Abstract
Idiopathic pulmonary fibrosis (IPF) is a disease with a poor prognosis and no curative therapies. Fibroblast activation by transforming growth factor β1 (TGFβ1) and disrupted metabolic pathways, including the arginine–polyamine pathway, play crucial roles in IPF development. Polyamines are agonists of the calcium/cation-sensing receptor (CaSR), activation of which is detrimental for asthma and pulmonary hypertension, but its role in IPF is unknown. To address this question, we evaluated polyamine abundance using metabolomic analysis of IPF patient saliva. Furthermore, we examined CaSR functional expression in human lung fibroblasts (HLFs), assessed the anti-fibrotic effects of a CaSR antagonist, NPS2143, in TGFβ1-activated normal and IPF HLFs by RNA sequencing and immunofluorescence imaging, respectively; and NPS2143 effects on polyamine synthesis in HLFs by immunoassays. Our results demonstrate that polyamine metabolites are increased in IPF patient saliva. Polyamines activate fibroblast CaSR in vitro, elevating intracellular calcium concentration. CaSR inhibition reduced TGFβ1-induced polyamine and pro-fibrotic factor expression in normal and IPF HLFs. TGFβ1 directly stimulated polyamine release by HLFs, an effect that was blocked by NPS2143. This suggests that TGFβ1 promotes CaSR activation through increased polyamine expression, driving a pro-fibrotic response. By halting some polyamine-induced pro-fibrotic changes, CaSR antagonists exhibit disease-modifying potential in IPF onset and development.
1. Introduction
Progressive pulmonary fibrosis occurs in the setting of various interstitial lung diseases and is associated with increased mortality rates [1]. Idiopathic pulmonary fibrosis (IPF) has a worse 5-year survival rate (25%) than many cancers [2]. Treatments such as oxygen therapy and anxiolytics/opiates have been used to reduce breathlessness and cough [3], but currently, the only disease-modifying therapeutic options are the anti-fibrotic agents, pirfenidone and nintedanib [4]. These agents can slow declining lung function in some patients [5], but do not directly target IPF comorbidities [5,6] or significantly improve key patient-reported outcomes, such as quality of life [7], likely due to the common side effects [8,9]. There is currently no cure for IPF, and lung transplantation still represents the best therapeutic option, which restores lung function and improves survival [5,6]. However, most patients are precluded from transplantation due to their advanced age/frailty and the presence of comorbidities, such as pulmonary hypertension (PH) and lung cancer [5,6,10]. Thus, there is an urgent need to develop new disease-modifying therapeutics with fewer side effects.
The development of lung fibrosis involves the activation of multiple cell types and a complex network of signaling cascades, many of which are associated with G protein-coupled receptors (GPCR) [11]. Although the initiating events leading to IPF are poorly understood, the consensus is that the disease occurs in genetically susceptible individuals subjected to certain environmental stressors [12]. The lungs are exposed to the external environment, containing multiple harmful agents including, but not limited to, pathogenic microorganisms, toxic fumes, and pollutants. This interaction is thought to establish a continuous cycle of unresolved epithelial injury, stimulating a pro-fibrotic milieu, at the center of which is the cytokine, transforming growth factor beta (TGFβ), which is essential to the repair process, attendant fibroblast activation and excessive extracellular matrix (ECM) deposition [13,14].
In IPF, activated fibroblasts require an upregulation of key metabolic pathways to meet the demands of increased pro-fibrotic activity [15]. Specifically, the glutamine and arginine metabolism pathways are responsible for the increased expression of non-essential amino acids, glycine and proline, required for collagen synthesis, and polyamines such as putrescine and spermidine, which promote cell survival and growth [16,17,18]. These changes in metabolism are likely due to increased arginase expression or activity, which have been previously reported in human IPF lung tissue and the bleomycin model of mouse lung fibrosis [19,20].
The extracellular calcium/cation-sensing receptor (CaSR) is a member of the group C GPCRs. Named after its main physiological ligand, extracellular Ca2+ (Ca2+o), the CaSR is highly expressed in calcitropic organs (such as the parathyroid glands, kidneys, and bone), where it detects changes in the extracellular free ionized extracellular Ca2+ concentration ([Ca2+]o) [19,20]. CaSR is also expressed by non-calcitropic organs, including, but not limited to, the skin [21], vasculature [22], heart [23], and lungs [24], where it regulates differentiation, proliferation, inflammation, secretion, and contractility.
During fetal development, the CaSR promotes lung expansion by regulating fluid secretion and branching morphogenesis [25,26,27]. In the adult lung, the CaSR acts as a multi-modal chemosensor for components of environmental pollutants (e.g., Ni2+ and Cd2+), in addition to bacterial and viral infections (e.g., polyamines) [24,28,29]. CaSR is also activated by various endogenous polycations (e.g., eosinophil cationic protein, major basic protein), polyamines (e.g., putrescine, spermine, and spermidine), glutathione, and basic amino acids (e.g., L-arginine and L-ornithine), which are particularly abundant in the lung [28,30] and whose expression is increased in IPF [18].
CaSR activation leads to the induction of various signaling pathways associated with IPF pathogenesis, including increases in intracellular Ca2+ concentration ([Ca2+]i) [28] and Rho kinase-mediated actin stress fiber assembly [31], activation of ERK1/2 and PI3K/Akt [32,33], and a reduction in intracellular cyclic adenosine monophosphate (cAMP) concentrations [24]. Previously, CaSR signaling has been implicated in the proliferative and remodeling processes of diseases, such as asthma [24], cardiac fibrosis [22], pulmonary arterial hypertension (PAH) [34], and chronic obstructive pulmonary disease (COPD) [35], some of which are common IPF comorbidities [2,36,37].
This study investigates the involvement of polyamines in IPF pathogenesis and the ability of NPS2143, a CaSR-negative allosteric modulator (NAM), to suppress TGFβ1-induced pro-fibrotic effects in normal and IPF human lung fibroblasts (HLFs).
2. Methods
2.1. Human Studies
Flow Infusion Electrospray High-Resolution Mass Spectrometry (FIE-HRMS)
The study “Novel Technologies for Diagnosing and Monitoring Respiratory Diseases” obtained regional ethical approval from Hywel Dda University Health Board. This study was conducted in accordance with the Helsinki principles. Written informed consent was obtained from all participants. Eligibility included a diagnosis of IPF/UIP at the time of sample collection, which was determined at regional respiratory multidisciplinary team meetings. Control saliva samples were collected from spouses accompanying patients attending clinics with no IPF/UIP diagnosis.
FIE-HRMS was used to evaluate the potential of saliva as a source of non-invasive polyamine biomarkers for IPF. Spontaneous saliva was collected and processed from 6 patients with IPF and 6 control participants. The clinical characteristics of sample donors are summarized in Table S1.
Samples were processed using methanol and chloroform (4:1) and analyzed by FIE-HRMS, as described previously [38]. FIE-HRMS was performed using the Q Executive Plus Mass Analyzer Instrument with the UHPLC System (ThermoFisher Scientific, Bremen, Germany), where mass/charge (m/z) were generated in positive and negative ionization mode in a single run. All samples were run in duplicate with a blank of the reagents; no significant differences in the results were obtained. Metabolomic data were log-transformed with MetaboAnalyst (v4.0), using R (v3.5.1) and Bioconductor packages (v3.7). Statistical significance was assessed, and box plots were generated using GraphPad Prism 10.0.
2.2. In Vitro Studies
2.2.1. Cell Culture
Primary human lung fibroblasts (HLFs) from healthy donors (CC-2612) were purchased from Lonza (Slough, UK).
Lung tissue samples from IPF patients undergoing lung transplantation were obtained from the International Institute for the Advancement of Medicine through collaborations with the UT Health Houston Pulmonary Center of Excellence biorepository led by Dr. Karmouty-Quintana (Houston, TX, USA). IPF primary human lung fibroblasts were isolated by explant outgrowth from cryopreserved distal airway-free lung tissue, i.e., parenchymal fibroblasts, according to published protocols [39,40].
HLFs were cultured in Lonza custom Fibroblast Growth Medium (FGM-2), containing basal medium (FBM) (CC-3131) and FGM SingleQuots supplements (CC-4126): 0.1% insulin, 0.1% human fibroblast growth factor-B (hFGF-B), 2% fetal bovine serum (FBS), and 0.1% gentamicin and amphotericin-B solution, according to the protocol provided by Lonza. Cells were maintained at 37 °C under humidified conditions in a 5% CO2 incubator, with media changes every 48–72 h.
2.2.2. HLF Stimulation and Treatment
Once HLFs reached ~60% confluence, the growth medium was replaced with an experimental medium (FBM, 0.1% insulin, 0.1% hFGF-B, and 0.1% FBS) for 24 h. Next, the medium was changed to an experimental medium supplemented with one of the following treatments: (1) vehicle control: 0.01% dimethyl sulfoxide (DMSO, #D2650; Sigma-Aldrich, Merck, Dorset, UK); (2) CaSR negative allosteric modulator (NAM): treated with 1 µM NPS2143 (#ab145050; Abcam, Cambridge, UK); (3) TGFβ1: treated with 5 ng/mL TGFβ1 (#T7039; Sigma-Aldrich); (4) TGFβ1 + NAM: pre-treated with 1 µM NPS2143 for 4 h, followed by 5 ng/mL TGFβ1. Cells were maintained at 37 °C under humidified conditions in a 5% CO2 incubator for 72 h. Cells in the vehicle control group were used for the measurements of [Ca2+]i.
2.2.3. Immunofluorescence Staining
HLFs were cultured using the protocol described above. Following treatment, cells were washed with PBS and fixed with 4% paraformaldehyde for 10 min at room temperature. The cells were permeabilized with 0.3% Triton X-100 for 10 min at room temperature. Non-specific antibody binding was prevented with blocking buffer (2% donkey serum and 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for 30 min. The cells were incubated with the relevant primary antibodies diluted in a blocking buffer at 4 °C overnight. Primary antibodies included mouse anti-human CaSR (5C10-ADD) monoclonal antibody, 1:500 (#ab19347; Abcam, Cambridge, UK); mouse anti-human αSMA monoclonal antibody, 1:500 (#ab7817; Abcam); or rabbit anti-human collagen I, 1:500 (#ab34710; Abcam). After washing with PBS, secondary antibodies raised in donkey (anti-mouse IgG AlexaFluor-488, 1:1000, #A21202 or anti-rabbit AlexaFluor-488, 1:1000, #A21206, ThermoFisher Scientific, Loughborough, UK) were applied for 1 h and incubated in the dark. After washing, cell nuclei were counterstained with DAPI (#62248; ThermoFisher Scientific) for 5 min and washed with PBS before mounting. Negative control staining was performed by omitting primary antibodies or replacing the CaSR antibody with an IgG2a isotype control (mouse, 1:100, #02-6200; ThermoFisher Scientific) (Figure S1). Images were captured using a Zeiss LSM 710 Confocal Fluorescence Microscope (Carl Zeiss Microscopy Ltd., Cambourne, UK).
2.2.4. Quantitative Immunofluorescence Microscopy
Fluorescence intensities were quantified using StrataQuest (v7.0)® (TissueGnostics GmbH, Vienna, Austria). Images were pre-processed to extract inputs using Gaussian smoothing, object boundary erosion, and background subtraction. To ensure only positively stained areas containing a nucleus were used for building measurement masks, nuclear object detection was performed first. This involved setting a mask to identify nuclei of approximately 42 µm2 from the DAPI channel of each image using the nuclear segmentation function within StrataQuest (v7.0). Measurement masks were then generated by overlaying the nuclei mask on the 488 nm channel of each image to permit the mask to “grow” in all directions, until encountering either a break in marker immunoreactivity or interaction with the measurement mask of an adjacent cell (maximum growing steps: 33 µm; skip steps: 0 µm), as illustrated in Figure S2. All objects and measurement masks were detected using thresholds, based on the greyscale values corresponding to the lowest and highest marker intensities confidently recognized as positive staining, with the observer blinded to the experimental conditions. Mean intensity values were calculated from greyscale fluorescence intensity values within the measurement masks for each image. All measurements were then normalized to the number of cells in that region of interest.
2.2.5. Measurements of Intracellular Free Ionized Calcium Concentration ([Ca2+]i)
Single-cell fluorescence Ca2+ imaging experiments were performed using a monochromator-based fluorimeter system (OptoFluor; Cairn Research, Faversham, UK) and the ratiometric dye, Fura-2 Acetoxymethyl (Fura-2 AM) ester. All intensities were background-subtracted before the 340:380 nm emission ratio was used to calculate maximal fold-change from baseline. CaSR-induced changes in [Ca2+]i were measured in Fura-2 loaded NHLFs, as described previously [24].
2.2.6. RNA Sequencing
Total RNA was extracted and purified using the RNeasy Mini Kit, including the optional on-column DNase digestion (Qiagen, Manchester, UK), according to the manufacturer’s protocol. RNA was quantitated using a Qubit RNA BR Assay Kit and Qubit® fluorometer (Fisher Fluorometer, ThermoFisher Scientific), and RNA integrity number (RIN) for each sample was measured using an Agilent 2200 TapeStation (Agilent Technologies, Stockport, UK), according to the manufacturer’s instructions. Only samples with RIN > 9.5 were used for library preparation.
Sequencing libraries were prepared using Illumina TruSeq Stranded Total RNA Kit (Illumina, Cambridge, UK). Dual Indexes were introduced using the Illumina Truseq RNA UD indexing plate to ensure accurate library sequencing. The resulting libraries were sequenced on the Illumina NextSeq 500 (Illumina) to generate 75 base-pair single-ended reads. A total of ~500 million reads were obtained across 12 samples. Each condition was sequenced in triplicate (different donors per replicate).
Reads were quality-checked with FastQC [41], trimmed with Trimmomatic [42], and aligned to the human reference genome version GRCh38.101 reference genome [43] using Spliced Transcripts Alignment to a Reference (STAR) package [44] version 2.7.3a. Duplicates were marked/removed using Picard (v2.12), and the raw reads were counted by FeatureCounts (v1.6.3) [45]. An additional quality control step was carried out using SortMeRNA (v4.0) [46], and differential gene expression was calculated using the SARTools (v1.7.3) pipeline [47] in DESeq2 (v1.24.0) [48]. ClusterProfiler (4.14.4) package in R (v3.5.1) was used to conduct the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentially expressed genes (DEGs) [49,50,51,52]. The ReactomePA (1.50.0) package was performed to identify comprehensive pathway information [53]. p adjusted values (padj) < 0.05 was used as a cut-off. Heatmaps were generated using the pheatmap (1.0.12) package in R (v3.5.1) [54].
The changes in gene expression were calculated for pairwise group comparisons, and the padj values were used to account for the false discovery rate using the Benjamini–Hochberg correction. Quantification, statistical analysis, and figure generation were performed using R (v3.5.1) [55] and Prism 10 (GraphPad Inc., La Jolla, CA, USA).
2.2.7. Polyamine Secretion
Ornithine ELISA
Ornithine was measured in supernatants of HLFs grown in culture using the competitive ELISA method, according to the manufacturer’s instructions (ImmuSmol, Bordeaux, France). Absorbance values were measured spectrophotometrically at a wavelength of 450 nm using a FLUOstar Omega Spectrophotometer Microplate Reader (BMG Labtech, Aylesbury, UK).
Spermine ELISA
Spermine was measured by using the competitive ELISA method in supernatants and cell lysates of HLFs, according to the manufacturer’s instructions (FineTest, Wuhan, China). Absorbance values were measured spectrophotometrically at a wavelength of 450 nm, as described above.
2.3. Statistics
Data were analyzed using the Prism 10 software package (GraphPad Inc.) and are expressed as the mean ± SD. Statistical comparisons were performed using the 2-tailed Student’s t-test or ANOVA (with Sidak’s post hoc analysis). p values of <0.05 were considered significant. N denotes the number of donors (biological replicates), while n represents the number of independent experiments (technical replicates). RNA sequencing data were analyzed using the R package, DESeq2 (v1.24.0). Adjusted p values (padj) accounted for the false discovery rate using the Benjamini–Hochberg correction; the significance cut-off used for padj values was 0.05. Quantification and statistical analysis were performed using R (v3.5.1) and Prism 10.
3. Results
3.1. Naturally Occurring Polyamines Are Enriched in the Salivary Metabolome of IPF Patients
Assessment of the variation between clusters indicated the prominence of the arginine–polyamine pathway in IPF (Figure 1A). Principal component analysis (PCA) on saliva samples demonstrated clear clustering of the IPF group, distinct from a more diffuse control group (Figure 1B). This result suggests a difference in the metabolome of IPF and controls. Plotting the loading vectors showed the importance of arginine and ornithine in driving the separation between the experimental groups; other polyamines were also relevant but showed smaller contributions (Figure S3). Box and whisker plotting of individual metabolites showed that IPF saliva samples contained elevated levels of arginine (p = 0.03), ornithine (p = 0.04), putrescine (p = 0.01), and spermidine (p = 0.06), compared with controls (Figure 1C). However, spermine enrichment levels were not significantly altered (p = 0.40).
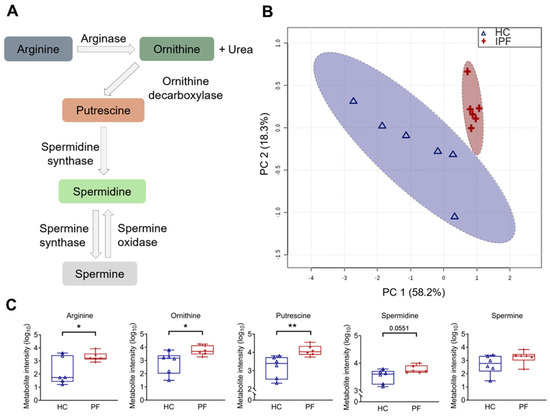
Figure 1.
Arginine pathway metabolites are increased in idiopathic pulmonary fibrosis (IPF) patient saliva samples. (A) Diagram representing the arginine–polyamine metabolic pathway. (B) Principal component analysis (PCA) score plot between the main principal components (PC1 and PC2) shows a tight cluster of the IPF samples based on their metabolic profile compared to controls, with a 95% confidence ellipse drawn for each group (shown by the dotted lines). Assessment of PC1 and PC2, which account for 76.4% of the total variation between the control (HC) and IPF clusters, highlights the prominence of polyamines. (C) Associated box and whisker plots of metabolites in the arginine–polyamine pathway highlight the differences between healthy and IPF profiles. Y-axis: Log10 normalization of metabolite intensity. Statistical analysis was performed with unpaired two-tailed Student t-test; * p < 0.05, ** p < 0.01. N = 6 controls; 6 IPF patients.
3.2. CaSR Is Functionally Expressed in Normal Human Lung Fibroblasts (NHLFs)
Since fibroblasts are key mediators of the pro-fibrotic response in IPF and intracellular Ca2+ signaling underpins several pro-fibrotic processes [56], we explored the ability of polyamines to activate the CaSR in NHLFs. Protein expression of CaSR in untreated NHLFs was confirmed by immunofluorescence (Figure 2A). The functionality of this CaSR expression was then confirmed by observing the response of these fibroblasts to increased [Ca2+]o from baseline 1.2 mM to 5 mM (Figure 2B,Ci). Figure 2Ci shows that, as expected, increasing [Ca2+]o evoked an increase in [Ca2+]i, an effect that was abrogated by a CaSR NAM (Figure 2B,Cii).
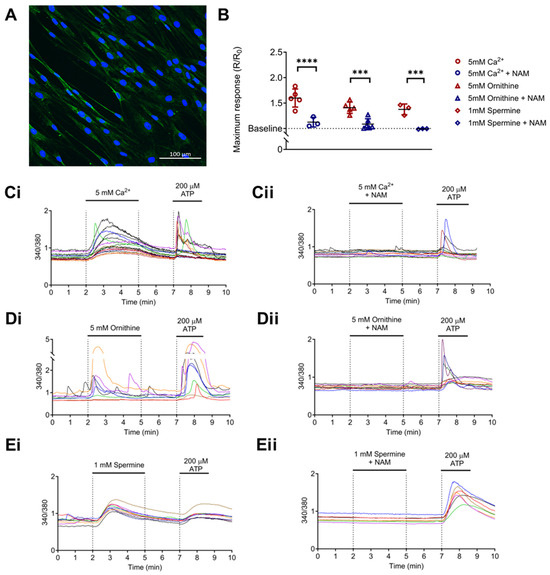
Figure 2.
Calcium-sensing receptor (CaSR) is functionally expressed in normal human lung fibroblasts (NHLFs) and is activated by polyamines upregulated in IPF. (A) Representative image of CaSR expression in NHLF (green), nuclei (blue). Scale bar: 100 µm. (B) Summary data of intracellular calcium levels ([Ca2+]i) in NHLF in response to: (Ci) 5 mM Ca2+ (divalent cation); (Di) 5 mM L-ornithine (basic amino acid); and (Ei) 1 mM spermine (polyamine). (B,Cii–Eii) Treatment with a CaSR negative allosteric modulator (CaSR NAM) prevents these increases in [Ca2+]i. (C–E) Representative traces of NHLF [Ca2+]i response to CaSR activators. Summary data are presented as mean ± SD. Statistical analysis was performed using one-way ANOVA (with Sidak’s post hoc test); *** p < 0.001, **** p < 0.0001. 5 mM Ca2+ (n = 3–6; 108 cells), 5 mM L-ornithine (n = 5–6; 109 cells), and 1 mM spermine (n = 3; 39 cells). n = 3–6 independent experiments. CaSR negative allosteric modulator: NAM, NPS2143 (1 μM).
As seen in Figure 2Di,Ei, both 5 mM ornithine (basic amino acid; Figure 2Di) and 1 mM spermine (polyamine; Figure 2Ei) evoked a significant rise in [Ca2+]i from baseline, effects that were again abolished in the presence of NAM (NPS2143, 1 µM; p = 0.0001 and p = 0.0005, respectively; Figure 2B,Dii,Eii). These results are consistent with the hypotheses that NHLFs express functional CaSR, which can be activated by arginine pathway metabolites, and this effect is abolished by the CaSR NAM, NPS2143.
3.3. NHLFs Treated with TGFβ1 Display a Pro-Fibrotic Genetic Profile and Co-Treatment with CaSR NAM Alters TGFβ1-Mediated Changes
TGFβ1 is used in vitro to induce fibrotic phenotypes in fibroblasts of various origins [57]. We used RNA sequencing to assess the suitability of TGFβ1 treatment of NHLFs as an in vitro model of IPF and examined the effects of the CaSR NAM, NPS2143, on relevant biological pathways.
TGFβ1 treatment significantly differentially expressed 4756 genes compared to unstimulated fibroblasts (vehicle control), with 2757 genes upregulated and 1999 genes downregulated (Figure 3A). Treating the fibroblasts with NAM alone did not induce any significant changes in gene expression (Figure 3B). Co-treating NHLFs with TGFβ1 and NAM significantly upregulated 2150 genes whilst downregulating 2608 genes when compared to TGFβ1-treated fibroblasts (Figure 3C).
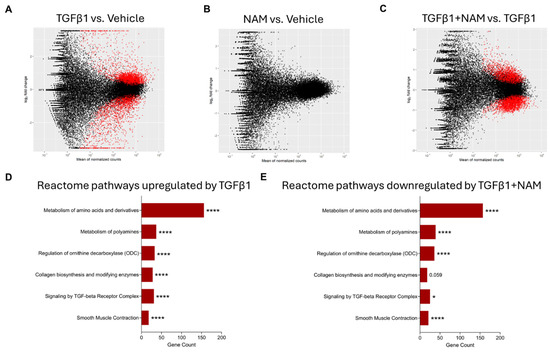
Figure 3.
Enrichment analysis of differentially expressed genes (DEGs) from RNA sequencing (RNAseq) in primary human lung fibroblasts treated with vehicle (0.01% DMSO), CaSR NAM (1 μM NPS2143), TGFβ1 (5 ng/mL), and TGFβ1+ CaSR NAM for 72 h. The MA plots show the log2-fold change between experimental groups (M) against the mean expression across all the samples (A) for each gene. Statistically significant differentially expressed genes (DEGs) for each comparison are shown in red. The x-axis shows the mean of normalized gene counts; the y-axis shows log2 fold-changes in expression (positive values represent upregulated genes, while negative values denote downregulation). (A) MA plot of RNAseq showing the DEGs in TGFβ1 treated cells relative to unstimulated cells (vehicle control) cells. Red dots represent statistically significant DEGs (Down-DEGs: 1999; Up-DEGs: 2757), while black dots represent non-significantly altered genes. (B) No significant difference in gene expression was observed when the CaSR NAM-treated cells were compared with vehicle control cells. (C) MA plot showing the DEGs in cells co-treated with TGFβ1 + CaSR NAM relative to TGFβ1-treated cells. Red dots represent statistically significant DEGs (Down-DEG: 2608; Up-DEG: 2150). (D,E) Annotation of key Reactome pathways based on the enrichment analysis of the statistically significant DEGs. (D) Key Reactome pathways upregulated by TGFβ1 relative to vehicle control. (E) Key Reactome pathways downregulated by CaSR NAM in the presence of the fibrotic stimulus, TGFβ1, relative to TGFβ1 treatment alone. Benjamini–Hochberg p-value adjustment was performed for all statistical tests; level of controlled false positive rate was set to 0.05. * p < 0.05, **** p < 0.0001. N = 3 donors. NAM: Calcium-sensing receptor (CaSR) Negative Allosteric Modulator treatment; TGFβ1: Transforming growth factor β1 (TGFβ1) treatment; TGFβ1 + NAM: TGFβ1 and CaSR Negative Allosteric Modulator co-treatment.
KEGG enrichment analysis of differentially expressed genes showed that TGFβ1 stimulation upregulated pathways associated with metabolic processes (such as metabolism of amino acids and derivatives, and polyamines), ECM organization, collagen biosynthesis, assembly and degradation, TGFβ signaling, and smooth muscle contraction (Figure S4). Co-treatment of fibroblasts with TGFβ1 and CaSR NAM downregulated pathways associated with amino acid and polyamine metabolism, collagen biosynthesis, TGFβ signaling, and smooth muscle contraction.
3.4. CaSR Regulates Polyamine Gene Expression in TGFβ1-Activated NHLF
In the presence of fibrotic stimuli, fibroblasts rely on several metabolic pathways to provide substrates (such as non-essential amino acids and polyamines) required for collagen synthesis [19,58] and growth [59]. We used RNA sequencing to examine the effects of TGFβ1 and the CaSR NAM, NPS2143, on polyamine metabolism gene expression.
TGFβ1 increased the expression of several amino acid and polyamine metabolic genes (Figure 4A,B) compared with the vehicle control. A panel of the full gene set is presented in Figure S5. Exposure of NHLF to TGFβ1 resulted in a significant increase in the expression of key genes involved in the proline–polyamine pathway, namely, glutaminase (GLS), pyrroline 5-carboxylate synthase (ALDH18A1), and ornithine aminotransferase (OAT), with concomitant downregulation of glutamine synthase (GLUL) and pyrroline 5-carboxylate dehydrogenase (ALDH4A1), which favor the conversion of pyrroline 5-carboxylate back into glutamine (Figure 4). In the presence of TGFβ1, CaSR NAM decreased GLS expression (padj value = 0.02) and restored baseline expression of pyrroline 5-carboxylate reductase (PYCR2; padj value = 0.003) and OAT (padj value = 0.003) (Figure 4B).
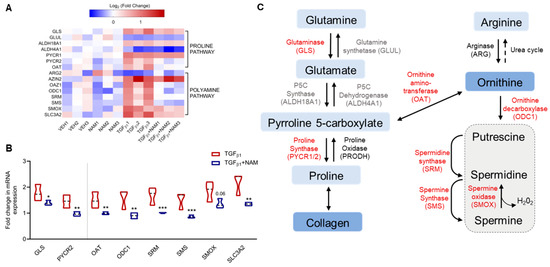
Figure 4.
TGFβ1 upregulates genes involved in proline and polyamine metabolism, an effect abrogated by the CaSR NAM, NPS2143, in normal human lung fibroblasts. Fibroblasts were treated with vehicle (0.01% DMSO), CaSR NAM (1 μM NPS2143), TGFβ1 (5 ng/mL), and TGFβ1 + CaSR NAM for 72 h. (A) Heatmap generated from RNA sequencing data showing the log2 ratio of differentially expressed genes (versus vehicle) implicated in these metabolic pathways. (B) RNA sequencing data show exogenous TGFβ1 treatment upregulates genes associated with proline synthesis and polyamine metabolism, while co-treatment with the CaSR NAM, NPS2143 downregulates gene expression, except for GLUL, ALDH4A1, ARG2, and ALDH18A1. (C) CaSR NAM restores TGFβ1-induced increase in polyamine metabolism to baseline. The genes in red are upregulated by TGFβ1 and downregulated by TGFβ1 and CaSR NAM co-treatment; genes in black were differentially expressed in the presence of TGFβ1, but CaSR NAM co-treatment had no effect on their expression; genes in grey were not affected by either treatment. Genes encoding glutaminase: GLS, glutamine synthase: GLUL, pyrroline 5-carboxylate synthase: ALDH18A1, pyrroline 5-carboxylate dehydrogenase: ALDH4A1, pyrroline 5-carboxylate reductase/proline synthase: PYCR, proline oxidase: PRODH, arginase: ARG, ornithine aminotransferase: OAT, antizyme inhibitor: AZIN, antizyme: OAZ1, ornithine decarboxylase: ODC, spermidine synthase: SRM, spermine synthase: SMS, and spermine oxidase: SMOX, polyamine transporter-Solute Carrier Family 3 Member 2 (SLC3A2) are involved in these pathways. Data in the violin plots represent median fold change, truncated at minimum and maximum values. Benjamini–Hochberg p-value adjustment was performed for all statistical tests; level of controlled false positive rate was set to 0.05. * p < 0.05, ** p < 0.01, *** p < 0.001. N = 3 donors. CaSR negative allosteric modulator (NAM); NPS2143 (1 μM); Vehicle (VEH).
Since ornithine is a precursor for polyamine synthesis, we explored this pathway in more detail. In the presence of TGFβ1, the NHLF exhibited a minimum 1.5-fold increase in expression of genes encoding the enzymes in the polyamine pathway, except for arginase (ARG2), which was not significantly altered (fold change = −0.5; padj value > 0.05). Baseline expression of these genes was restored in the presence of the CaSR NAM and TGFβ1: ornithine decarboxylase 1 (ODC1; padj value = 0.002), ornithine decarboxylase antizyme 1 (OAZ1; padj value = 0.0002), spermidine synthase (SRM; padj value = 0.0003), and spermine synthase (SMS; padj value = 0.0001) (Figure 4A,B).
The expression of mRNA encoding spermine oxidase (SMOX), which converts spermine back to spermidine, was also significantly elevated in the presence of TGFβ1 (fold change = 1.9; padj value = 0.0002), as was that of mRNA encoding antizyme inhibitor (AZIN2) (fold change = 1.4; padj value < 0.0001; Figure 4A,B), which positively regulates ornithine decarboxylase activity. Co-treatment with the CaSR NAM restored the baseline expression of SMOX (padj value = 0.06); however, AZIN2 expression was unchanged. Expression of the polyamine transporter (SLC3A2) increased with TGFβ1 treatment (padj value < 0.0001; Figure 4A,B), while co-treatment with CaSR NAM reduced SLC3A2 gene expression (padj value = 0.002; Figure 4B). Treatment with CaSR NAM alone had no significant effect on the expression of the aforementioned genes (Figure S5).
3.5. CaSR Regulates Pro-Fibrotic Gene Expression in TGFβ1-Activated NHLF
Cytoskeletal changes and excessive collagen and ECM deposition are central to IPF pathophysiology [60]. Our RNAseq data demonstrated that genes implicated in these pathways are enriched by TGFβ1 treatment in vitro (Figure 5A). Exogenous TGFβ1 treatment upregulated TGFB1 gene expression (fold change = 1.4; padj value < 0.0001), while co-treatment with CaSR NAM reduced its expression (fold change = −0.6; padj value < 0.0001; Figure 5A,B). Expression of the cytoskeletal gene encoding α-smooth muscle actin (ACTA2) was doubled in the presence of TGFβ1 (padj value < 0.0001; Figure 5B). Co-treatment with CaSR NAM completely abolished this pro-fibrotic effect (padj < 0.0001; Figure 5B). Several collagen subtypes and genes that maintain collagen fiber integrity and stability were upregulated by TGFβ1 application, e.g., collagens COL1A1, COL3A1, COL5A1, and prolyl 4-hydroxylase, alpha polypeptide II (P4HA2) (padj value < 0.05; Figure 5A,B). COL4A1 and COL7A1, which are involved in basement membrane integrity, were also upregulated by TGFβ1. Lastly, key ECM-related genes were upregulated by pro-fibrotic stimuli, including COL6A2, fibronectin (FN1), elastin (ELN), matrix-metalloproteinases (MMP1, MMP2, MMP14), tissue inhibitors of metalloproteinases (TIMP1, TIMP2, TIMP3), and serine protease inhibitor E1 (SERPINE1) (padj value < 0.05; Figure 5A and Figure S6). Co-treatment with CaSR NAM reduced the expression of these genes except COL4A1 (padj value < 0.05; Figure 5A,B).
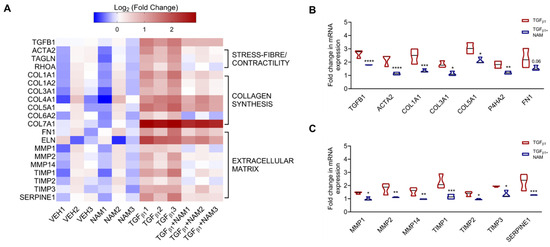
Figure 5.
CaSR negative allosteric modulator, NPS2143, attenuates TGFβ1-induced upregulation of pro-fibrotic genes in normal human lung fibroblasts. NHLFs were treated with vehicle (0.01% DMSO), CaSR NAM (1 μM NPS2143), TGFβ1 (5 ng/mL), and TGFβ1 + CaSR NAM for 72 h. (A) Heatmap generated from RNA sequencing data showing the log2 ratio of differentially expressed genes (versus vehicle). Exogenous TGFβ1 application upregulates genes related to fibroblast contractility, collagen synthesis, and extracellular matrix (ECM) remodeling. (B) CaSR NAM reduces the expression of genes associated with fibroblast contractility, collagen synthesis and assembly; and (C) ECM remodeling and maintenance. Genes encoding Transforming Growth Factor-β1: TGFB1, α-smooth muscle actin: ACTA2, smooth muscle-22α: TAGLN, ras homolog family member A: RHOA, collagen: COL, prolyl 4-hydroxylase, alpha polypeptide II: P4HA2, fibronectin: FN1, elastin: ELN, matrix-metalloproteinases: MMP, tissue inhibitors of metalloproteinases: TIMP, and serine protease inhibitor E1: SERPINE1 are involved in these pathways. Data in the violin plots represent median fold change, truncated at minimum and maximum values. Benjamini–Hochberg p-value adjustment was performed in all statistical tests; level of controlled false positive rate was set to 0.05. * padj <0.05, ** padj <0.01, *** padj <0.001, **** padj <0.0001. N = 3 donors. CaSR negative allosteric modulator: NAM, NPS2143 (1 μM).
3.6. Effects of TGFβ1 on CaSR Expression in Normal and IPF Primary Lung Fibroblasts
Next, we investigated the effects of TGFβ1 and NAM on CaSR protein expression in normal and IPF HLFs (Figure 6). In the fibroblasts from normal control subjects, CaSR expression doubled in the presence of TGFβ1 (p = 0.03), while receptor expression was reduced when co-treated with CaSR NAM. In IPF HLFs, CaSR expression was maintained at a comparable level across all treatment conditions. These results suggest that CaSR expression is upregulated in response to pro-fibrotic stimulus in healthy HLF, an effect prevented by a CaSR NAM. Baseline CaSR expression was unaffected by NAM treatment in the absence of TGFβ1 co-stimulation.
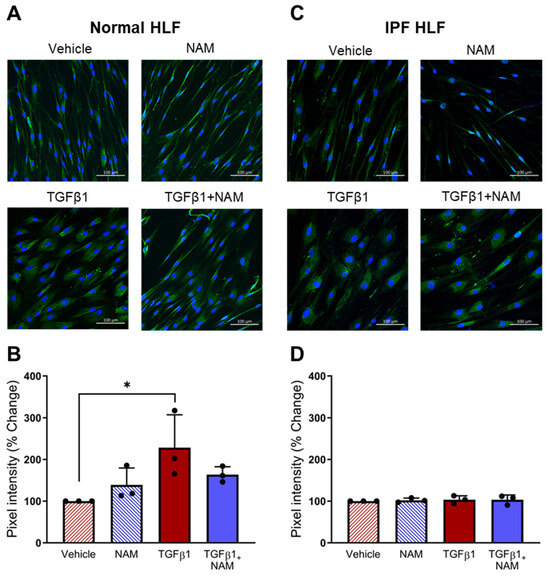
Figure 6.
CaSR is expressed in normal human lung fibroblasts (NHLFs) and IPF human lung fibroblasts (IPF HLFs). (A) Representative images showing CaSR expression when NHLFs were treated with vehicle (0.01% DMSO), CaSR NAM (1 μM NPS2143), TGFβ1 (5 ng/mL), and TGFβ1 + CaSR NAM for 72 h. (B) Image quantification analysis with bespoke software, StrataQuest, shows that TGFβ1 treatment increases CaSR expression in NHLFs. (C) Representative images showing CaSR expression in IPF HLFs in the same treatment conditions. (D) Image quantification with StrataQuest shows CaSR expression is maintained in IPF HLFs in all treatment conditions. Data are presented as mean ± SD. Statistical analysis was performed using one-way ANOVA (with Sidak’s post hoc test); * p < 0.05. N = 3 donors; n = 3 independent experiments. Scale bar: 100 µm. CaSR negative allosteric modulator: NAM.
3.7. TGFβ1 Upregulates Polyamine Production in Normal and IPF HLF, and CaSR Antagonism Abrogates These Responses
Given that polyamine metabolites are enriched in IPF saliva samples and their gene transcripts are increased in the presence of pro-fibrotic stimuli, we investigated whether exposure of normal and IPF fibroblasts to TGFβ1 could directly increase the expression of polyamines.
Exogenous treatment of NHLFs and IPF HLFs with TGFβ1 induced ornithine secretion; CaSR NAM co-treatment reduced ornithine secretion in IPF fibroblasts by 65% (p = 0.0003) (Figure 7A,B). Due to the low concentration of spermine secreted in all treatment conditions (Figure 7C,D), we investigated the intracellular concentration of spermine in normal and IPF HLFs. TGFβ1 increased spermine concentration in both normal and diseased fibroblasts (p < 0.05). In both circumstances, co-treatment with CaSR NAM restored baseline levels of spermine (p < 0.05; Figure 7E,F).
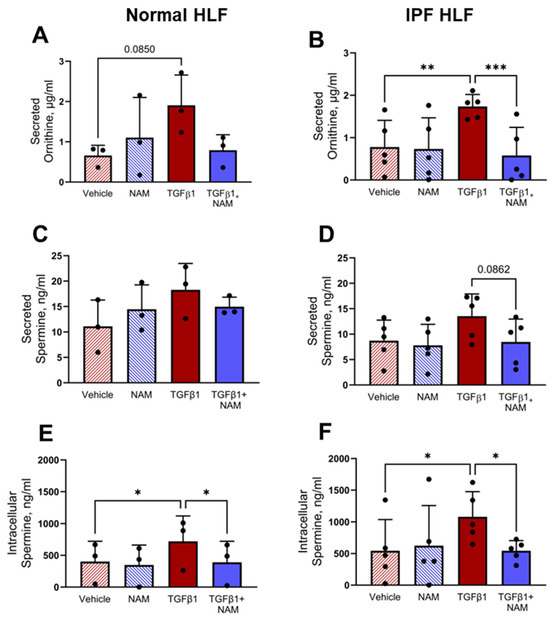
Figure 7.
TGFβ1-induced polyamine expression is abrogated by CaSR NAM in normal and IPF human lung fibroblasts (HLFs). (A,B) Ornithine secretion by normal HLFs and IPF HLFs is increased by TGFβ1 treatment, and the response is attenuated in the presence of CaSR NAM. (C,D) Low concentration of spermine is secreted by normal HLFs and IPF HLFs, with levels remaining relatively unchanged across the treatment conditions. (E,F) Intracellular spermine concentration is increased by exogenous TGFβ1 treatment in normal HLFs and IPF HLFs; co-treatment with CaSR NAM restores vehicle control levels of the polyamine. Data are presented as mean ± SD. Statistical analysis was performed using one-way ANOVA (with Sidak’s post hoc test); * p < 0.05, ** p < 0.01, *** p < 0.001. N = 3 donors; n = 3–6 independent experiments. CaSR negative allosteric modulator: NAM, NPS2143 (1 μM).
3.8. CaSR Antagonism Reduces TGFβ1 Pro-Fibrotic Responses in IPF Lung Fibroblasts
We also assessed the expression of pro-fibrotic markers implicated in IPF. TGFβ1 treatment increased αSMA expression (p < 0.0001) and collagen expression (p = 0.03), compared with vehicle control (Figure 8). CaSR NAM co-treatment reduced αSMA protein expression (p < 0.0001; Figure 8A,B). While CaSR NAM treatment did not reduce total collagen expression, it altered its expression pattern from diffuse to concentrated (Figure 8C,D) and modified the morphology of the cells from large and extended (associated with more secretory and contractile functions) to smaller, spindle-shaped morphology, typical of inactive fibroblasts [61] (Figure S7).
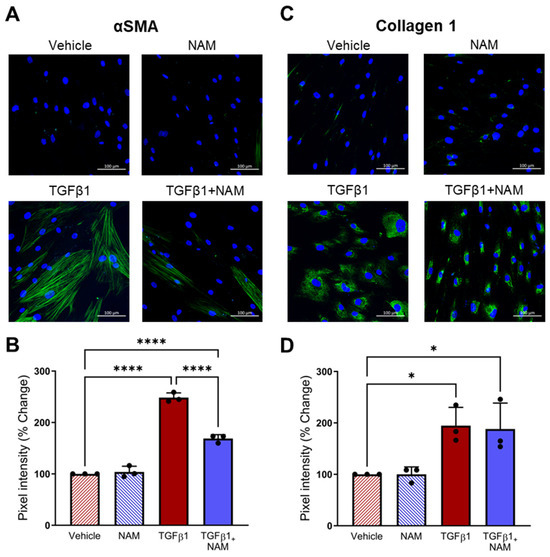
Figure 8.
CaSR NAM reduces TGFβ1-induced pro-fibrotic changes in IPF human lung fibroblasts (HLFs). IPF HLFs were treated with vehicle (0.01% DMSO), CaSR NAM (1 μM NPS2143), TGFβ1 (5 ng/mL), and TGFβ1 + CaSR NAM for 72 h. (A) Representative images showing that TGFβ1 treatment increases alpha smooth muscle actin (αSMA) expression and stress-fibre formation (green), a response abrogated by the CaSR NAM. (B) Image quantification analysis shows that TGFβ1-induced increase in αSMA expression is halved in the presence of CaSR NAM. (C) Representative images showing diffuse Collagen 1 expression in IPF HLFs treated with TGFβ1. Co-treatment with CaSR NAM appears to reduce cell area (see Figure S7). (D) Image quantification confirms increased collagen 1 expression in IPF HLFs treated with TGFβ1. Data are presented as mean ± SD. Statistical analysis was performed using one-way ANOVA (with Sidak’s post hoc test); * p < 0.05, **** p < 0.0001. N = 3 donors; n = 3 independent experiments. Scale bar: 100 µm. CaSR negative allosteric modulator: NAM.
4. Discussion
In recent years, arginine pathway metabolites have emerged as important biomarkers in the pathogenesis of lung disease [62,63,64,65,66], but whether polyamines actively participate in IPF pathogenesis or are mainly bystanders remains unclear. Our studies show that: (1) polyamine levels are increased in the saliva of IPF patients; (2) polyamines directly act at the CaSR, a receptor expressed in normal and IPF human lung fibroblasts, leading to a rise in intracellular calcium concentration; (3) TGFβ1 upregulates the expression of both polyamines and CaSR in NHLFs, and polyamines only in IPF fibroblasts; and (4) CaSR inhibition with NAM abrogates the key effects of pro-fibrotic stimuli in both normal and IPF HLFs, suggesting that the CaSR and NAM might hold therapeutic potential for the treatment of IPF. Furthermore, our studies indicate that the levels of expression of arginine pathway-derived polyamines could provide valuable non-invasive tools in the diagnosis and management of IPF.
Our findings show that the saliva of IPF patients is enriched in arginine pathway metabolites compared to healthy controls. This aligns with studies from other groups that report increased levels of circulating ornithine and ornithine-derived metabolites, putrescine and spermidine in IPF lung tissue [18,67]. These natural polyamines are established activators of the CaSR [28], which, in many cell types, elicit an increase in intracellular calcium [Ca2+]i via the activation of G protein signaling pathways [24]. Here, our results demonstrate that CaSR is functionally expressed by normal human lung fibroblasts, as confirmed by the use of the classical ligand for the receptor, calcium ions, which induced robust Ca2+ mobilization in HLFs. This response was abolished by co-treatment with NAM. Pertinently, this study shows that ornithine and spermine also activate the CaSR in fibroblasts, evoking similar increases in [Ca2+]i. Since increases in [Ca2+]i underpin key fibrotic processes, such as activating genes responsible for the production of TGFβ, collagen, and α-smooth muscle actin (αSMA) in fibroblasts [56], our findings suggest a potential pathophysiological interplay between polyamines, the CaSR and Ca2+i, which may contribute to IPF development.
Using TGFβ1-treated human lung fibroblasts as an in vitro model of IPF, we have shown that pro-fibrotic stimuli upregulate the expression of gene pathways associated with increases of cytosolic calcium concentrations and increased metabolism of amino acids. Polyamine expression and secretion are also increased in the presence of pro-fibrotic stimuli, which is consistent with our metabolomic data from patient saliva. Although ARG2 (encoding arginase) expression remained unchanged in our model, we observed significant alterations in the key genes involved in metabolizing all the polyamines in this pathway, i.e., ornithine, putrescine, spermidine, and spermine, which are increased in IPF patients [18]. These findings suggest that arginase activity may still be relevant, and future studies could investigate this at the protein level to clarify its potential role in this context. An alternative source of polyamines precursor, ornithine, can be synthetized from glutamate, and upregulated OAT (encoding ornithine aminotransferase) is likely to fulfill this role. Notably, in the pro-fibrotic context, CaSR negative allosteric modulation abrogates this increase in polyamines and the polyamine exporter, SLC3A2, at the transcriptomic level in lung fibroblasts from healthy subjects, and at the molecular level in lung fibroblasts from patients with IPF. Furthermore, treating fibroblasts with NAM on its own did not result in differential gene expression, which is consistent with NAMs acting as allosteric modulators and being inactive in the absence of ligands [68]. Since polyamines play an essential role in cellular growth, survival, and proliferation, they are attractive therapeutic targets not only in such highly proliferative diseases as cancer [69], but also in several unrelated lung diseases, such as asthma [24], pulmonary hypertension [70], and respiratory viral infections [66].
In asthma and pulmonary hypertension, direct activation of CaSR with the polyamines and/or polycationic proteins increases cytosolic calcium signaling, smooth muscle cell proliferation, and hyperresponsiveness, all of which are abolished by genetic or pharmacological inhibition of CaSR [24,35,70]. Notably, increased levels of polyamines are essential for virus–host interaction, viral invasion, and replication [66,71]. In addition, several pathogenic bacteria use polyamines to avoid host immune defenses and improve survival [72]. Several studies have also shown that cells infected by viruses associated with IPF exhibit increased glutamine utilization [73,74,75,76]. Upregulation of the glutamine metabolic pathway is crucial for collagen synthesis in IPF [17], with lower circulating levels of glutamine being associated with an increased risk of IPF [77]. Arguably, this decrease in circulating glutamine levels could be a result of the upregulation of downstream metabolic pathways such as proline and polyamine metabolism. Our results demonstrate that treatment with TGFβ1 upregulates genes associated with the glutamine degradation pathway, such as glutaminase, pyrroline 5-carboxylate synthase, and ornithine aminotransferase, while downregulating genes associated with glutamine synthesis, such as glutamine synthase and pyrroline 5-carboxylate dehydrogenase, thus favoring proline and ornithine biosynthesis instead of glutamine. Importantly, treatment with CaSR NAM restored the baseline expression of these genes in our in vitro IPF model, suggesting a potential role for the abovementioned metabolic dysregulation in IPF through the activation of CaSR. Furthermore, the polyamine-CaSR pathway might augment pro-fibrotic processes and facilitate infections, leading to a vicious cycle.
Fibrotic remodeling involves the perturbation of the balance between ECM deposition and degradation, with the scales tipped in favor of deposition [78]. Furthermore, TGFβ1 acts as a ‘master switch’ for the secretion of collagen and other ECM proteins from αSMA-expressing myofibroblasts [79]. Our RNAseq data demonstrates that exogenous TGFβ1 autoregulates its expression at the transcriptomic level, which can lead to an amplification of the pro-fibrotic signal. Several TGFβ1-mediated pathways are implicated in αSMA stress fiber formation, including ERK, Akt, and Rho-kinase activation [80], which are all signaling pathways downstream of the CaSR. The RNAseq data from our in vitro model of IPF demonstrate upregulation of TGFβ1-signaling pathways, collagen biosynthetic pathways, and genes implicated in smooth muscle contractility. Notably, the negative allosteric modulator of the CaSR, NPS2143, attenuated the upregulation of all the above pathways, targeting the key pathological hallmarks of IPF: transition to myofibroblast and contractility, collagen synthesis, ECM regulation, and altered cell morphology, suggesting the receptor plays a role in IPF pathogenesis.
Maintaining the structural integrity of the ECM also involves matrix proteinases, MMPs, and their inhibitor proteins, TIMPs, both of which are known to be increased in IPF [81]. Our results highlight enhanced expression of MMP1, responsible for fibrillar collagen, fibronectin, and elastin degradation, and MMP2, which participates in the degradation of the basement membrane [82]. The upregulation of MMPs (especially MMP1) may contribute to cleavage and activation of a plethora of cytokines, including pro-TNFα, pro-1L-1β, and IGFBP3 [83,84], which play a key role in IPF. Interestingly, extracellular MMP1 and 2 also activate their pro-peptides, creating a positive feedback loop, whilst intracellular MMP1 accumulates during mitosis, and inhibiting this enzyme resulted in a faster induction of apoptosis compared to untreated cells, suggesting that it might be important in maintaining the balance between cell division and cell death [85]. Lastly, the concomitant increase in the gene transcripts of protease inhibitors, TIMP1, TIMP3, and SERPINE1, suggest that CaSR may facilitate a profibrotic environment promoting a non-degrading collagen-rich ECM, which is a defining feature of human and animal models of lung fibrosis [86,87].
5. Conclusions
Our findings demonstrate that CaSR contributes to the pro-fibrotic responses in human lung fibroblasts, suggesting novel therapeutic tools for IPF management. Given that polyamines, elevated in the saliva of IPF patients and in the in vitro model of IPF, are the physiological CaSR ligands, this study also provides a potential mechanistic link between the receptor and the TGFβ1-induced upregulation of polyamine metabolism. Notably, as early diagnosis remains critical, our findings, together with patient preference for non-invasive disease monitoring methods, provide a rationale to explore whether polyamines in patient saliva could be used as a viable biomarker for disease detection, patient stratification, and prediction of prophylactic and therapeutic responses to CaSR NAM treatment for IPF.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biom15040509/s1, Table S1. Clinical and histopathological characteristics of metabolomic samples. Figure S1. Representative images showing immunofluorescence negative control staining; Figure S2. Representative images of in vitro immunofluorescent mask labelling protocol using StrataQuest. Figure S3. Metabolomic fingerprint of idiopathic pulmonary fibrosis (IPF) patient saliva samples. Figure S4. Annotation of key Reactome pathways upregulated by TGFβ1 in human primary lung fibroblasts based on the enrichment analysis of the obtained DEGs. Figure S5. CaSR negative allosteric modulator, NPS2143 attenuates TGFβ1-induced up-regulation of genes associated with amino acid metabolism and polyamines in normal human lung fibroblasts. Figure S6. CaSR negative allosteric modulator, NPS2143 attenuates TGFβ1-induced up-regulation of pro-fibrotic genes in normal human lung fibroblasts. Figure S7. CaSR NAM reduces TGFβ1-induced increase in cell area in IPF lung fibroblasts.
Author Contributions
Conceptualization, K.W., P.L.Y., and D.R.; Methodology, J.D., R.M., R.E., P.L.Y., and L.A.J.M.; Software, R.T.B., and D.A.P.; Validation, R.L., B.M., and P.L.Y.; Formal Analysis, K.W., R.L., R.T.B., B.S.H., L.A.J.M., and P.L.Y.; Investigation, K.W., R.L., R.P.d.A., B.M., P.H., and P.L.Y.; Resources, L.A.J.M., K.E.L., J.D., R.M., H.K.-Q., R.Z.J., A.J.S., and P.L.Y.; Data Curation, K.W.; Writing—Original Draft Preparation, K.W., D.R., and P.L.Y.; Writing—Review and Editing, J.D., R.M., R.T.B., D.A.P., L.A.J.M., R.P.d.A., K.E.L., H.K.-Q., C.J.C., R.Z.J., D.R., A.J.S., and P.L.Y.; Visualization, K.W., B.S.H., L.A.J.M., D.R., and P.L.Y.; Supervision, D.R., R.M., R.Z.J., J.P.T.W., and C.J.C. and P.L.Y.; Project Administration, K.W., R.Z.J., A.J.S., and P.L.Y.; Funding Acquisition, B.D.H.-G., A.J.S., P.L.Y., and D.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the King’s Commercialisation Institute (grant number: 150902, to D.R.), the Saunders Legacy Lung Research (grant number: 9471 to B.H-G. and D.R.), the Marie Curie ETN “CaSR Biomedicine” (grant number: 675228 to D.R.), the Wellcome Trust (grant number: 221678/Z/20/Z to P.L.Y.], and the Newcastle University Research Excellence Development award (grant number: NU-018860 to P.Y.). L.A.J.M. is partially supported by the Shandong Province “Double-Hundred Talent Plan” Teams (grant number: WSR2023049). The APC was funded by Conselho Nacional de Desenvolvimento Científico e tecnológico—CNPq.
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki. The loco-regional ethical approval for the saliva samples used for metabolomic analysis in this study was obtained from Hywel Dda University Health Board (Novel Technologies for Diagnosing and Monitoring Respiratory Diseases–ref: 16/WA/0036). All patients gave written informed consent before inclusion in the study, and all data were link-anonymized before analysis. The protocol for IPF lung tissue collection was approved by the institutional review board of the University of Texas Health Science Center at Houston (HSC-MS-08-0354, latest approval date: 16 January 2025; HSC-MS-15-1049, latest approval date: 21 January 2025) and followed the guidelines of the Declaration of Helsinki.
Informed Consent Statement
Written informed consent was obtained from all participants involved in this study.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author(s). Bioinformatics data presented in this study are openly available in NCBI Sequence Read Archive (SRA) at https://www.ncbi.nlm.nih.gov/sra/ (accessed on 18 March 2025) or accession number: PRJNA1213828.
Acknowledgments
The researchers would like to thank Rahat Hussain, Manish Patel, and Bindu Akkanti from the University of Texas Health Science Center in Houston Texas, USA, who were instrumental in obtaining lung tissue samples from IPF patients undergoing lung transplantation for this project. John Simpson is a National Institute for Health and Care Research (NIHR) Senior Investigator. The views expressed in this article are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.
Conflicts of Interest
Rupert Ecker is co-founder and CEO of the for-profit company TissueGnostics GmbH, which used to be a beneficiary in the EU-funded Marie–Sklodowska–Curie Innovative Training Network “CaSR Biomedicine” (Grant Agreement No 675228), which contributed to this publication. John Simpson is the lead investigator on grants with Clinigen, Aerogen, and Partner Therapeutics as co-applicants, and a co-applicant on a grant with Partner Therapeutics as a co-applicant. He has previously been an applicant on grants with Becton Dickinson as co-applicants. He is the Director of an NIHR Healthtech Research Centre that works with companies across the diagnostics industry.
References
- Adegunsoye, A.; Oldham, J.M.; Bellam, S.K.; Montner, S.; Churpek, M.M.; Noth, I.; Vij, R.; Strek, M.E.; Chung, J.H. Computed Tomography Honeycombing Identifies a Progressive Fibrotic Phenotype with Increased Mortality across Diverse Interstitial Lung Diseases. Ann. Am. Thorac. Soc. 2019, 16, 580–588. [Google Scholar] [CrossRef]
- Vancheri, C.; Failla, M.; Crimi, N.; Raghu, G. Idiopathic Pulmonary Fibrosis: A Disease with Similarities and Links to Cancer Biology. Eur. Respir. J. 2010, 35, 496–504. [Google Scholar] [CrossRef]
- Somogyi, V.; Chaudhuri, N.; Torrisi, S.E.; Kahn, N.; Müller, V.; Kreuter, M. The Therapy of Idiopathic Pulmonary Fibrosis: What Is Next? Eur. Respir. Rev. 2019, 28, 190021. [Google Scholar] [CrossRef] [PubMed]
- Raghu, G. Pharmacotherapy for Idiopathic Pulmonary Fibrosis: Current Landscape and Future Potential. Eur. Respir. Rev. 2017, 26, 170071. [Google Scholar] [CrossRef] [PubMed]
- Raghu, G.; Rochwerg, B.; Zhang, Y.; Garcia, C.A.C.; Azuma, A.; Behr, J.; Brozek, J.L.; Collard, H.R.; Cunningham, W.; Homma, S.; et al. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline: Treatment of Idiopathic Pulmonary Fibrosis. An Update of the 2011 Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 2015, 192, e3–e19. [Google Scholar] [CrossRef] [PubMed]
- Tarride, J.-E.; Hopkins, R.B.; Burke, N.; Guertin, J.R.; O’Reilly, D.; Fell, C.D.; Dion, G.; Kolb, M. Clinical and Economic Burden of Idiopathic Pulmonary Fibrosis in Quebec, Canada. Clin. Outcomes Res. 2018, 10, 127–137. [Google Scholar] [CrossRef]
- Van Manen, M.J.G.; Geelhoed, J.J.M.; Tak, N.C.; Wijsenbeek, M.S. Optimizing Quality of Life in Patients with Idiopathic Pulmonary Fibrosis. Ther. Adv. Respir. Dis. 2017, 11, 157–169. [Google Scholar] [CrossRef]
- Chianese, M.; Screm, G.; Salton, F.; Confalonieri, P.; Trotta, L.; Barbieri, M.; Ruggero, L.; Mari, M.; Reccardini, N.; Geri, P.; et al. Pirfenidone and Nintedanib in Pulmonary Fibrosis: Lights and Shadows. Pharmaceuticals 2024, 17, 709. [Google Scholar] [CrossRef]
- Kou, M.; Jiao, Y.; Li, Z.; Wei, B.; Li, Y.; Cai, Y.; Wei, W. Real-World Safety and Effectiveness of Pirfenidone and Nintedanib in the Treatment of Idiopathic Pulmonary Fibrosis: A Systematic Review and Meta-Analysis. Eur. J. Clin. Pharmacol. 2024, 80, 1445–1460. [Google Scholar] [CrossRef]
- Rajagopal, K.; Bryant, A.J.; Sahay, S.; Wareing, N.; Zhou, Y.; Pandit, L.M.; Karmouty-Quintana, H. Idiopathic Pulmonary Fibrosis and Pulmonary Hypertension: Heracles Meets the Hydra. Br. J. Pharmacol. 2021, 178, 172–186. [Google Scholar] [CrossRef]
- Haak, A.J.; Ducharme, M.T.; Diaz Espinosa, A.M.; Tschumperlin, D.J. Targeting GPCR Signaling for Idiopathic Pulmonary Fibrosis Therapies. Trends Pharmacol. Sci. 2020, 41, 172–182. [Google Scholar] [CrossRef]
- Mikolasch, T.A.; Garthwaite, H.S.; Porter, J.C. Update in Diagnosis and Management of Interstitial Lung Disease. Clin. Med. 2017, 17, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Hewlett, J.C.; Kropski, J.A.; Blackwell, T.S. Idiopathic Pulmonary Fibrosis: Epithelial-Mesenchymal Interactions and Emerging Therapeutic Targets. Matrix Biol. 2018, 71–72, 112. [Google Scholar] [CrossRef]
- Pardo, A.; Selman, M. The Interplay of the Genetic Architecture, Aging, and Environmental Factors in the Pathogenesis of Idiopathic Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2021, 64, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Roque, W.; Romero, F. Cellular Metabolomics of Pulmonary Fibrosis, from Amino Acids to Lipids. Am. J. Physiol. Physiol. 2021, 320, C689–C695. [Google Scholar] [CrossRef] [PubMed]
- Bernard, K.; Logsdon, N.J.; Benavides, G.A.; Sanders, Y.; Zhang, J.; Darley-Usmar, V.M.; Thannickal, V.J. Glutaminolysis Is Required for Transforming Growth Factor-Β1–Induced Myofibroblast Differentiation and Activation. J. Biol. Chem. 2018, 293, 1218–1228. [Google Scholar] [CrossRef]
- Hamanaka, R.B.; O’Leary, E.M.; Witt, L.J.; Tian, Y.; Gökalp, G.A.; Meliton, A.Y.; Dulin, N.O.; Mutlu, G.M. Glutamine Metabolism Is Required for Collagen Protein Synthesis in Lung Fibroblasts. Am. J. Respir. Cell Mol. Biol. 2019, 61, 597–606. [Google Scholar] [CrossRef]
- Zhao, Y.D.; Yin, L.; Archer, S.; Lu, C.; Zhao, G.; Yao, Y.; Wu, L.; Hsin, M.; Waddell, T.K.; Keshavjee, S.; et al. Metabolic Heterogeneity of Idiopathic Pulmonary Fibrosis: A Metabolomic Study. BMJ Open Respir. Res. 2017, 4, e000183. [Google Scholar] [CrossRef]
- Kitowska, K.; Zakrzewicz, D.; Königshoff, M.; Chrobak, I.; Grimminger, F.; Seeger, W.; Bulau, P.; Eickelberg, O. Functional Role and Species-Specific Contribution of Arginases in Pulmonary Fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 2008, 294, L34–L45. [Google Scholar] [CrossRef]
- Endo, M.; Oyadomari, S.; Terasaki, Y.; Takeya, M.; Suga, M.; Mori, M.; Gotoh, T. Induction of Arginase I and II in Bleomycin-Induced Fibrosis of Mouse Lung. Am. J. Physiol. Lung Cell Mol. Physiol. 2003, 285, L313–L321. [Google Scholar] [CrossRef]
- Komuves, L.; Oda, Y.; Tu, C.L.; Chang, W.H.; Ho-Pao, C.L.; Mauro, T.; Bikle, D.D. Epidermal Expression of the Full-Length Extracellular Calcium-Sensing Receptor Is Required for Normal Keratinocyte Differentiation. J. Cell Physiol. 2002, 192, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Schepelmann, M.; Yarova, P.L.; Lopez-Fernandez, I.; Davies, T.S.; Brennan, S.C.; Edwards, P.J.; Aggarwal, A.; Graça, J.; Rietdorf, K.; Matchkov, V.; et al. The Vascular Ca2+-Sensing Receptor Regulates Blood Vessel Tone and Blood Pressure. Am. J. Physiol. Cell Physiol. 2016, 310, C193–C204. [Google Scholar] [CrossRef] [PubMed]
- Sundararaman, S.S.; van der Vorst, E.P.C. Calcium-Sensing Receptor (Casr), Its Impact on Inflammation and the Consequences on Cardiovascular Health. Int. J. Mol. Sci. 2021, 22, 2478. [Google Scholar] [CrossRef] [PubMed]
- Yarova, P.L.; Stewart, A.L.; Sathish, V.; Britt, R.D.; Thompson, M.A.; P Lowe, A.P.; Freeman, M.; Aravamudan, B.; Kita, H.; Brennan, S.C.; et al. Calcium-Sensing Receptor Antagonists Abrogate Airway Hyperresponsiveness and Inflammation in Allergic Asthma. Sci. Transl. Med. 2015, 7, 284ra60. [Google Scholar] [CrossRef]
- Brennan, S.C.; Wilkinson, W.J.; Tseng, H.-E.; Finney, B.; Monk, B.; Dibble, H.; Quilliam, S.; Warburton, D.; Galietta, L.J.; Kemp, P.J.; et al. The Extracellular Calcium-Sensing Receptor Regulates Human Fetal Lung Development via CFTR. Sci. Rep. 2016, 6, 21975. [Google Scholar] [CrossRef]
- Finney, B.A.; Del Moral, P.M.; Wilkinson, W.J.; Cayzac, S.; Cole, M.; Warburton, D.; Kemp, P.J.; Riccardi, D. Regulation of Mouse Lung Development by the Extracellular Calcium-Sensing Receptor, CaR. J. Physiol. 2008, 586, 6007–6019. [Google Scholar] [CrossRef]
- Riccardi, D.; Brennan, S.C.; Chang, W. The Extracellular Calcium-Sensing Receptor, CaSR, in Fetal Development. Best. Pr. Res. Clin. Endocrinol. Metab. 2013, 27, 443–453. [Google Scholar] [CrossRef]
- Brown, E.M.; MacLeod, R.J. Extracellular Calcium Sensing and Extracellular Calcium Signaling. Physiol. Rev. 2001, 81, 239–297. [Google Scholar] [CrossRef]
- Quinn, S.J.; Ye, C.-P.; Diaz, R.; Kifor, O.; Bai, M.; Vassilev, P.; Brown, E. The Ca2+-sensing receptor: A target for polyamines. Am. J. Physiol. 1997, 273, C1315–C1323. [Google Scholar] [CrossRef] [PubMed]
- Hoet, P.H.M.; Nemery, B. Polyamines in the Lung: Polyamine Uptake and Polyamine-Linked Pathological or Toxicological Conditions. Am. J. Physiol. Lung Cell Mol. Physiol. 2000, 278, L417–L433. [Google Scholar] [CrossRef]
- Davies, S.L.; Gibbons, C.E.; Vizard, T.; Ward, D.T. Ca2+ -Sensing Receptor Induces Rho Kinase-Mediated Actin Stress Fiber Assembly and Altered Cell Morphology, but Not in Response to Aromatic Amino Acids. Am. J. Physiol. Cell Physiol. 2006, 290, C1543–C1551. [Google Scholar] [CrossRef] [PubMed]
- Rybchyn, M.S.; Slater, M.; Conigrave, A.D.; Mason, R.S. An Akt-Dependent Increase in Canonical Wnt Signaling and a Decrease in Sclerostin Protein Levels Are Involved in Strontium Ranelate-Induced Osteogenic Effects in Human Osteoblasts. J. Biol. Chem. 2011, 286, 23771–23779. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, Z.; Zheng, L.; You, C.; Chi, H.; Zhang, T.; Xu, G. Calcium-Sensing Receptor Activating ERK1/2 and PI3K-Akt Pathways to Induce the Proliferation of Osteosarcoma Cells. Clin. Exp. Pharmacol. Physiol. 2020, 47, 517–519. [Google Scholar] [CrossRef]
- Yamamura, A.; Guo, Q.; Yamamura, H.; Zimnicka, A.M.; Pohl, N.M.; Smith, K.A.; Fernandez, R.A.; Zeifman, A.; Makino, A.; Dong, H.; et al. Enhanced Ca(2+)-Sensing Receptor Function in Idiopathic Pulmonary Arterial Hypertension. Circ. Res. 2012, 111, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Yarova, P.; Davies, C.; Price, S.A.; Huang, Q.; Graca, J.A.; Maleki-Toyserkani, S.; Lowe, A.P.P.; Kidd, E.J.; Ford, W.R.; Broadley, K.J.; et al. Inhaled Calcilytics: Effects on Airway Inflammation and Remodeling. Respir. Drug Deliv. 2016, 1, 1–12. [Google Scholar]
- Buendía-Roldán, I.; Mejía, M.; Navarro, C.; Selman, M. Idiopathic Pulmonary Fibrosis: Clinical Behavior and Aging Associated Comorbidities. Respir. Med. 2017, 129, 46–52. [Google Scholar] [CrossRef]
- Ruffenach, G.; Hong, J.; Vaillancourt, M.; Medzikovic, L.; Eghbali, M. Pulmonary Hypertension Secondary to Pulmonary Fibrosis: Clinical Data, Histopathology and Molecular Insights. Respir. Res. 2020, 21, 303. [Google Scholar] [CrossRef]
- de Araújo, R.P.; Bertoni, N.; Seneda, A.L.; Felix, T.F.; Carvalho, M.; Lewis, K.E.; Hasimoto, É.N.; Beckmann, M.; Drigo, S.A.; Reis, P.P.; et al. Defining Metabolic Rewiring in Lung Squamous Cell Carcinoma. Metabolites 2019, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, U.; Llamazares Prada, M.; Pohl, S.T.; Richter, M.; Tamas, R.; Schuler, M.; Keller, C.; Mijosek, V.; Muley, T.; Schneider, M.A.; et al. High-resolution Transcriptomic and Epigenetic Profiling Identifies Novel Regulators of COPD. EMBO J. 2023, 42, e111272. [Google Scholar] [CrossRef]
- Llamazares-Prada, M.; Espinet, E.; Mijošek, V.; Schwartz, U.; Lutsik, P.; Tamas, R.; Richter, M.; Behrendt, A.; Pohl, S.T.; Benz, N.P.; et al. Versatile Workflow for Cell Type-Resolved Transcriptional and Epigenetic Profiles from Cryopreserved Human Lung. JCI Insight 2021, 6, e140443. [Google Scholar] [CrossRef]
- Andrews, S. Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 2 November 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic. Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and Accurate Filtering of Ribosomal RNAs in Metatranscriptomic Data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G. Thirteen Years of ClusterProfiler. Innovation 2024, 5, 100722. [Google Scholar] [CrossRef]
- Xu, S.; Hu, E.; Cai, Y.; Xie, Z.; Luo, X.; Zhan, L.; Tang, W.; Wang, Q.; Liu, B.; Wang, R.; et al. Using ClusterProfiler to Characterize Multiomics Data. Nat. Protoc. 2024, 19, 3292–3320. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; He, Q.Y. ReactomePA: An R/Bioconductor Package for Reactome Pathway Analysis and Visualization. Mol. Biosyst. 2016, 12, 477–479. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps. Available online: https://github.com/raivokolde/pheatmap (accessed on 13 February 2025).
- R Core Team R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 22 November 2021).
- Janssen, L.J.; Mukherjee, S.; Ask, K. Calcium Homeostasis and Ionic Mechanisms in Pulmonary Fibroblasts. Am. J. Respir. Cell Mol. Biol. 2015, 53, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. Transforming Growth Factor–β in Tissue Fibrosis. J. Exp. Med. 2020, 217, e20190103. [Google Scholar] [CrossRef]
- Hamanaka, R.B.; Mutlu, G.M. Metabolic Requirements of Pulmonary Fibrosis: Role of Fibroblast Metabolism. FEBS J. 2021, 288, 6331–6352. [Google Scholar] [CrossRef]
- Possemato, R.; Marks, K.M.; Shaul, Y.D.; Pacold, M.E.; Kim, D.; Birsoy, K.; Sethumadhavan, S.; Woo, H.K.; Jang, H.G.; Jha, A.K.; et al. Functional Genomics Reveal That the Serine Synthesis Pathway Is Essential in Breast Cancer. Nature 2011, 476, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Fear, M.W.; Suk Choi, Y.; Wood, F.M.; Allahham, A.; Mutsaers, S.E.; Prêle, C.M. The Extracellular Matrix and Mechanotransduction in Pulmonary Fibrosis. Int. J. Biochem. Cell Biol. 2020, 126, 105802. [Google Scholar] [CrossRef]
- D’Urso, M.; Kurniawan, N.A. Mechanical and Physical Regulation of Fibroblast-Myofibroblast Transition: From Cellular Mechanoresponse to Tissue Pathology. Front. Bioeng. Biotechnol. 2020, 8, 609653. [Google Scholar] [CrossRef]
- Zimmermann, N.; Rothenberg, M.E. The Arginine-Arginase Balance in Asthma and Lung Inflammation. Eur. J. Pharmacol. 2006, 533, 253–262. [Google Scholar] [CrossRef]
- Hasan, C.M.; Pottenger, S.; Green, A.E.; Cox, A.A.; White, J.S.; Jones, T.; Winstanley, C.; Kadioglu, A.; Wright, M.H.; Neill, D.R.; et al. Pseudomonas Aeruginosa Utilizes the Host-Derived Polyamine Spermidine to Facilitate Antimicrobial Tolerance. JCI Insight 2022, 7, e158879. [Google Scholar] [CrossRef]
- Grasemann, H.; Dhaliwal, R.; Ivanovska, J.; Kantores, C.; McNamara, P.J.; Scott, J.A.; Belik, J.; Jankov, R.P. Arginase Inhibition Prevents Bleomycin-Induced Pulmonary Hypertension, Vascular Remodeling, and Collagen Deposition in Neonatal Rat Lungs. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L503–L510. [Google Scholar] [CrossRef]
- He, Y.Y.; Yan, Y.; Jiang, X.; Zhao, J.H.; Wang, Z.; Wu, T.; Wang, Y.; Guo, S.S.; Ye, J.; Lian, T.Y.; et al. Spermine Promotes Pulmonary Vascular Remodelling and Its Synthase Is a Therapeutic Target for Pulmonary Arterial Hypertension. Eur. Respir. J. 2020, 56, 2000522. [Google Scholar] [CrossRef] [PubMed]
- Firpo, M.R.; Mounce, B.C. Diverse Functions of Polyamines in Virus Infection. Biomolecules 2020, 10, 628. [Google Scholar] [CrossRef] [PubMed]
- Summer, R.; Todd, J.L.; Neely, M.L.; Lobo, L.J.; Namen, A.; Newby, L.K.; Shafazand, S.; Suliman, S.; Hesslinger, C.; Keller, S.; et al. Circulating Metabolic Profile in Idiopathic Pulmonary Fibrosis: Data from the IPF-PRO Registry. Respir. Res. 2024, 25, 58. [Google Scholar] [CrossRef] [PubMed]
- Christopoulos, A. Advances in G Protein-Coupled Receptor Allostery: From Function to Structure. Mol. Pharmacol. 2014, 86, 463–478. [Google Scholar] [CrossRef]
- Minois, N.; Carmona-Gutierrez, D.; Madeo, F. Polyamines in Aging and Disease. Aging 2011, 3, 716–732. [Google Scholar] [CrossRef]
- Zhu, L.; Xiao, R.; Zhang, X.; Lang, Y.; Liu, F.; Yu, Z.; Zhang, J.; Su, Y.; Lu, Y.; Wang, T.; et al. Spermine on Endothelial Extracellular Vesicles Mediates Smoking-Induced Pulmonary Hypertension Partially through Calcium-Sensing Receptor. Arter. Thromb. Vasc. Biol. 2019, 39, 482–495. [Google Scholar] [CrossRef]
- Zhou, Y.; Frey, T.K.; Yang, J.J. Viral Calciomics: Interplays between Ca2+ and Virus. Cell Calcium 2009, 46, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, M.L.; Campilongo, R.; Casalino, M.; Micheli, G.; Colonna, B.; Prosseda, G. Polyamines: Emerging Players in Bacteria–Host Interactions. Int. J. Med. Microbiol. 2013, 303, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Sheng, G.; Chen, P.; Wei, Y.; Yue, H.; Chu, J.; Zhao, J.; Wang, Y.; Zhang, W.; Zhang, H.L. Viral Infection Increases the Risk of Idiopathic Pulmonary Fibrosis: A Meta-Analysis. Chest 2020, 157, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Pulkkinen, V.; Salmenkivi, K.; Kinnula, V.L.; Sutinen, E.; Halme, M.; Hodgson, U.; Lehto, J.; Jääskelinen, A.; Piiparinen, H.; Kere, J.; et al. A Novel Screening Method Detects Herpesviral DNA in the Idiopathic Pulmonary Fibrosis Lung. Ann. Med. 2012, 44, 178–186. [Google Scholar] [CrossRef]
- Egan, J.J.; Woodcock, A.A.; Stewart, J.P. Viruses and Idiopathic Pulmonary Fibrosis. Eur. Respir. J. 1997, 10, 1433–1437. [Google Scholar] [CrossRef]
- Bharadwaj, S.; Singh, M.; Kirtipal, N.; Kang, S.G. SARS-CoV-2 and Glutamine: SARS-CoV-2 Triggered Pathogenesis via Metabolic Reprograming of Glutamine in Host Cells. Front. Mol. Biosci. 2021, 7, 627842. [Google Scholar] [CrossRef]
- Xu, T.; Liu, C.; Ning, X.; Gao, Z.; Li, A.; Wang, S.; Leng, L.; Kong, P.; Liu, P.; Zhang, S.; et al. Causal Relationship between Circulating Glutamine Levels and Idiopathic Pulmonary Fibrosis: A Two-Sample Mendelian Randomization Study. BMC Pulm. Med. 2024, 24, 451. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, J.; Sun, H.; Zhang, Y.; Zou, D. New Insights into Fibrosis from the ECM Degradation Perspective: The Macrophage-MMP-ECM Interaction. Cell Biosci. 2022, 12, 117. [Google Scholar] [CrossRef]
- Upagupta, C.; Shimbori, C.; Alsilmi, R.; Kolb, M. Matrix Abnormalities in Pulmonary Fibrosis. Eur. Respir. Rev. 2018, 27, 180033. [Google Scholar] [CrossRef]
- Fernandez, I.E.; Eickelberg, O. The Impact of TGF-β on Lung Fibrosis: From Targeting to Biomarkers. Proc. Am. Thorac. Soc. 2012, 9, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Pardo, A.; Cabrera, S.; Maldonado, M.; Selman, M. Role of Matrix Metalloproteinases in the Pathogenesis of Idiopathic Pulmonary Fibrosis. Respir. Res. 2016, 17, 23. [Google Scholar] [CrossRef] [PubMed]
- Pardo, A.; Selman, M.; Kaminski, N. Approaching the Degradome in Idiopathic Pulmonary Fibrosis. Int. J. Biochem. Cell Biol. 2008, 40, 1141–1155. [Google Scholar] [CrossRef]
- Pardo, A.; Selman, M. Matrix Metalloproteases in Aberrant Fibrotic Tissue Remodeling. Proc. Am. Thorac. Soc. 2006, 3, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Manicone, A.M.; McGuire, J.K. Matrix Metalloproteinases as Modulators of Inflammation. Semin. Cell Dev. Biol. 2008, 19, 34–41. [Google Scholar] [CrossRef]
- Limb, G.A.; Matter, K.; Murphy, G.; Cambrey, A.D.; Bishop, P.N.; Morris, G.E.; Khaw, P.T. Matrix Metalloproteinase-1 Associates with Intracellular Organelles and Confers Resistance to Lamin A/C Degradation during Apoptosis. Am. J. Pathol. 2005, 166, 1555–1563. [Google Scholar] [CrossRef]
- Selman, M.; Ruiz, V.; Cabrera, S.; Segura, L.; Ramírez, R.; Barrios, R.; Pardo, A. TIMP-1, -2, -3, and -4 in Idiopathic Pulmonary Fibrosis. A Prevailing Nondegradative Lung Microenvironment? Am. J. Physiol. Lung Cell Mol. Physiol. 2000, 279, L562–L574. [Google Scholar] [CrossRef]
- Bergeron, S.; Lemieux, E.; Durand, V.; Cagnol, S.; Carrier, J.C.; Lussier, J.G.; Boucher, M.J.; Rivard, N. The Serine Protease Inhibitor SerpinE2 Is a Novel Target of ERK Signaling Involved in Human Colorectal Tumorigenesis. Mol. Cancer 2010, 9, 271. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).