and in vivo antitumor activity by disrupting the HIF-1α/p300 complex in a preclinical model of prostate cancer. Mol. Cancer 2014, 13, 91. [Google Scholar] [CrossRef]
- Yeo, E.J.; Ryu, J.H.; Cho, Y.S.; Chun, Y.S.; Huang, L.E.; Kim, M.S.; Park, J.W. Amphotericin B blunts erythropoietin response to hypoxia by reinforcing FIH-mediated repression of HIF-1. Blood 2006, 107, 916–923. [Google Scholar] [CrossRef]
- Chen, W.; Hill, H.; Christie, A.; Kim, M.S.; Holloman, E.; Pavia-Jimenez, A.; Homayoun, F.; Ma, Y.; Patel, N.; Yell, P.; et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 2016, 539, 112–117. [Google Scholar] [CrossRef]
- Tuveson, D.; Clevers, H. Cancer modeling meets human organoid technology. Science 2019, 364, 952–955. [Google Scholar] [CrossRef]
- Banijamali, M.; Höjer, P.; Nagy, A.; Hååg, P.; Gomero, E.P.; Stiller, C.; Kaminskyy, V.O.; Ekman, S.; Lewensohn, R.; Karlström, A.E.; et al. Characterizing single extracellular vesicles by droplet barcode sequencing for protein analysis. J. Extracell. Vesicles 2022, 11, e12277. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Yi, Z.; Shen, Y.; Lin, L.; Chen, F.; Xu, Y.; Wu, Z.; Tang, H.; Zhang, X.; Tian, F.; et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 2022, 185, 1728–1744.e1716. [Google Scholar] [CrossRef] [PubMed]

| NcRNAs | Expression | Target Genes | Functions | Cancers | Reference |
|---|---|---|---|---|---|
| lncRNA IGFL2- AS1 | Upregulate | HIF-1α | Promotes CRC cell proliferation and invasion | CRC | [54] |
| lncRNA NBR2 | Downregulate | HIF-1α | Suppresses the progression | CRC | [55] |
| LINC00525 | Upregulate | HIF-1α | Regulates the Warburg effect | CRC | [44] |
| lncRNA NORAD | Upregulate | HIF-1α | Enhances vasculogenic mimicry and contributes to resistance against 5-fluorouracil | CRC | [60] |
| lncRNA STEAP3-AS1 | Upregulate | STEAP3 | Facilitates CRC progression by inhibiting m(6)A-mediated STEAP3 mRNA degradation. | CRC | [58] |
| Lnc-RP11-536 K7.3 | Upregulate | HIF-1α | Enhances progression and chemoresistance via the SOX2/USP7/HIF-1α signaling pathway. | CRC | [59] |
| LncRNA SNHG11 | Upregulate | TCEB1 | Facilitates metastasis through the downregulation of TCEB1 mRNA. | CRC | [57] |
| lncRNA HITT | Downregulate | HIF-1α | Inhibits cancer cell adaptation to hypoxia by collaborating with Ezh2 to repress HIF-1α transcription. | CRC | [64] |
| LncRNA CPS1-IT1 | Downregulate | HIF-1α | Inhibits EMT and metastasis by suppressing autophagy through the inactivation of HIF-1α. | CRC | [53] |
| lncRNA/HIF1A-AS1 | Upregulate | Facilitates tumor proliferation and metastasis | CRC | [63] | |
| miR-4299 | Upregulate | ZBTB4 | Enhances the proliferation and metastasis by modulating the HIF-1α/miR-4299/ZBTB4 pathway | CRC | [65] |
| miR-21-5p | Upregulate | HIF-1α | Facilitates the proliferation and migration of CRC cells | CRC | [66] |
| miR-210 | Upregulate | P53 | Induces EMT and drug resistance via activation of p53 | CRC | [67] |
| MicroRNA-148a | Downregulate | HIF-1α | Triggers apoptosis and inhibits angiogenesis with bevacizumab by suppressing ROCK1/c-Met through HIF-1α | CRC | [68] |
| miR-6887-3p | Downregulate | RAP1/MAPK | Facilitates oncogenesis via the RAP1/MAPK signaling pathway and is suppressed by miR-6887-3p. | CRC | [69] |
| miR-24 | Upregulate | HIF-1α | Promotes CRC progression via reprogramming metabolism | CRC | [70] |
| miR-210-3p | Downregulate | HIF-1α | Facilitates metabolic adaptation and maintains DNA damage repair in CRC cells resistant to 5-fluorouracil treatment. | CRC | [71] |
| miR-148a | Downregulate | Inhibits VEGF production and early relapse by targeting HIF-1α | CRC | [72] | |
| miR-338-5p | Downregulate | IL-6 | Triggers drug resistance via the HIF-1α/miR-338-5p/IL-6 feedback loop. | CRC | [73] |
| miR-206 | Upregulate | HIF-1α | inhibits the Met/ERK/Elk-1/HIF-1α/VEGF-A pathway to prevent angiogenesis. | CRC | [74] |
| miR-200b | Downregulate | HIF-1α | Affects the EMT-MET plasticity | CRC | [75] |
| miR-210 | Upregulate | Bcl-2 | Decreases radiosensitivity via the HIF-1α/miR-210/Bcl-2 pathway | CRC | [76] |
| miR-199a | Downregulate | HIF-1α | Inhibits CRC progression via the HIF-1α/VEGF pathway. | CRC | [77] |
| MicroRNA-22 | Downregulate | HIF-1α | Suppresses angiogenesis | CRC | [78] |
| NCT Number | NcRNAs | Sample | Biomarker | Study Status | Disease | Study Type |
|---|---|---|---|---|---|---|
| NCT06351384 | miRNA | Peripheral blood | Diagnostic | Recruiting | CRC | Observational |
| NCT02635087 | miRNA | Postoperative specimen | Therapeutic response | Recruiting | CRC | Observational |
| NCT02466113 | miRNA | Postoperative specimen | Therapeutic response | NA | CRC | Interventional |
| NCT01828918 | miRNA | Peripheral blood | Diagnostic | Phase 1 | CRC | Interventional |
| NCT04269746 | lncRNA | Peripheral blood | Diagnostic | Complete | CRC | Observational |
| NCT06432413 | lncRNA | Peripheral blood | Diagnostic | Complete | CRC | Observational |
| NCT06307249 | lncRNA | Blood or tissue | Diagnostic | Phase 1 | CRC | Interventional |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, L.; Zhang, H.; Liu, Y.; Wang, X.; Xia, R. Interactions Between Non-Coding RNAs and HIF-1alpha in the Context of Colorectal Cancer. Biomolecules 2025, 15, 510. https://doi.org/10.3390/biom15040510
Gong L, Zhang H, Liu Y, Wang X, Xia R. Interactions Between Non-Coding RNAs and HIF-1alpha in the Context of Colorectal Cancer. Biomolecules. 2025; 15(4):510. https://doi.org/10.3390/biom15040510
Chicago/Turabian StyleGong, Lianfeng, Haixia Zhang, Ying Liu, Xianwang Wang, and Ruohan Xia. 2025. "Interactions Between Non-Coding RNAs and HIF-1alpha in the Context of Colorectal Cancer" Biomolecules 15, no. 4: 510. https://doi.org/10.3390/biom15040510
APA StyleGong, L., Zhang, H., Liu, Y., Wang, X., & Xia, R. (2025). Interactions Between Non-Coding RNAs and HIF-1alpha in the Context of Colorectal Cancer. Biomolecules, 15(4), 510. https://doi.org/10.3390/biom15040510






