G-Quadruplex Conformational Switching for miR-155-3p Detection Using a Ligand-Based Fluorescence Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Oligonucleotide Sequences
2.2. Fluorescence Studies
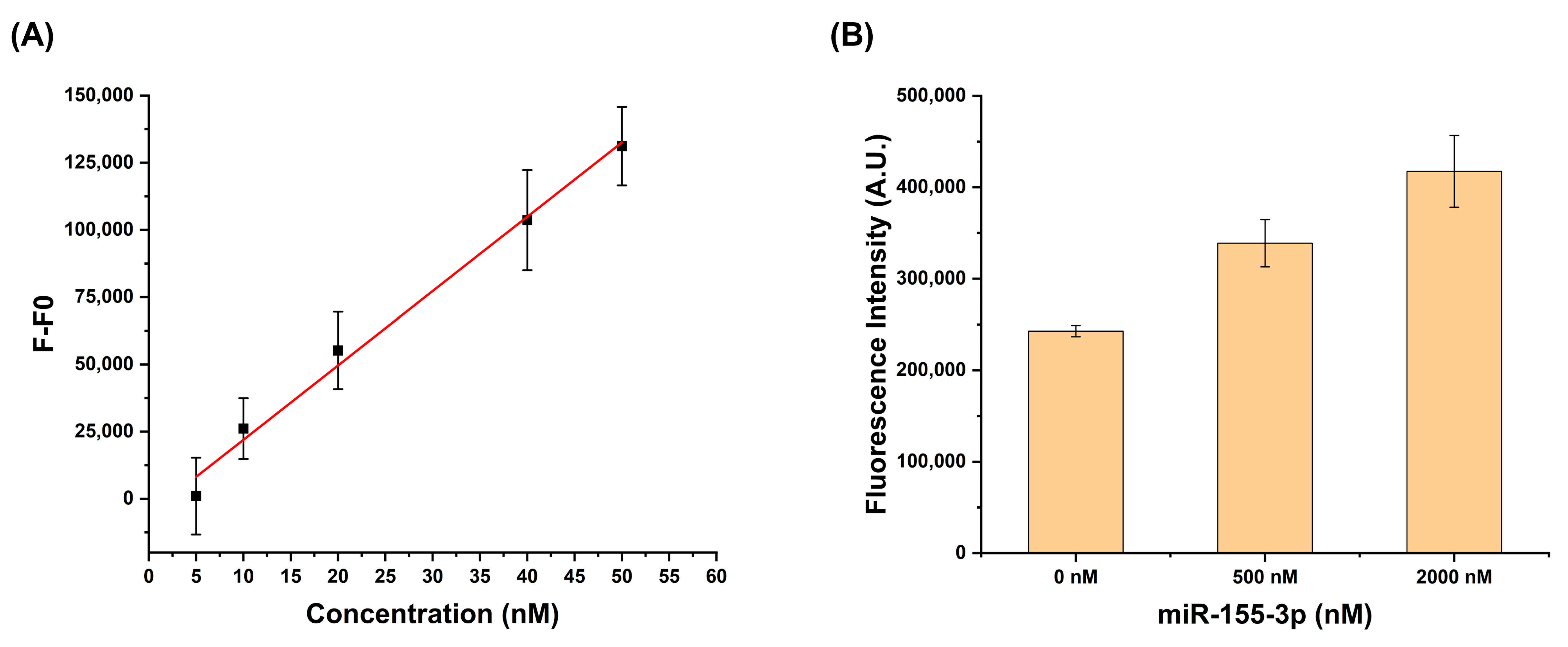
2.2.1. Limit of Detection
2.2.2. Total RNA Detection
2.3. Circular Dichroism (CD) Spectroscopy
2.4. Non-Denaturing Polyacrylamide Gel Electrophoresis (PAGE) Analysis
2.5. Cell Culture and Total RNA Extraction
3. Results
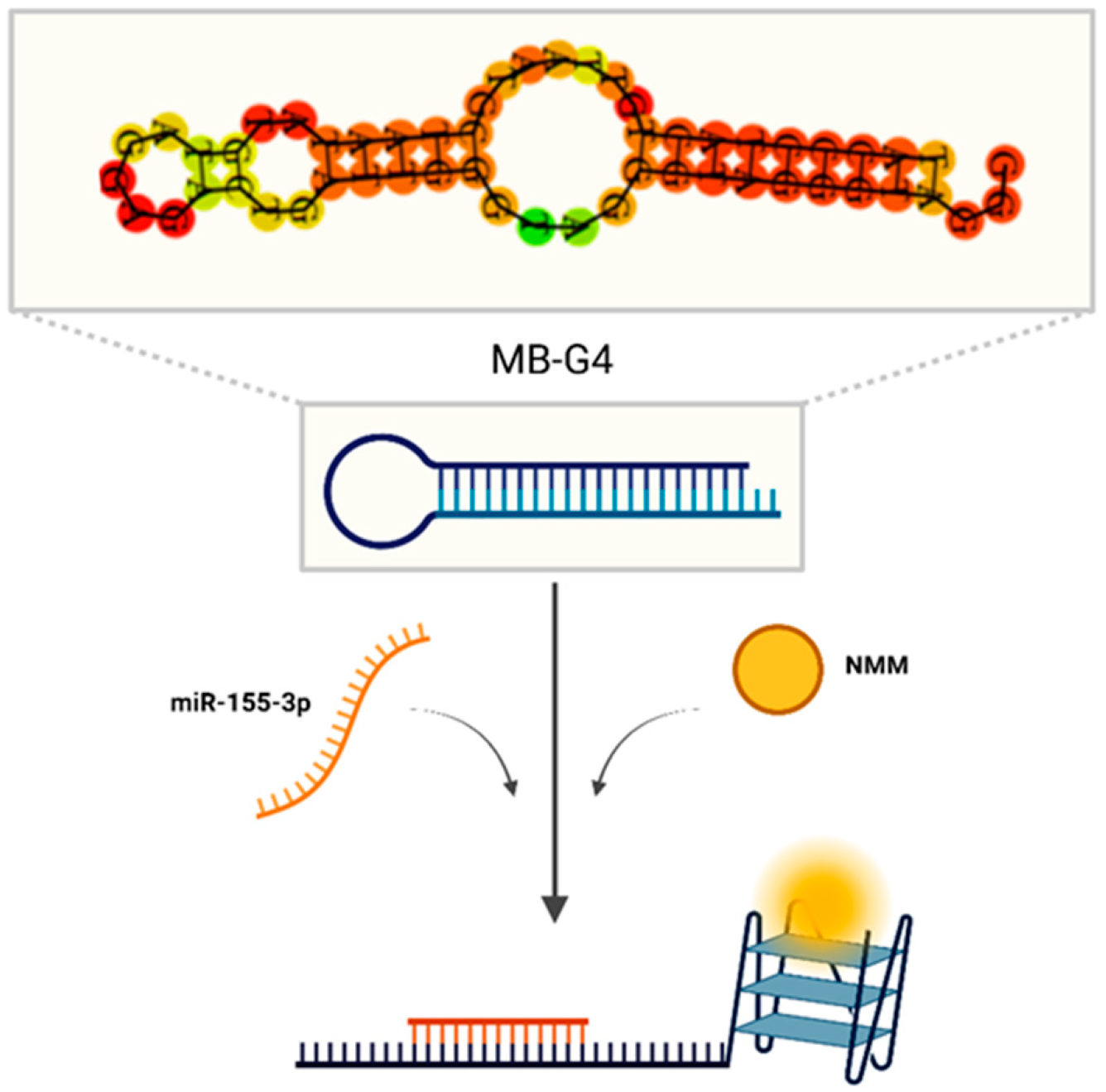
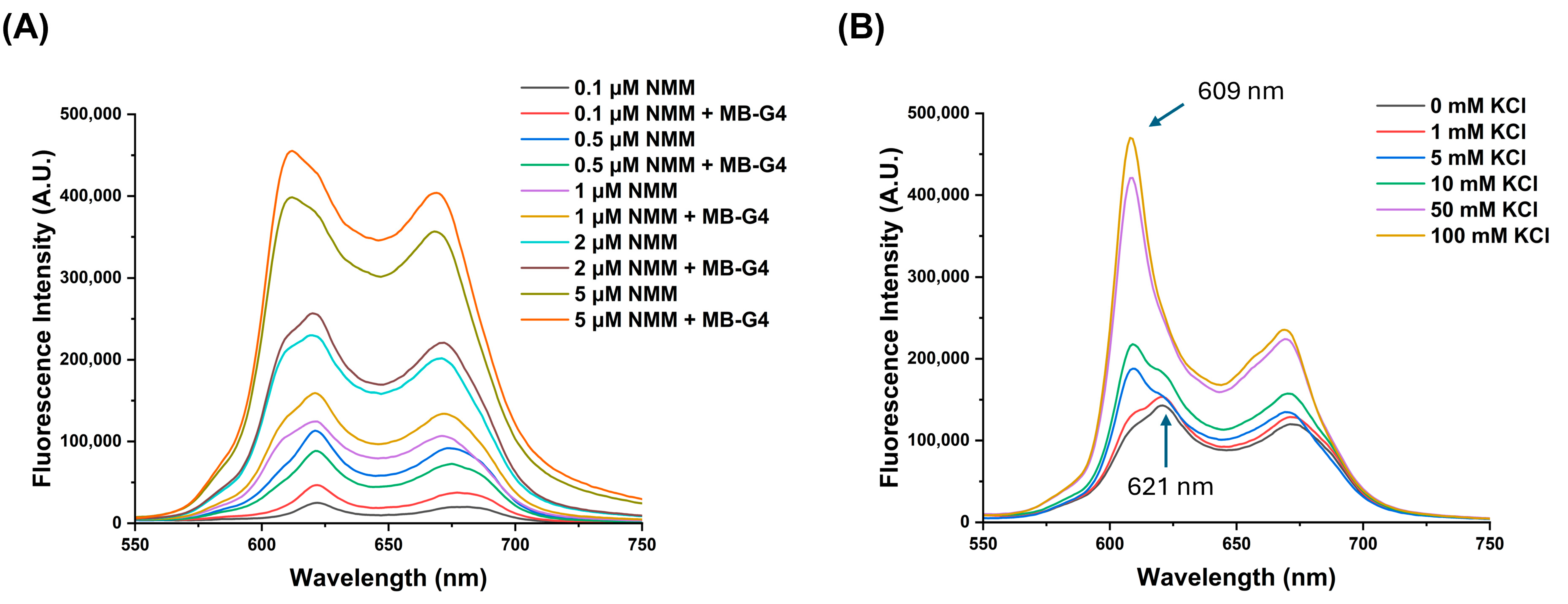
3.1. Assessment of G4 Formation by MB-G4
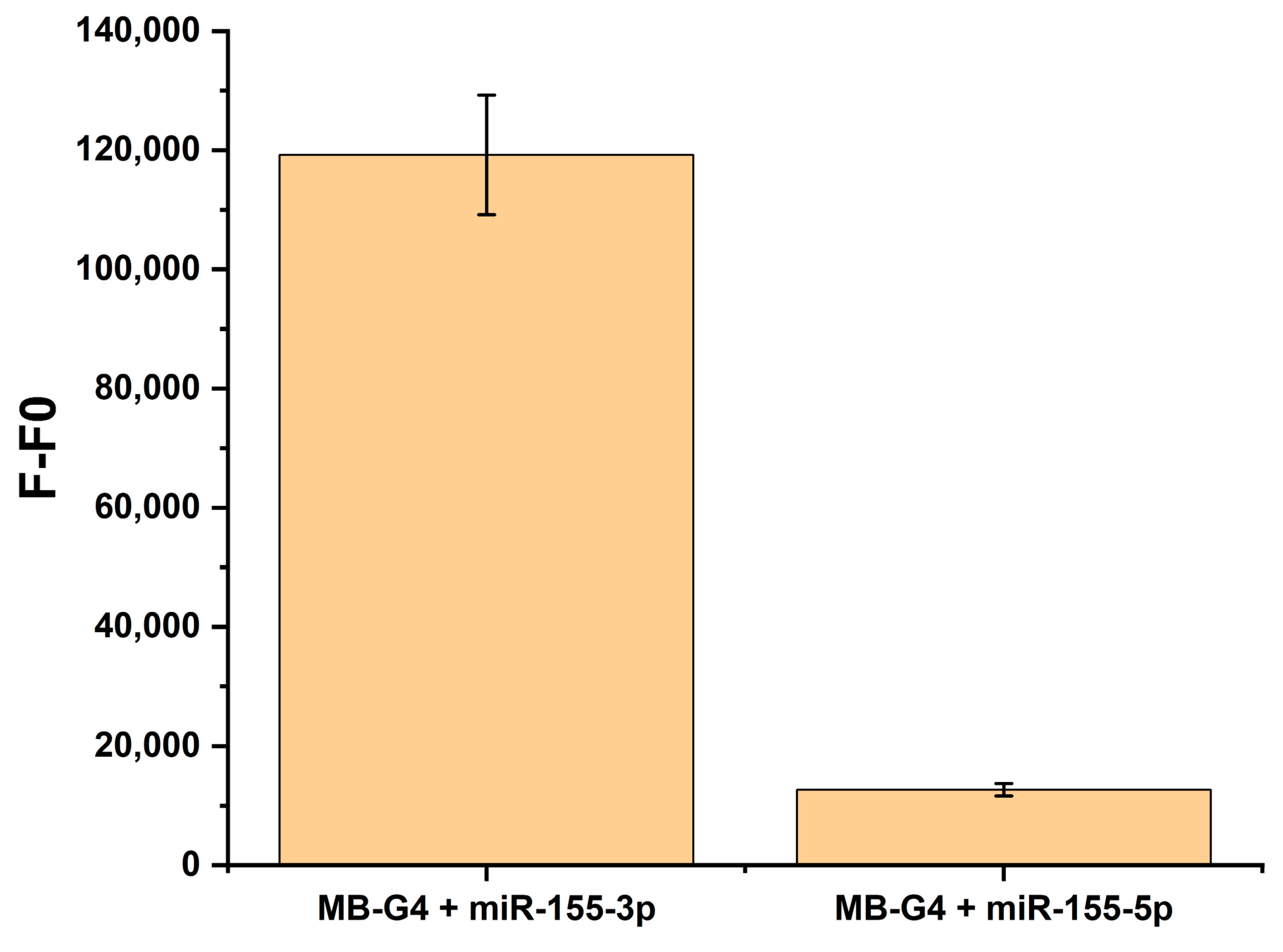
3.2. MB-G4 and miR-155-3p Interaction Studies
3.3. Specificity and Detection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Araghi, M.; Mannani, R.; Heidarnejad Maleki, A.; Hamidi, A.; Rostami, S.; Safa, S.H.; Faramarzi, F.; Khorasani, S.; Alimohammadi, M.; Tahmasebi, S.; et al. Recent advances in non-small cell lung cancer targeted therapy; an update review. Cancer Cell Int. 2023, 23, 162. [Google Scholar] [CrossRef] [PubMed]
- Cavazzoni, A.; Digiacomo, G.; Volta, F.; Alfieri, R.; Giovannetti, E.; Gnetti, L.; Bellini, L.; Galetti, M.; Fumarola, C.; Xu, G.; et al. PD-L1 overexpression induces STAT signaling and promotes the secretion of pro-angiogenic cytokines in non-small cell lung cancer (NSCLC). Lung Cancer 2024, 187, 107438. [Google Scholar] [CrossRef]
- Charpidou, A.; Hardavella, G.; Boutsikou, E.; Panagiotou, E.; Simsek, G.Ö.; Verbeke, K.; Xhemalaj, D.; Domagała-Kulawik, J. Unravelling the diagnostic pathology and molecular biomarkers in lung cancer. Breathe 2024, 20, 230192. [Google Scholar] [CrossRef] [PubMed]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef]
- Seyhan, A.A. Trials and Tribulations of MicroRNA Therapeutics. Int. J. Mol. Sci. 2024, 25, 1469. [Google Scholar] [CrossRef]
- Abdul Manap, A.S.; Wisham, A.A.; Wong, F.W.; Ahmad Najmi, H.R.; Ng, Z.F.; Diba, R.S. Mapping the function of MicroRNAs as a critical regulator of tumor-immune cell communication in breast cancer and potential treatment strategies. Front. Cell Dev. Biol. 2024, 12, 1390704. [Google Scholar] [CrossRef]
- Raisch, J.; Darfeuille-Michaud, A.; Nguyen, H.T.T. Role of microRNAs in the immune system, inflammation and cancer. World J. Gastroenterol. 2013, 19, 2985. [Google Scholar] [CrossRef]
- Jafari, N.; Abediankenari, S. Role of microRNAs in immunoregulatory functions of epithelial cells. BMC Immunol. 2024, 25, 84. [Google Scholar] [CrossRef]
- Xu, S.-J.; Chen, J.-H.; Chang, S.; Li, H.-L. The role of miRNAs in T helper cell development, activation, fate decisions and tumor immunity. Front. Immunol. 2023, 14, 1320305. [Google Scholar] [CrossRef]
- Chakrabortty, A.; Patton, D.J.; Smith, B.F.; Agarwal, P. miRNAs: Potential as Biomarkers and Therapeutic Targets for Cancer. Genes 2023, 14, 1375. [Google Scholar] [CrossRef] [PubMed]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Baylie, T.; Kasaw, M.; Getinet, M.; Getie, G.; Jemal, M.; Nigatu, A.; Ahmed, H.; Bogale, M. The role of miRNAs as biomarkers in breast cancer. Front. Oncol. 2024, 14, 1374821. [Google Scholar] [CrossRef]
- Mahesh, G.; Biswas, R. MicroRNA-155: A Master Regulator of Inflammation. J. Interf. Cytokine Res. 2019, 39, 321. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Huang, S.; Liu, X.; Zhang, Y.; Wei, S.; Hu, X. miR-155: An Important Role in Inflammation Response. J. Immunol. Res. 2022, 2022, 7437281. [Google Scholar] [CrossRef]
- Zanoaga, O.; Braicu, C.; Chiroi, P.; Andreea, N.; Al Hajjar, N.; Mărgărit, S.; Korban, S.S.; Berindan-Neagoe, I. The Role of miR-155 in Nutrition: Modulating Cancer-Associated Inflammation. Nutrients 2021, 13, 2245. [Google Scholar] [CrossRef]
- Shao, C.; Yang, F.; Qin, Z.; Jing, X.; Shu, Y.; Shen, H. The value of miR-155 as a biomarker for the diagnosis and prognosis of lung cancer: A systematic review with meta-analysis. BMC Cancer 2019, 19, 1103. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Xu, M.; Tian, X.; Cai, S.; Zeng, S. Research advances in the detection of miRNA. J. Pharm. Anal. 2019, 9, 217–226. [Google Scholar] [CrossRef]
- Git, A.; Dvinge, H.; Salmon-Divon, M.; Osborne, M.; Kutter, C.; Hadfield, J.; Bertone, P.; Caldas, C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 2010, 16, 991–1006. [Google Scholar] [CrossRef]
- Yang, D. G-Quadruplex DNA and RNA. Methods Mol. Biol. 2019, 2035, 1–24. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef]
- Santos, T.; Salgado, G.F.; Cabrita, E.J.; Cruz, C. G-quadruplexes and their ligands: Biophysical methods to unravel g-quadruplex/ligand interactions. Pharmaceuticals 2021, 14, 769. [Google Scholar] [CrossRef]
- Bednarz, A.; Rosendal, R.T.; Lund, L.M.; Birkedal, V. Probing G-quadruplex-ligand binding using DNA intrinsic fluorescence. Biochimie 2024, 227, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Mitteaux, J.; Wang, Z.; Wheeler, E.; Tandon, N.; Jung, S.Y.; Hudson, R.H.; Monchaud, D.; Tsvetkov, A.S. Small molecule-based regulation of gene expression in human astrocytes switching on and off the G-quadruplex control systems. J. Biol. Chem. 2025, 301, 108040. [Google Scholar] [CrossRef]
- Li, Y.-S.; Feng, C.-F.; Chen, H.-R.; Yang, W.-G.; Liu, F.; Su, M.-L.; Yuan, R.; Zhang, L.-Q.; Liang, W.-B. Concentration and activation biresponsive strategy in one analysis system with simultaneous use of G4 structure-specific signal probe and enzyme-catalyzed reaction. Anal. Chim. Acta 2024, 1329, 343246. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Granzhan, A.; Marquevielle, J.; Cucchiarini, A.; Lacroix, L.; Amrane, S.; Verga, D.; Mergny, J.-L. Guidelines for G-quadruplexes: I. In vitro characterization. Biochimie 2023, 214, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Kypr, J.; Kejnovska, I.; Renciuk, D.; Vorlickova, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef]
- Alexandre, D.; Fernandes, A.R.; Baptista, P.V.; Cruz, C. Evaluation of miR-155 silencing using a molecular beacon in human lung adenocarcinoma cell line. Talanta 2024, 274, 126052. [Google Scholar] [CrossRef]
- Lourenço, P.; Miranda, A.; Campello, M.P.C.; Paulo, A.; Louis-Mergny, J.; Cruz, C. Targeting proto-oncogene B-MYB G-quadruplex with a nucleic acid–based fluorescent probe. Int. J. Biol. Macromol. 2024, 266, 131055. [Google Scholar] [CrossRef]
- Genna, V.; Iglesias-Fernández, J.; Reyes-Fraile, L.; Villegas, N.; Guckian, K.; Seth, P.; Wan, B.; Cabrero, C.; Terrazas, M.; Brun-Heath, I.; et al. Controlled sulfur-based engineering confers mouldability to phosphorothioate antisense oligonucleotides. Nucleic Acids Res. 2023, 51, 4713–4725. [Google Scholar] [CrossRef]
- Chauca-Diaz, A.M.; Choi, Y.J.; Resendiz, M.J.E. Biophysical properties and thermal stability of oligonucleotides of RNA containing 7,8-dihydro-8-hydroxyadenosine. Biopolymers 2015, 103, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Persano, S.; Guevara, M.L.; Wolfram, J.; Blanco, E.; Shen, H.; Ferrari, M.; Pompa, P.P. Label-Free Isothermal Amplification Assay for Specific and Highly Sensitive Colorimetric miRNA Detection. ACS Omega 2016, 1, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Ning, W.; Chen, M.; Zhang, C.; Li, Q.; Bai, J. High performance electrochemical biosensor based on 3D nitrogen-doped reduced graphene oxide electrode and tetrahedral DNA nanostructure. Talanta 2019, 194, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Moreira, D.; Alexandre, D.; Miranda, A.; Lourenço, P.; Baptista, P.V.; Tomaz, C.; Lu, Y.; Cruz, C. Detecting mir-155-3p through a Molecular Beacon Bead-Based Assay. Molecules 2024, 29, 3182. [Google Scholar] [CrossRef]

| Sequence Name | Sequence (5′→ 3′) | ntNumber | Ɛ(L·mol−1·cm−1) at 260 nm |
|---|---|---|---|
| Molecular Beacon(MB-G4) | TACCCTACTGTTAATGCTAATATGTAGGAGACTGATTGGGTAGGGTAGGGTAGGG | 55 | 553,900 |
| miR-155-3p | CUCCUACAUAUUAGCAUUAACA | 22 | 206,500 |
| miR-155-5p | TTAATGCTAATCGTGATAGGGGT | 23 | 230,300 |
| Detection Method | LOD | Target | Ref. |
|---|---|---|---|
| Colorimetric Detection with Gold Nanoparticles | 100 aM | miRNA-10b | [32] |
| Gold and silver nanorod/thionine/complementary DNA composite | 1 pM | miRNA-155 | [33] |
| Fluorophore and quencher dyes conjugated molecular beacon | 0.66 nM | miR-155-3p | [28] |
| Molecular Beacon Bead-Based | 42 nM | miR-155-3p | [34] |
| MB-G4 | 10.85 nM | miR-155-3p | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lourenço, P.; Cruz, C. G-Quadruplex Conformational Switching for miR-155-3p Detection Using a Ligand-Based Fluorescence Approach. Biomolecules 2025, 15, 410. https://doi.org/10.3390/biom15030410
Lourenço P, Cruz C. G-Quadruplex Conformational Switching for miR-155-3p Detection Using a Ligand-Based Fluorescence Approach. Biomolecules. 2025; 15(3):410. https://doi.org/10.3390/biom15030410
Chicago/Turabian StyleLourenço, Pedro, and Carla Cruz. 2025. "G-Quadruplex Conformational Switching for miR-155-3p Detection Using a Ligand-Based Fluorescence Approach" Biomolecules 15, no. 3: 410. https://doi.org/10.3390/biom15030410
APA StyleLourenço, P., & Cruz, C. (2025). G-Quadruplex Conformational Switching for miR-155-3p Detection Using a Ligand-Based Fluorescence Approach. Biomolecules, 15(3), 410. https://doi.org/10.3390/biom15030410







