The Osgin Gene Family: Underexplored Yet Essential Mediators of Oxidative Stress
Abstract
1. Introduction
2. Methods
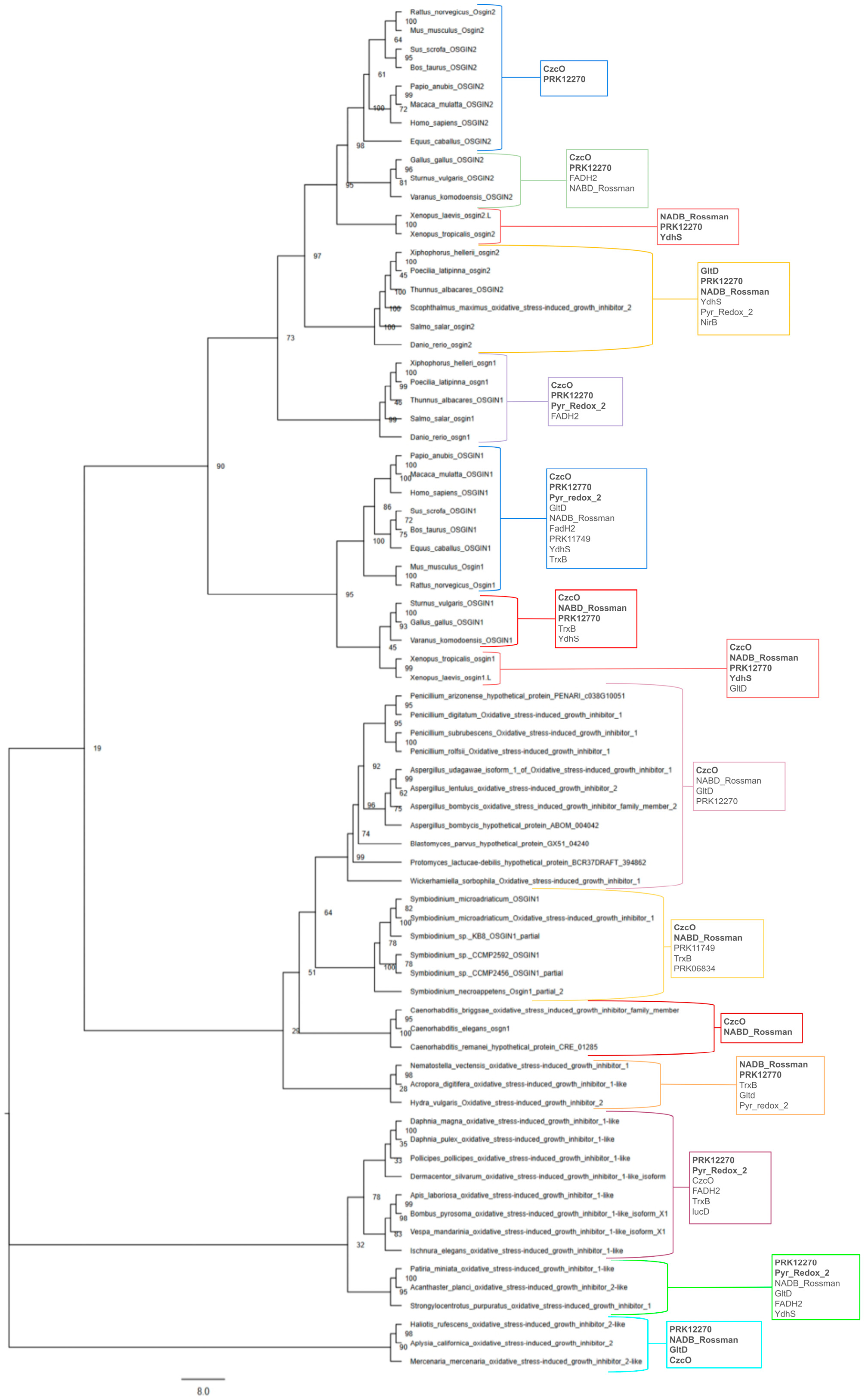
3. Phylogeny and Evolution of Osgin Genes
4. Structure and Biochemical Function of OSGIN Proteins
4.1. Genomic Structure
4.2. Differential Splicing
4.3. Protein Structure
4.4. Biochemical Function
5. Physiological Function of OSGIN Proteins
5.1. Role of OSGIN in Detecting and Countering Oxidative Stress
5.2. Role of OSGIN in Regulating Cell Proliferation
5.3. Role of OSGIN in Mediating Apoptosis
5.4. Role of OSGIN in Mediating Ferroptosis
5.5. Role of OSGIN in Mediating Autophagy
5.6. Regulation of Osgin Expression
6. Expression of Osgin Genes
6.1. Osgin Gene Expression in Invertebrates
6.2. Osgin Gene Expression During Vertebrate Development
6.3. Osgin Gene Expression in Adult Vertebrate Organisms
7. The Role of Osgin in Disease
7.1. The Role of Osgin in Breast Cancer
7.2. The Role of Osgin in Hepatocellular Cancer
7.3. The Role of Osgin in Ovarian Cancer
7.4. The Role of Osgin in Other Cancers
7.5. The Role of Osgin in Other Conditions and Diseases
| Pathology | Method | Osgin1 Expression | Osgin2 Expression |
|---|---|---|---|
| Breast Cancer | RNA-seq | ↓ | ↑ |
| Cholangiocarcinoma | RNA-seq | ↓ | ↑ |
| Hepatocellular Carcinoma | RNA-seq | ↑ | ↑ |
| Lung Carcinoma | RNA-seq/ microarray | ↑ | ↓ |
| Osteosarcoma | RNA-seq/ microarray | ↓ | ↑ |
| Glioma | RNA-seq/ microarray | ↓ | ↑ |
| Nevus Sebaceous of Jadassohn | RNA-seq | ↑ | ↓ |
| Sepsis | RNA-seq | ↑ | ↓ |
| Graft vs. Host Disease | RNA-seq | ↓ | ↑ |
8. Conclusions
9. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMPK | Adenosine monophosphate-activated protein kinase |
| ARE | Antioxidant response element |
| ATM | Ataxia-telangiectasia mutated |
| BAC | Bacterial artificial chromosome |
| BDGI | Bone marrow stromal cell-derived growth inhibitor |
| BYA | Billion years ago |
| C8orf1 | Chromosome 8 open reading frame 1 |
| CDD | Conserved Domain Database |
| CG | Chorionic gonadotropin |
| COPD | Chronic obstructive pulmonary disease |
| DHA | Docosahexaenoic acid |
| DTNB | 5,5′-dithiobis(2-nitrobenzoate) |
| ER | Endoplasmic reticulum |
| ETC | Electron transport chain |
| FAD | Flavin adenine dinucleotide |
| FMO | Flavin-containing monooxygenase |
| GCLM | Glutamate-cysteine ligase modifier subunit |
| GSH | Glutathione |
| HBEC | Human bronchial epithelial cell |
| hT41 | Human testis 4.1-kb transcript |
| ICC | Immunocytochemistry |
| IHC | Immunohistochemistry |
| IK2 | Ikaros factor 2 |
| ISH | In situ hybridization |
| MDA | Malondialdehyde |
| MEGA | Molecular Evolutionary Genetics Analysis |
| MGI | Mouse Genome Informatics |
| ML | Maximum likelihood |
| MSL | Mesenchymal stem-like |
| NAD(P)H | Nicotinamide adenine dinucleotide phosphate hydrogen |
| NBS | Nijmegen breakage syndrome |
| NCBI | National Center for Biotechnology Information |
| NGF | Nerve growth factor |
| NMR | Nuclear magnetic resonance |
| NSCLC | Non-small cell lung cancer |
| OKL38 | Ovary, kidney, and liver protein 38 |
| ORF | Open reading frame |
| Osgin | Oxidative stress-induced growth inhibitor |
| Osgin1 | Oxidative stress-induced growth inhibitor 1 |
| Osgin2 | Oxidative stress-induced growth inhibitor 2 |
| OXEG | Oxidative stress-related gene |
| OxPAPC | Oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero3-phosphorylcholine |
| P1 | Promoter 1 |
| P2 | Promoter 2 |
| PCOS | Polycystic ovary syndrome |
| PCR | Polymerase chain reaction |
| PDAC | Pancreatic ductal adenocarcinoma cells |
| PLOOH | Phospholipid peroxidases |
| PM2.5 | Fine (diameter of 2.5 μm or less) particulate matter |
| qRT-PCR | Quantitative reverse transcription polymerase chain reaction |
| RACE | Rapid amplification of cDNA ends |
| RGD | Rat Genome Database |
| RNA-seq | RNA sequencing |
| ROS | Reactive oxygen species |
| RPKM | Reads per kilobase million |
| SAR | Stramenopiles, Alveolates, and Rhizaria |
| SLC2A3 | Solute carrier family 2 member 3 |
| TCGA-STAD | The Cancer Genome Atlas Stomach Adenocarcinoma |
| TNB | Nitro-5-thiobenzoate |
| TNBC | Triple-negative breast cancer |
| TPE | Transcripts per embryo |
| TPM | Transcripts per million |
| UTR | Untranslated region |
References
- Preiser, J.C. Oxidative stress. JPEN J. Parenter. Enteral Nutr. 2012, 36, 147–154. [Google Scholar] [CrossRef]
- Hamanaka, R.B.; Chandel, N.S. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem. Sci. 2010, 35, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef] [PubMed]
- Krumova, K.; Cosa, G. Overview of Reactive Oxygen Species. In Singlet Oxygen: Applications in Biosciences and Nanosciences; Nonell, S., Flors, C., Nonell, S., Flors, C., Eds.; The Royal Society of Chemistry: London, UK, 2016; pp. 1–21. [Google Scholar]
- Ahmed, O.M.; Mohammed, M.T. Oxidative Stress: The Role of Reactive Oxygen Species (ROS) and Antioxidants in Human Diseases. Plant Arch. 2020, 20, 4089–4096. [Google Scholar]
- Jomova, K.; Raptova, R.; Alomar, S.Y.; Alwasel, S.H.; Nepovimova, E.; Kuca, K.; Valko, M. Reactive oxygen species, toxicity, oxidative stress, and antioxidants: Chronic diseases and aging. Arch. Toxicol. 2023, 97, 2499–2574. [Google Scholar] [CrossRef]
- Ray, P.D.; Huang, B.W.; Tsuji, Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal 2012, 24, 981–990. [Google Scholar] [CrossRef]
- Khorsandi, K.; Hosseinzadeh, R.; Esfahani, H.; Zandsalimi, K.; Shahidi, F.K.; Abrahamse, H. Accelerating skin regeneration and wound healing by controlled ROS from photodynamic treatment. Inflamm. Regen. 2022, 42, 40. [Google Scholar] [CrossRef]
- Hong, Y.; Boiti, A.; Vallone, D.; Foulkes, N.S. Reactive Oxygen Species Signaling and Oxidative Stress: Transcriptional Regulation and Evolution. Antioxidants 2024, 13, 312. [Google Scholar] [CrossRef]
- Nakamura, H.; Takada, K. Reactive oxygen species in cancer: Current findings and future directions. Cancer Sci. 2021, 112, 3945–3952. [Google Scholar] [CrossRef]
- Pizzino, G.; Irrera, N.; Cucinotta, M.; Pallio, G.; Mannino, F.; Arcoraci, V.; Squadrito, F.; Altavilla, D.; Bitto, A. Oxidative Stress: Harms and Benefits for Human Health. Oxid. Med. Cell Longev. 2017, 2017, 8416763. [Google Scholar] [CrossRef]
- Thannickal, V.J.; Fanburg, B.L. Reactive oxygen species in cell signaling. Am. J. Physiol. Lung Cell Mol. Physiol. 2000, 279, L1005–L1028. [Google Scholar] [CrossRef]
- Siauciunaite, R.; Foulkes, N.S.; Calabro, V.; Vallone, D. Evolution Shapes the Gene Expression Response to Oxidative Stress. Int. J. Mol. Sci. 2019, 20, 3040. [Google Scholar] [CrossRef]
- Lennicke, C.; Cocheme, H.M. Redox metabolism: ROS as specific molecular regulators of cell signaling and function. Mol. Cell 2021, 81, 3691–3707. [Google Scholar] [CrossRef]
- Ruder, E.H.; Hartman, T.J.; Blumberg, J.; Goldman, M.B. Oxidative stress and antioxidants: Exposure and impact on female fertility. Hum. Reprod. Update 2008, 14, 345–357. [Google Scholar] [CrossRef]
- Guerin, P.; El Mouatassim, S.; Menezo, Y. Oxidative stress and protection against reactive oxygen species in the pre-implantation embryo and its surroundings. Hum. Reprod. Update 2001, 7, 175–189. [Google Scholar] [CrossRef]
- Nimse, S.B.; Pal, D. Free radicals, natural antioxidants, and their reaction mechanisms. RSC Adv. 2015, 5, 27986–28006. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Huynh, H.; Ng, C.Y.; Ong, C.K.; Lim, K.B.; Chan, T.W. Cloning and characterization of a novel pregnancy-induced growth inhibitor in mammary gland. Endocrinology 2001, 142, 3607–3615. [Google Scholar] [CrossRef]
- Li, R.; Chen, W.; Yanes, R.; Lee, S.; Berliner, J.A. OKL38 is an oxidative stress response gene stimulated by oxidized phospholipids. J. Lipid Res. 2007, 48, 709–715. [Google Scholar] [CrossRef]
- Tauchi, H.; Matsuura, S.; Isomura, M.; Kinjo, T.; Nakamura, A.; Sakamoto, S.; Kondo, N.; Endo, S.; Komatsu, K.; Nakamura, Y. Sequence analysis of an 800-kb genomic DNA region on chromosome 8q21 that contains the Nijmegen breakage syndrome gene, NBS1. Genomics 1999, 55, 242–247. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.4.4, a Graphical Viewer of Phylogenetic Trees; University of Edinburgh, Institute of Evolutionary Biology: Edinburgh, UK, 2018. [Google Scholar]
- Goupil, E.; Lacroix, L.; Briere, J.; Guga, S.; Saba-El-Leil, M.K.; Meloche, S.; Labbe, J.C. OSGN-1 is a conserved flavin-containing monooxygenase required to stabilize the intercellular bridge in late cytokinesis. Proc. Natl. Acad. Sci. USA 2024, 121, e2308570121. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010. [Google Scholar] [CrossRef]
- Ong, C.K.; Ng, C.Y.; Leong, C.; Ng, C.P.; Ong, C.S.; Nguyen, T.T.; Huynh, H. Structural characterization of three novel rat OKL38 transcripts, their tissue distributions, and their regulation by human chorionic gonadotropin. Endocrinology 2004, 145, 4763–4774. [Google Scholar] [CrossRef]
- Satta, S.; Beal, R.; Smith, R.; Luo, X.; Ferris, G.R.; Langford-Smith, A.; Teasdale, J.; Ajime, T.T.; Serre, J.; Hazell, G.; et al. A Nrf2-OSGIN1&2-HSP70 axis mediates cigarette smoke-induced endothelial detachment: Implications for plaque erosion. Cardiovasc. Res. 2023, 119, 1869–1882. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Y. OSGIN1 (oxidative stress induced growth inhibitor 1). Atlas Genet. Cytogenet. Oncol. Haematol. 2014, 19, 117–120. [Google Scholar] [CrossRef]
- Tegenfeldt, F.; Kuznetsov, D.; Manni, M.; Berkeley, M.; Zdobnov, E.M.; Kriventseva, E.V. OrthoDB and BUSCO update: Annotation of orthologs in the widest sampling of organismal diversity. Nucleic Acids Res. 2025, 51, D516–D522. [Google Scholar] [CrossRef]
- Altenhoff, A.M.; Warwick Vesztrocy, A.; Bernard, C.; Train, C.M.; Nicheperovich, A.; Prieto Banos, S.; Julca, I.; Moi, D.; Nevers, Y.; Majidian, S.; et al. OMA orthology in 2024: Improved prokaryote coverage, ancestral and extant GO enrichment, a revamped synteny viewer and more in the OMA Ecosystem. Nucleic Acids Res. 2024, 52, D513–D521. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Parkinson, J.; Blaxter, M. SimiTri--visualizing similarity relationships for groups of sequences. Bioinformatics 2003, 19, 390–395. [Google Scholar] [CrossRef]
- Coghlan, A. Nematode genome evolution. In WormBook: The Online Review of C. elegans Biology; WormBook: Pasadena, CA, USA, 2005. [Google Scholar]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Pandurangan, A.P.; Stahlhacke, J.; Oates, M.E.; Smithers, B.; Gough, J. The SUPERFAMILY 2.0 database: A significant proteome update and a new webserver. Nucleic Acids Res. 2019, 47, D490–D494. [Google Scholar] [CrossRef]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–D215. [Google Scholar] [CrossRef]
- Ong, C.K.; Ng, C.Y.; Leong, C.; Ng, C.P.; Foo, K.T.; Tan, P.H.; Huynh, H. Genomic structure of human OKL38 gene and its differential expression in kidney carcinogenesis. J. Biol. Chem. 2004, 279, 743–754. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef]
- Ong, C.K.; Huynh, H. Interaction of Nuclear Factor 1 with promoter P1 of OSGIN1 represses transcription of OSGIN-1a variant. Cancer Res. 2008, 68, 165. [Google Scholar]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Martin, F.J.; Amode, M.R.; Aneja, A.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2023. Nucleic Acids Res. 2023, 51, D933–D941. [Google Scholar] [CrossRef]
- Hu, J.; Yao, H.; Gan, F.; Tokarski, A.; Wang, Y. Interaction of OKL38 and p53 in regulating mitochondrial structure and function. PLoS ONE 2012, 7, e43362. [Google Scholar] [CrossRef] [PubMed]
- Suzukawa, K.; Miura, K.; Mitsushita, J.; Resau, J.; Hirose, K.; Crystal, R.; Kamata, T. Nerve growth factor-induced neuronal differentiation requires generation of Rac1-regulated reactive oxygen species. J. Biol. Chem. 2000, 275, 13175–13178. [Google Scholar] [CrossRef] [PubMed]
- Hammerstad, M.; Gudim, I.; Hersleth, H.P. The Crystal Structures of Bacillithiol Disulfide Reductase Bdr (YpdA) Provide Structural and Functional Insight into a New Type of FAD-Containing NADPH-Dependent Oxidoreductase. Biochemistry 2020, 59, 4793–4798. [Google Scholar] [CrossRef] [PubMed]
- Goupil, E.; Amini, R.; Hall, D.H.; Labbe, J.C. Actomyosin contractility regulators stabilize the cytoplasmic bridge between the two primordial germ cells during Caenorhabditis elegans embryogenesis. Mol. Biol. Cell 2017, 28, 3789–3800. [Google Scholar] [CrossRef]
- Eswaramoorthy, S.; Bonanno, J.B.; Burley, S.K.; Swaminathan, S. Mechanism of action of a flavin-containing monooxygenase. Proc. Natl. Acad. Sci. USA 2006, 103, 9832–9837. [Google Scholar] [CrossRef]
- Huijbers, M.M.; Montersino, S.; Westphal, A.H.; Tischler, D.; van Berkel, W.J. Flavin dependent monooxygenases. Arch. Biochem. Biophys. 2014, 544, 2–17. [Google Scholar] [CrossRef]
- Reis, R.A.G.; Li, H.; Johnson, M.; Sobrado, P. New frontiers in flavin-dependent monooxygenases. Arch. Biochem. Biophys. 2021, 699, 108765. [Google Scholar] [CrossRef]
- Liu, M.; Li, Y.; Chen, L.; Chan, T.H.; Song, Y.; Fu, L.; Zeng, T.T.; Dai, Y.D.; Zhu, Y.H.; Li, Y.; et al. Allele-specific imbalance of oxidative stress-induced growth inhibitor 1 associates with progression of hepatocellular carcinoma. Gastroenterology 2014, 146, 1084–1096. [Google Scholar] [CrossRef]
- Yao, H.; Li, P.; Venters, B.J.; Zheng, S.; Thompson, P.R.; Pugh, B.F.; Wang, Y. Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J. Biol. Chem. 2008, 283, 20060–20068. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L.; Xu, J.; Tang, Y.; Huang, B.; Chen, Z.; Zhang, T.; Shen, H.M.; Wu, Y.; Xia, D. Bone marrow stromal cell-derived growth inhibitor serves as a stress sensor to induce autophagy. FEBS Lett. 2020, 594, 1248–1260. [Google Scholar] [CrossRef]
- Xie, X.; Laster, K.V.; Li, J.; Nie, W.; Yi, Y.W.; Liu, K.; Seong, Y.S.; Dong, Z.; Kim, D.J. OSGIN1 is a novel TUBB3 regulator that promotes tumor progression and gefitinib resistance in non-small cell lung cancer. Cell Mol. Life Sci. 2023, 80, 272. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Zhang, X.; Cai, Y.; Yu, H.; Cao, G.; Dai, E.; Kang, R.; Tang, D.; Hu, N.; Han, L. OSGIN1 promotes ferroptosis resistance by directly enhancing GCLM activity. Biochem. Biophys. Res. Commun. 2024, 740, 151015. [Google Scholar] [CrossRef]
- Deng, M.; Tang, F.; Chang, X.; Zhang, Y.; Liu, P.; Ji, X.; Zhang, Y.; Yang, R.; Jiang, J.; He, J.; et al. A targetable OSGIN1-AMPK-SLC2A3 axis controls the vulnerability of ovarian cancer to ferroptosis. NPJ Precis. Oncol. 2025, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Brennan, M.S.; Matos, M.F.; Richter, K.E.; Li, B.; Scannevin, R.H. The NRF2 transcriptional target, OSGIN1, contributes to monomethyl fumarate-mediated cytoprotection in human astrocytes. Sci. Rep. 2017, 7, 42054. [Google Scholar] [CrossRef]
- Hammad, S.M.; Twal, W.O.; Barth, J.L.; Smith, K.J.; Saad, A.F.; Virella, G.; Argraves, W.S.; Lopes-Virella, M.F. Oxidized LDL immune complexes and oxidized LDL differentially affect the expression of genes involved with inflammation and survival in human U937 monocytic cells. Atherosclerosis 2009, 202, 394–404. [Google Scholar] [CrossRef]
- Ong, C.K.; Leong, C.; Tan, P.H.; Van, T.; Huynh, H. The role of 5′ untranslated region in translational suppression of OKL38 mRNA in hepatocellular carcinoma. Oncogene 2007, 26, 1155–1165. [Google Scholar] [CrossRef]
- Yuan, Q.; Zhu, H.; Liu, H.; Wang, M.; Chu, H.; Zhang, Z. METTL3 regulates PM2.5-induced cell injury by targeting OSGIN1 in human airway epithelial cells. J. Hazard. Mater. 2021, 415, 125573. [Google Scholar] [CrossRef]
- Yamashita, N.; Uchiyama, M.; Yamagata, R.; Hwang, G.W. Methylmercury Induces Apoptosis in Mouse C17.2 Neural Stem Cells through the Induction of OSGIN1 Expression by NRF2. Int. J. Mol. Sci. 2024, 25, 3886. [Google Scholar] [CrossRef]
- Wang, G.; Zhou, H.; Strulovici-Barel, Y.; Al-Hijji, M.; Ou, X.; Salit, J.; Walters, M.S.; Staudt, M.R.; Kaner, R.J.; Crystal, R.G. Role of OSGIN1 in mediating smoking-induced autophagy in the human airway epithelium. Autophagy 2017, 13, 1205–1220. [Google Scholar] [CrossRef]
- Khoi, C.S.; Xiao, C.Q.; Hung, K.Y.; Lin, T.Y.; Chiang, C.K. Oxidative Stress-Induced Growth Inhibitor (OSGIN1), a Target of X-Box-Binding Protein 1, Protects Palmitic Acid-Induced Vascular Lipotoxicity through Maintaining Autophagy. Biomedicines 2022, 10, 992. [Google Scholar] [CrossRef]
- Zheng, Y.; Sun, W.; Wang, Z.; Liu, J.; Shan, C.; He, C.; Li, B.; Hu, X.; Zhu, W.; Liu, L.; et al. Activation of Pancreatic Acinar FXR Protects against Pancreatitis via Osgin1-Mediated Restoration of Efficient Autophagy. Research 2022, 2022, 9784081. [Google Scholar] [CrossRef] [PubMed]
- Defamie, V.; Cursio, R.; Le Brigand, K.; Moreilhon, C.; Saint-Paul, M.C.; Laurens, M.; Crenesse, D.; Cardinaud, B.; Auberger, P.; Gugenheim, J.; et al. Gene expression profiling of human liver transplants identifies an early transcriptional signature associated with initial poor graft function. Am. J. Transplant. 2008, 8, 1221–1236. [Google Scholar] [CrossRef]
- Kessler, J.; Rot, S.; Bache, M.; Kappler, M.; Wurl, P.; Vordermark, D.; Taubert, H.; Greither, T. miR-199a-5p regulates HIF-1alpha and OSGIN2 and its expression is correlated to soft-tissue sarcoma patients’ outcome. Oncol. Lett. 2016, 12, 5281–5288. [Google Scholar] [CrossRef]
- Wang, P.; Zhu, Y.; Jia, X.; Ying, X.; Sun, L.; Ruan, S. Clinical prognostic value of OSGIN2 in gastric cancer and its proliferative effect in vitro. Sci. Rep. 2023, 13, 5775. [Google Scholar] [CrossRef]
- Shuai, Y.; Liu, B.; Rong, L.; Shao, B.; Chen, B.; Jin, L. OSGIN2 regulates osteogenesis of jawbone BMSCs in osteoporotic rats. BMC Mol. Cell Biol. 2022, 23, 22. [Google Scholar] [CrossRef]
- Raharijaona, M.; Le Pennec, S.; Poirier, J.; Mirebeau-Prunier, D.; Rouxel, C.; Jacques, C.; Fontaine, J.F.; Malthiery, Y.; Houlgatte, R.; Savagner, F. PGC-1-related coactivator modulates mitochondrial-nuclear crosstalk through endogenous nitric oxide in a cellular model of oncocytic thyroid tumours. PLoS ONE 2009, 4, e7964. [Google Scholar] [CrossRef]
- Romanoski, C.E.; Che, N.; Yin, F.; Mai, N.; Pouldar, D.; Civelek, M.; Pan, C.; Lee, S.; Vakili, L.; Yang, W.P.; et al. Network for activation of human endothelial cells by oxidized phospholipids: A critical role of heme oxygenase 1. Circ. Res. 2011, 109, e27–e41. [Google Scholar] [CrossRef]
- Boscaro, C.; Schimdt, G.; Cignarella, A.; Dal Maso, L.; Bolego, C.; Trevisi, L. The antiangiogenic effect of digitoxin is dependent on a ROS-elicited RhoA/ROCK pathway activation. Biochem. Pharmacol. 2024, 222, 116049. [Google Scholar] [CrossRef]
- Hobbs, G.A.; Zhou, B.; Cox, A.D.; Campbell, S.L. Rho GTPases, oxidation, and cell redox control. Small GTPases 2014, 5, e28579. [Google Scholar] [CrossRef]
- Hurst, M.; McGarry, D.J.; Olson, M.F. Rho GTPases: Non-canonical regulation by cysteine oxidation. Bioessays 2022, 44, e2100152. [Google Scholar] [CrossRef]
- Liang, D.; Minikes, A.M.; Jiang, X. Ferroptosis at the intersection of lipid metabolism and cellular signaling. Mol. Cell 2022, 82, 2215–2227. [Google Scholar] [CrossRef] [PubMed]
- Baird, L.; Yamamoto, M. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol. Cell. Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef]
- Yan, X.; Lee, S.; Gugiu, B.G.; Koroniak, L.; Jung, M.E.; Berliner, J.; Cheng, J.; Li, R. Fatty acid epoxyisoprostane E2 stimulates an oxidative stress response in endothelial cells. Biochem. Biophys. Res. Commun. 2014, 444, 69–74. [Google Scholar] [CrossRef]
- Tsai, C.H.; Shen, Y.C.; Chen, H.W.; Liu, K.L.; Chang, J.W.; Chen, P.Y.; Lin, C.Y.; Yao, H.T.; Li, C.C. Docosahexaenoic acid increases the expression of oxidative stress-induced growth inhibitor 1 through the PI3K/Akt/Nrf2 signaling pathway in breast cancer cells. Food Chem. Toxicol. 2017, 108, 276–288. [Google Scholar] [CrossRef]
- Sternberg, P.W.; Van Auken, K.; Wang, Q.; Wright, A.; Yook, K.; Zarowiecki, M.; Arnaboldi, V.; Becerra, A.; Brown, S.; Cain, S.; et al. WormBase 2024: Status and transitioning to Alliance infrastructure. Genetics 2024, 227, iyae050. [Google Scholar] [CrossRef]
- Wolkow, C.A.; Hall, D.H. Introduction to the Dauer Larva, Overview. WormAtlas 2015. [Google Scholar] [CrossRef]
- Kudtarkar, P.; Cameron, R.A. Echinobase: An expanding resource for echinoderm genomic information. Database 2017, 2017, bax074. [Google Scholar] [CrossRef]
- Giraldo-Calderon, G.I.; Emrich, S.J.; MacCallum, R.M.; Maslen, G.; Dialynas, E.; Topalis, P.; Ho, N.; Gesing, S.; the VectorBase Consortium; Madey, G.; et al. VectorBase: An updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015, 43, D707–D713. [Google Scholar] [CrossRef]
- Bastian, F.B.; Cammarata, A.B.; Carsanaro, S.; Detering, H.; Huang, W.T.; Joye, S.; Niknejad, A.; Nyamari, M.; Mendes de Farias, T.; Moretti, S.; et al. Bgee in 2024: Focus on curated single-cell RNA-seq datasets, and query tools. Nucleic Acids Res. 2025, 53, D878–D885. [Google Scholar] [CrossRef]
- Karimi, K.; Fortriede, J.D.; Lotay, V.S.; Burns, K.A.; Wang, D.Z.; Fisher, M.E.; Pells, T.J.; James-Zorn, C.; Wang, Y.; Ponferrada, V.G.; et al. Xenbase: A genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res. 2018, 46, D861–D868. [Google Scholar] [CrossRef]
- Thisse, C.; Thisse, B. High Throughput Expression Analysis of ZF-Models Consortium Clones. Available online: http://zfin.org. (accessed on 4 March 2025).
- Diez-Roux, G.; Banfi, S.; Sultan, M.; Geffers, L.; Anand, S.; Rozado, D.; Magen, A.; Canidio, E.; Pagani, M.; Peluso, I.; et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011, 9, e1000582. [Google Scholar] [CrossRef] [PubMed]
- Bottoni, L.; Minetti, A.; Realini, G.; Pio, E.; Giustarini, D.; Rossi, R.; Rocchio, C.; Franci, L.; Salvini, L.; Catona, O.; et al. NRF2 activation by cysteine as a survival mechanism for triple-negative breast cancer cells. Oncogene 2024, 43, 1701–1713. [Google Scholar] [CrossRef] [PubMed]
- Arslan, A.A.; Tuminello, S.; Yang, L.; Zhang, Y.; Durmus, N.; Snuderl, M.; Heguy, A.; Zeleniuch-Jacquotte, A.; Shao, Y.; Reibman, J. Genome-Wide DNA Methylation Profiles in Community Members Exposed to the World Trade Center Disaster. Int. J. Environ. Res. Public Health 2020, 17, 5493. [Google Scholar] [CrossRef]
- Trichopoulos, D.; Hsieh, C.C.; MacMahon, B.; Lin, T.M.; Lowe, C.R.; Mirra, A.P.; Ravnihar, B.; Salber, E.J.; Valaoras, V.G.; Yuasa, S. Age at any birth and breast cancer risk. Int. J. Cancer 1983, 31, 701–704. [Google Scholar] [CrossRef] [PubMed]
- Russo, I.H.; Koszalka, M.; Russo, J. Human chorionic gonadotropin and rat mammary cancer prevention. J. Natl. Cancer Inst. 1990, 82, 1286–1289. [Google Scholar] [CrossRef]
- Tsai, C.H.; Lii, C.K.; Wang, T.S.; Liu, K.L.; Chen, H.W.; Huang, C.S.; Li, C.C. Docosahexaenoic acid promotes the formation of autophagosomes in MCF-7 breast cancer cells through oxidative stress-induced growth inhibitor 1 mediated activation of AMPK/mTOR pathway. Food Chem. Toxicol. 2021, 154, 112318. [Google Scholar] [CrossRef]
- Hojo, H.; Enya, S.; Arai, M.; Suzuki, Y.; Nojiri, T.; Kangawa, K.; Koyama, S.; Kawaoka, S. Remote reprogramming of hepatic circadian transcriptome by breast cancer. Oncotarget 2017, 8, 34128–34140. [Google Scholar] [CrossRef]
- Chen, Z.; Mei, K.; Xiao, Y.; Xiong, Y.; Long, W.; Wang, Q.; Zhong, J.; Di, D.; Ge, Y.; Luo, Y.; et al. Prognostic Assessment of Oxidative Stress-Related Genes in Colorectal Cancer and New Insights into Tumor Immunity. Oxid. Med. Cell Longev. 2022, 2022, 2518340. [Google Scholar] [CrossRef]
- Yetkin Yildirim, G.; Tola, E.N.; Dag, I. A novel biochemical marker -OKL38- with its apoptotic and antioxidant properties for the development of PCOS and its related clinical implications. Gynecol. Endocrinol. 2020, 36, 673–677. [Google Scholar] [CrossRef]
- Tang, X.; Zhu, H.; Zhou, M.; Zhang, H.; Xiao, Q.; Yuan, Q.; Sun, G.; Zhang, Z.; Chu, H. OSGIN1 regulates PM2.5-induced fibrosis via mediating autophagy in an in vitro model of COPD. Toxicol. Lett. 2024, 401, 35–43. [Google Scholar] [CrossRef]
- Eswaran, J.; Cyanam, D.; Mudvari, P.; Reddy, S.D.; Pakala, S.B.; Nair, S.S.; Florea, L.; Fuqua, S.A.; Godbole, S.; Kumar, R. Transcriptomic landscape of breast cancers through mRNA sequencing. Sci. Rep. 2012, 2, 264. [Google Scholar] [CrossRef] [PubMed]
- Papatheodorou, I.; Moreno, P.; Manning, J.; Fuentes, A.M.; George, N.; Fexova, S.; Fonseca, N.A.; Fullgrabe, A.; Green, M.; Huang, N.; et al. Expression Atlas update: From tissues to single cells. Nucleic Acids Res. 2020, 48, D77–D83. [Google Scholar] [CrossRef]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Sheng, J.; Gao, D.; Li, F.; Durrans, A.; Ryu, S.; Lee, S.B.; Narula, N.; Rafii, S.; Elemento, O.; et al. Transcriptome analysis of individual stromal cell populations identifies stroma-tumor crosstalk in mouse lung cancer model. Cell Rep. 2015, 10, 1187–1201. [Google Scholar] [CrossRef]
- Straessler, K.M.; Jones, K.B.; Hu, H.; Jin, H.; van de Rijn, M.; Capecchi, M.R. Modeling clear cell sarcomagenesis in the mouse: Cell of origin differentiation state impacts tumor characteristics. Cancer Cell 2013, 23, 215–227. [Google Scholar] [CrossRef]
- Jones, K.B.; Salah, Z.; Del Mare, S.; Galasso, M.; Gaudio, E.; Nuovo, G.J.; Lovat, F.; LeBlanc, K.; Palatini, J.; Randall, R.L.; et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012, 72, 1865–1877. [Google Scholar] [CrossRef]
- Linsley, P.S.; Speake, C.; Whalen, E.; Chaussabel, D. Copy number loss of the interferon gene cluster in melanomas is linked to reduced T cell infiltrate and poor patient prognosis. PLoS ONE 2014, 9, e109760. [Google Scholar] [CrossRef]
- Wu, M.; Chen, Y.; Xia, H.; Wang, C.; Tan, C.Y.; Cai, X.; Liu, Y.; Ji, F.; Xiong, P.; Liu, R.; et al. Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc. Natl. Acad. Sci. USA 2020, 117, 28336–28343. [Google Scholar] [CrossRef]
- Cho, J.S.; Guo, Y.; Ramos, R.I.; Hebroni, F.; Plaisier, S.B.; Xuan, C.; Granick, J.L.; Matsushima, H.; Takashima, A.; Iwakura, Y.; et al. Neutrophil-derived IL-1beta is sufficient for abscess formation in immunity against Staphylococcus aureus in mice. PLoS Pathog. 2012, 8, e1003047. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Bult, C.J.; Blake, J.A.; Smith, C.L.; Kadin, J.A.; Richardson, J.E.; Mouse Genome Database, G. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 2019, 47, D801–D806. [Google Scholar] [CrossRef] [PubMed]
- White, R.J.; Collins, J.E.; Sealy, I.M.; Wali, N.; Dooley, C.M.; Digby, Z.; Stemple, D.L.; Murphy, D.N.; Billis, K.; Hourlier, T.; et al. A high-resolution mRNA expression time course of embryonic development in zebrafish. eLife 2017, 6, e30860. [Google Scholar] [CrossRef] [PubMed]
- Owens, N.D.L.; Blitz, I.L.; Lane, M.A.; Patrushev, I.; Overton, J.D.; Gilchrist, M.J.; Cho, K.W.Y.; Khokha, M.K. Measuring Absolute RNA Copy Numbers at High Temporal Resolution Reveals Transcriptome Kinetics in Development. Cell Rep. 2016, 14, 632–647. [Google Scholar] [CrossRef]
- Lindsay, S.J.; Xu, Y.; Lisgo, S.N.; Harkin, L.F.; Copp, A.J.; Gerrelli, D.; Clowry, G.J.; Talbot, A.; Keogh, M.J.; Coxhead, J.; et al. HDBR Expression: A Unique Resource for Global and Individual Gene Expression Studies during Early Human Brain Development. Front. Neuroanat. 2016, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium, Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef]
- Pipes, L.; Li, S.; Bozinoski, M.; Palermo, R.; Peng, X.; Blood, P.; Kelly, S.; Weiss, J.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; et al. The non-human primate reference transcriptome resource (NHPRTR) for comparative functional genomics. Nucleic Acids Res. 2013, 41, D906–D914. [Google Scholar] [CrossRef]
- Steen, R.G.; Kwitek-Black, A.E.; Glenn, C.; Gullings-Handley, J.; Van Etten, W.; Atkinson, O.S.; Appel, D.; Twigger, S.; Muir, M.; Mull, T.; et al. A high-density integrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res. 1999, 9, AP1–AP8. [Google Scholar] [CrossRef]
- Allen Institute for Brain Science. Allen Developing Mouse Brain Atlas. 2025. Available online: https://developingmouse.brain-map.org/ (accessed on 4 March 2025).
- Session, A.M.; Uno, Y.; Kwon, T.; Chapman, J.A.; Toyoda, A.; Takahashi, S.; Fukui, A.; Hikosaka, A.; Suzuki, A.; Kondo, M.; et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 2016, 538, 336–343. [Google Scholar] [CrossRef]
- Cardoso-Moreira, M.; Halbert, J.; Valloton, D.; Velten, B.; Chen, C.; Shao, Y.; Liechti, A.; Ascencao, K.; Rummel, C.; Ovchinnikova, S.; et al. Gene expression across mammalian organ development. Nature 2019, 571, 505–509. [Google Scholar] [CrossRef]
- Mirzamohammadi, F.; Kozlova, A.; Papaioannou, G.; Paltrinieri, E.; Ayturk, U.M.; Kobayashi, T. Distinct molecular pathways mediate Mycn and Myc-regulated miR-17-92 microRNA action in Feingold syndrome mouse models. Nat. Commun. 2018, 9, 1352. [Google Scholar] [CrossRef]
- Delpretti, S.; Montavon, T.; Leleu, M.; Joye, E.; Tzika, A.; Milinkovitch, M.; Duboule, D. Multiple enhancers regulate Hoxd genes and the Hotdog LncRNA during cecum budding. Cell Rep. 2013, 5, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Tallack, M.R.; Magor, G.W.; Dartigues, B.; Sun, L.; Huang, S.; Fittock, J.M.; Fry, S.V.; Glazov, E.A.; Bailey, T.L.; Perkins, A.C. Novel roles for KLF1 in erythropoiesis revealed by mRNA-seq. Genome Res. 2012, 22, 2385–2398. [Google Scholar] [CrossRef] [PubMed]
- Dupacova, N.; Antosova, B.; Paces, J.; Kozmik, Z. Meis homeobox genes control progenitor competence in the retina. Proc. Natl. Acad. Sci. USA 2021, 118, e2013136118. [Google Scholar] [CrossRef] [PubMed]
- Kraiczy, J.; Nayak, K.M.; Howell, K.J.; Ross, A.; Forbester, J.; Salvestrini, C.; Mustata, R.; Perkins, S.; Andersson-Rolf, A.; Leenen, E.; et al. DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development. Gut 2019, 68, 49–61. [Google Scholar] [CrossRef]
- Yamaji, D.; Kang, K.; Robinson, G.W.; Hennighausen, L. Sequential activation of genetic programs in mouse mammary epithelium during pregnancy depends on STAT5A/B concentration. Nucleic Acids Res. 2013, 41, 1622–1636. [Google Scholar] [CrossRef]
- Kistler, W.S.; Baas, D.; Lemeille, S.; Paschaki, M.; Seguin-Estevez, Q.; Barras, E.; Ma, W.; Duteyrat, J.L.; Morle, L.; Durand, B.; et al. RFX2 Is a Major Transcriptional Regulator of Spermiogenesis. PLoS Genet. 2015, 11, e1005368. [Google Scholar] [CrossRef]
- Merkin, J.; Russell, C.; Chen, P.; Burge, C.B. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 2012, 338, 1593–1599. [Google Scholar] [CrossRef]
- Chang, A.J.; Ortega, F.E.; Riegler, J.; Madison, D.V.; Krasnow, M.A. Oxygen regulation of breathing through an olfactory receptor activated by lactate. Nature 2015, 527, 240–244. [Google Scholar] [CrossRef]
- Suzuki, T.; Kikuguchi, C.; Nishijima, S.; Nagashima, T.; Takahashi, A.; Okada, M.; Yamamoto, T. Postnatal liver functional maturation requires Cnot complex-mediated decay of mRNAs encoding cell cycle and immature liver genes. Development 2019, 146, dev168146. [Google Scholar] [CrossRef]
- Jain, M.; Kaiser, R.W.J.; Bohl, K.; Hoehne, M.; Gobel, H.; Bartram, M.P.; Habbig, S.; Muller, R.U.; Fogo, A.B.; Benzing, T.; et al. Inactivation of Apoptosis Antagonizing Transcription Factor in tubular epithelial cells induces accumulation of DNA damage and nephronophthisis. Kidney Int. 2019, 95, 846–858. [Google Scholar] [CrossRef]
- Fuhrmann, D.; Mernberger, M.; Nist, A.; Stiewe, T.; Elsasser, H.P. Miz1 Controls Schwann Cell Proliferation via H3K36(me2) Demethylase Kdm8 to Prevent Peripheral Nerve Demyelination. J. Neurosci. 2018, 38, 858–877. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Vitting-Seerup, K.; Waage, J.; Tang, C.; Ge, Y.; Porse, B.T.; Yan, W. UPF2-Dependent Nonsense-Mediated mRNA Decay Pathway Is Essential for Spermatogenesis by Selectively Eliminating Longer 3′UTR Transcripts. PLoS Genet. 2016, 12, e1005863. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, D.; Yanagiya, A.; Georgiadou, E.; Wu, Y.; Stylianides, T.; Rutter, G.A.; Suzuki, T.; Yamamoto, T. Loss of beta-cell identity and diabetic phenotype in mice caused by disruption of CNOT3-dependent mRNA deadenylation. Commun. Biol. 2020, 3, 476. [Google Scholar] [CrossRef]
- Mishra, V.; Bose, A.; Kiran, S.; Banerjee, S.; Shah, I.A.; Chaukimath, P.; Reshi, M.M.; Srinivas, S.; Barman, A.; Visweswariah, S.S. Gut-associated cGMP mediates colitis and dysbiosis in a mouse model of an activating mutation in GUCY2C. J. Exp. Med. 2021, 218, e20210479. [Google Scholar] [CrossRef]
- Anuar, N.D.; Kurscheid, S.; Field, M.; Zhang, L.; Rebar, E.; Gregory, P.; Buchou, T.; Bowles, J.; Koopman, P.; Tremethick, D.J.; et al. Gene editing of the multi-copy H2A.B gene and its importance for fertility. Genome Biol. 2019, 20, 23. [Google Scholar] [CrossRef]
- Martin-Alonso, M.; Iqbal, S.; Vornewald, P.M.; Lindholm, H.T.; Damen, M.J.; Martinez, F.; Hoel, S.; Diez-Sanchez, A.; Altelaar, M.; Katajisto, P.; et al. Smooth muscle-specific MMP17 (MT4-MMP) regulates the intestinal stem cell niche and regeneration after damage. Nat. Commun. 2021, 12, 6741. [Google Scholar] [CrossRef]
- Takahashi, A.; Takaoka, S.; Kobori, S.; Yamaguchi, T.; Ferwati, S.; Kuba, K.; Yamamoto, T.; Suzuki, T. The CCR4—NOT Deadenylase Complex Maintains Adipocyte Identity. Int. J. Mol. Sci. 2019, 20, 5274. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, Y.K.; Lee, K.P.; Lee, S.M.; Kang, T.W.; Kim, H.J.; Dho, S.H.; Kim, S.Y.; Kwon, K.S. Genome-wide profiling of the microRNA-mRNA regulatory network in skeletal muscle with aging. Aging 2014, 6, 524–544. [Google Scholar] [CrossRef]
- Patial, S.; Stumpo, D.J.; Young, W.S., 3rd; Ward, J.M.; Flake, G.P.; Blackshear, P.J. Effects of Combined Tristetraprolin/Tumor Necrosis Factor Receptor Deficiency on the Splenic Transcriptome. Mol. Cell Biol. 2016, 36, 1395–1411. [Google Scholar] [CrossRef]
- Haering, C.; Kanageswaran, N.; Bouvain, P.; Scholz, P.; Altmuller, J.; Becker, C.; Gisselmann, G.; Waring-Bischof, J.; Hatt, H. Ion transporter NKCC1, modulator of neurogenesis in murine olfactory neurons. J. Biol. Chem. 2015, 290, 9767–9779. [Google Scholar] [CrossRef]
- West, D.B.; Engelhard, E.K.; Adkisson, M.; Nava, A.J.; Kirov, J.V.; Cipollone, A.; Willis, B.; Rapp, J.; de Jong, P.J.; Lloyd, K.C. Transcriptome Analysis of Targeted Mouse Mutations Reveals the Topography of Local Changes in Gene Expression. PLoS Genet. 2016, 12, e1005691. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.; Zidek, L.M.; Ackermann, T.; de Jong, T.; Liu, P.; Kliche, V.; Zaini, M.A.; Kortman, G.; Harkema, L.; Verbeek, D.S.; et al. Reduced expression of C/EBPβ-LIP extends health and lifespan in mice. eLife 2018, 7, e34985. [Google Scholar] [CrossRef] [PubMed]
- Guccini, I.; Revandkar, A.; D’Ambrosio, M.; Colucci, M.; Pasquini, E.; Mosole, S.; Troiani, M.; Brina, D.; Sheibani-Tezerji, R.; Elia, A.R.; et al. Senescence Reprogramming by TIMP1 Deficiency Promotes Prostate Cancer Metastasis. Cancer Cell 2021, 39, 68–82.e69. [Google Scholar] [CrossRef]
- Munger, S.C.; Raghupathy, N.; Choi, K.; Simons, A.K.; Gatti, D.M.; Hinerfeld, D.A.; Svenson, K.L.; Keller, M.P.; Attie, A.D.; Hibbs, M.A.; et al. RNA-Seq alignment to individualized genomes improves transcript abundance estimates in multiparent populations. Genetics 2014, 198, 59–73. [Google Scholar] [CrossRef]
- Bond, S.T.; King, E.J.; Henstridge, D.C.; Tran, A.; Moody, S.C.; Yang, C.; Liu, Y.; Mellett, N.A.; Nath, A.P.; Inouye, M.; et al. Deletion of Trim28 in committed adipocytes promotes obesity but preserves glucose tolerance. Nat. Commun. 2021, 12, 74. [Google Scholar] [CrossRef]
- O’Rourke, J.G.; Bogdanik, L.; Muhammad, A.; Gendron, T.F.; Kim, K.J.; Austin, A.; Cady, J.; Liu, E.Y.; Zarrow, J.; Grant, S.; et al. C9orf72 BAC Transgenic Mice Display Typical Pathologic Features of ALS/FTD. Neuron 2015, 88, 892–901. [Google Scholar] [CrossRef]
- Ishimura, R.; Nagy, G.; Dotu, I.; Chuang, J.H.; Ackerman, S.L. Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. eLife 2016, 5, e14295. [Google Scholar] [CrossRef]
- Brooks, M.J.; Rajasimha, H.K.; Roger, J.E.; Swaroop, A. Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl−/− retinal transcriptomes. Mol. Vis. 2011, 17, 3034–3054. [Google Scholar]
- Narayanan, M.; Huynh, J.L.; Wang, K.; Yang, X.; Yoo, S.; McElwee, J.; Zhang, B.; Zhang, C.; Lamb, J.R.; Xie, T.; et al. Common dysregulation network in the human prefrontal cortex underlies two neurodegenerative diseases. Mol. Syst. Biol. 2014, 10, 743. [Google Scholar] [CrossRef]
- Aldunate, E.Z.; Di Foggia, V.; Di Marco, F.; Hervas, L.A.; Ribeiro, J.C.; Holder, D.L.; Patel, A.; Jannini, T.B.; Thompson, D.A.; Martinez-Barbera, J.P.; et al. Conditional Dicer1 depletion using Chrnb4-Cre leads to cone cell death and impaired photopic vision. Sci. Rep. 2019, 9, 2314. [Google Scholar] [CrossRef]
- Isaac, J.; Erthal, J.; Gordon, J.; Duverger, O.; Sun, H.W.; Lichtler, A.C.; Stein, G.S.; Lian, J.B.; Morasso, M.I. DLX3 regulates bone mass by targeting genes supporting osteoblast differentiation and mineral homeostasis in vivo. Cell Death Differ. 2014, 21, 1365–1376. [Google Scholar] [CrossRef] [PubMed]
- Rousset, F.; Nacher-Soler, G.; Coelho, M.; Ilmjarv, S.; Kokje, V.B.C.; Marteyn, A.; Cambet, Y.; Perny, M.; Roccio, M.; Jaquet, V.; et al. Redox activation of excitatory pathways in auditory neurons as mechanism of age-related hearing loss. Redox Biol. 2020, 30, 101434. [Google Scholar] [CrossRef] [PubMed]
- Jena, K.K.; Mehto, S.; Nath, P.; Chauhan, N.R.; Sahu, R.; Dhar, K.; Das, S.K.; Kolapalli, S.P.; Murmu, K.C.; Jain, A.; et al. Autoimmunity gene IRGM suppresses cGAS-STING and RIG-I-MAVS signaling to control interferon response. EMBO Rep. 2020, 21, e50051. [Google Scholar] [CrossRef]
- Huntley, M.A.; Lou, M.; Goldstein, L.D.; Lawrence, M.; Dijkgraaf, G.J.; Kaminker, J.S.; Gentleman, R. Complex regulation of ADAR-mediated RNA-editing across tissues. BMC Genom. 2016, 17, 61. [Google Scholar] [CrossRef]
- Zheng, C.L.; Wilmot, B.; Walter, N.A.; Oberbeck, D.; Kawane, S.; Searles, R.P.; McWeeney, S.K.; Hitzemann, R. Splicing landscape of the eight collaborative cross founder strains. BMC Genom. 2015, 16, 52. [Google Scholar] [CrossRef]
- Kao, C.S.; van Bruggen, R.; Kim, J.R.; Chen, X.X.L.; Chan, C.; Lee, J.; Cho, W.I.; Zhao, M.; Arndt, C.; Maksimovic, K.; et al. Selective neuronal degeneration in MATR3 S85C knock-in mouse model of early-stage ALS. Nat. Commun. 2020, 11, 5304. [Google Scholar] [CrossRef]
- Lipiec, M.A.; Bem, J.; Kozinski, K.; Chakraborty, C.; Urban-Ciecko, J.; Zajkowski, T.; Dabrowski, M.; Szewczyk, L.M.; Toval, A.; Ferran, J.L.; et al. TCF7L2 regulates postmitotic differentiation programmes and excitability patterns in the thalamus. Development 2020, 147, dev190181. [Google Scholar] [CrossRef]
- Klaus, C.; Hansen, J.N.; Ginolhac, A.; Gerard, D.; Gnanapragassam, V.S.; Horstkorte, R.; Rossdam, C.; Buettner, F.F.R.; Sauter, T.; Sinkkonen, L.; et al. Reduced sialylation triggers homeostatic synapse and neuronal loss in middle-aged mice. Neurobiol. Aging 2020, 88, 91–107. [Google Scholar] [CrossRef]
- Alen, F.; Gomez-Redondo, I.; Rivera, P.; Suarez, J.; Ramos-Ibeas, P.; Pericuesta, E.; Fernandez-Gonzalez, R.; Perez-Cerezales, S.; Horiuchi, K.; Orio, L.; et al. Sex-Dimorphic Behavioral Alterations and Altered Neurogenesis in U12 Intron Splicing-Defective Zrsr1 Mutant Mice. Int. J. Mol. Sci. 2019, 20, 3543. [Google Scholar] [CrossRef]
- Yu, Y.; Fuscoe, J.C.; Zhao, C.; Guo, C.; Jia, M.; Qing, T.; Bannon, D.I.; Lancashire, L.; Bao, W.; Du, T.; et al. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat. Commun. 2014, 5, 3230. [Google Scholar] [CrossRef]
- Langfelder, P.; Cantle, J.P.; Chatzopoulou, D.; Wang, N.; Gao, F.; Al-Ramahi, I.; Lu, X.H.; Ramos, E.M.; El-Zein, K.; Zhao, Y.; et al. Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat. Neurosci. 2016, 19, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.; Godfrey, A.K.; Hughes, J.F.; Goodheart, M.L.; Mitchell, R.N.; Page, D.C. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science 2019, 365, eaaw7317. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Samaras, P.; Frejno, M.; Gessulat, S.; Barnert, M.; Kienegger, H.; Krcmar, H.; Schlegl, J.; Ehrlich, H.C.; Aiche, S.; et al. ProteomicsDB. Nucleic Acids Res. 2018, 46, D1271–D1281. [Google Scholar] [CrossRef] [PubMed]
- Darbellay, F.; Necsulea, A. Comparative Transcriptomics Analyses across Species, Organs, and Developmental Stages Reveal Functionally Constrained lncRNAs. Mol. Biol. Evol. 2020, 37, 240–259. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallstrom, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Barbosa-Morais, N.L.; Irimia, M.; Pan, Q.; Xiong, H.Y.; Gueroussov, S.; Lee, L.J.; Slobodeniuc, V.; Kutter, C.; Watt, S.; Colak, R.; et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 2012, 338, 1587–1593. [Google Scholar] [CrossRef]
- Brawand, D.; Soumillon, M.; Necsulea, A.; Julien, P.; Csardi, G.; Harrigan, P.; Weier, M.; Liechti, A.; Aximu-Petri, A.; Kircher, M.; et al. The evolution of gene expression levels in mammalian organs. Nature 2011, 478, 343–348. [Google Scholar] [CrossRef]
- Soumillon, M.; Necsulea, A.; Weier, M.; Brawand, D.; Zhang, X.; Gu, H.; Barthes, P.; Kokkinaki, M.; Nef, S.; Gnirke, A.; et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013, 3, 2179–2190. [Google Scholar] [CrossRef]
- Duff, M.O.; Olson, S.; Wei, X.; Garrett, S.C.; Osman, A.; Bolisetty, M.; Plocik, A.; Celniker, S.E.; Graveley, B.R. Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature 2015, 521, 376–379. [Google Scholar] [CrossRef]
- Madhavan, S.; Zenklusen, J.C.; Kotliarov, Y.; Sahni, H.; Fine, H.A.; Buetow, K. Rembrandt: Helping personalized medicine become a reality through integrative translational research. Mol. Cancer Res. 2009, 7, 157–167. [Google Scholar] [CrossRef]
- Birks, D.K.; Donson, A.M.; Patel, P.R.; Sufit, A.; Algar, E.M.; Dunham, C.; Kleinschmidt-DeMasters, B.K.; Handler, M.H.; Vibhakar, R.; Foreman, N.K. Pediatric rhabdoid tumors of kidney and brain show many differences in gene expression but share dysregulation of cell cycle and epigenetic effector genes. Pediatr. Blood Cancer 2013, 60, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Kanth, P.; Bronner, M.P.; Boucher, K.M.; Burt, R.W.; Neklason, D.W.; Hagedorn, C.H.; Delker, D.A. Gene Signature in Sessile Serrated Polyps Identifies Colon Cancer Subtype. Cancer Prev. Res. 2016, 9, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hong, Y.; Jia, L.; Wu, J.; Xia, J.; Wang, J.; Hu, Q.; Cheng, B. Modulation of IL-1beta reprogrammes the tumor microenvironment to interrupt oral carcinogenesis. Sci. Rep. 2016, 6, 20208. [Google Scholar] [CrossRef]
- Griesinger, A.M.; Josephson, R.J.; Donson, A.M.; Mulcahy Levy, J.M.; Amani, V.; Birks, D.K.; Hoffman, L.M.; Furtek, S.L.; Reigan, P.; Handler, M.H.; et al. Interleukin-6/STAT3 Pathway Signaling Drives an Inflammatory Phenotype in Group A Ependymoma. Cancer Immunol. Res. 2015, 3, 1165–1174. [Google Scholar] [CrossRef]
- Madan, V.; Kanojia, D.; Li, J.; Okamoto, R.; Sato-Otsubo, A.; Kohlmann, A.; Sanada, M.; Grossmann, V.; Sundaresan, J.; Shiraishi, Y.; et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun. 2015, 6, 6042. [Google Scholar] [CrossRef]
- Woodward, W.A.; Krishnamurthy, S.; Yamauchi, H.; El-Zein, R.; Ogura, D.; Kitadai, E.; Niwa, S.; Cristofanilli, M.; Vermeulen, P.; Dirix, L.; et al. Genomic and expression analysis of microdissected inflammatory breast cancer. Breast Cancer Res. Treat. 2013, 138, 761–772. [Google Scholar] [CrossRef]
- Marcinkiewicz, K.M.; Gudas, L.J. Altered epigenetic regulation of homeobox genes in human oral squamous cell carcinoma cells. Exp. Cell Res. 2014, 320, 128–143. [Google Scholar] [CrossRef][Green Version]
- Haberman, Y.; Tickle, T.L.; Dexheimer, P.J.; Kim, M.O.; Tang, D.; Karns, R.; Baldassano, R.N.; Noe, J.D.; Rosh, J.; Markowitz, J.; et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Investig. 2014, 124, 3617–3633. [Google Scholar] [CrossRef]
- Voisin, G.; Bouvet, G.F.; Legendre, P.; Dagenais, A.; Masse, C.; Berthiaume, Y. Oxidative stress modulates the expression of genes involved in cell survival in DeltaF508 cystic fibrosis airway epithelial cells. Physiol. Genom. 2014, 46, 634–646. [Google Scholar] [CrossRef]
- Spurlock, C.F., 3rd; Tossberg, J.T.; Guo, Y.; Sriram, S.; Crooke, P.S., 3rd; Aune, T.M. Defective structural RNA processing in relapsing-remitting multiple sclerosis. Genome Biol. 2015, 16, 58. [Google Scholar] [CrossRef]
- Grunert, M.; Dorn, C.; Schueler, M.; Dunkel, I.; Schlesinger, J.; Mebus, S.; Alexi-Meskishvili, V.; Perrot, A.; Wassilew, K.; Timmermann, B.; et al. Rare and private variations in neural crest, apoptosis and sarcomere genes define the polygenic background of isolated Tetralogy of Fallot. Hum. Mol. Genet. 2014, 23, 3115–3128. [Google Scholar] [CrossRef] [PubMed]
- Roedder, S.; Kimura, N.; Okamura, H.; Hsieh, S.C.; Gong, Y.; Sarwal, M.M. Significance and suppression of redundant IL17 responses in acute allograft rejection by bioinformatics based drug repositioning of fenofibrate. PLoS ONE 2013, 8, e56657. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sharma, S.M.; Choi, D.; Planck, S.R.; Harrington, C.A.; Austin, C.R.; Lewis, J.A.; Diebel, T.N.; Martin, T.M.; Smith, J.R.; Rosenbaum, J.T. Insights in to the pathogenesis of axial spondyloarthropathy based on gene expression profiles. Arthritis Res. Ther. 2009, 11, R168. [Google Scholar] [CrossRef]
- Spiess, A.N.; Feig, C.; Schulze, W.; Chalmel, F.; Cappallo-Obermann, H.; Primig, M.; Kirchhoff, C. Cross-platform gene expression signature of human spermatogenic failure reveals inflammatory-like response. Hum. Reprod. 2007, 22, 2936–2946. [Google Scholar] [CrossRef]
- Ulmasov, B.; Oshima, K.; Rodriguez, M.G.; Cox, R.D.; Neuschwander-Tetri, B.A. Differences in the degree of cerulein-induced chronic pancreatitis in C57BL/6 mouse substrains lead to new insights in identification of potential risk factors in the development of chronic pancreatitis. Am. J. Pathol. 2013, 183, 692–708. [Google Scholar] [CrossRef]
- Chen, Y.W.; Shi, R.; Geraci, N.; Shrestha, S.; Gordish-Dressman, H.; Pachman, L.M. Duration of chronic inflammation alters gene expression in muscle from untreated girls with juvenile dermatomyositis. BMC Immunol. 2008, 9, 43. [Google Scholar] [CrossRef]
- Canna, S.W.; de Jesus, A.A.; Gouni, S.; Brooks, S.R.; Marrero, B.; Liu, Y.; DiMattia, M.A.; Zaal, K.J.; Sanchez, G.A.; Kim, H.; et al. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat. Genet. 2014, 46, 1140–1146. [Google Scholar] [CrossRef]
- Frades, I.; Andreasson, E.; Mato, J.M.; Alexandersson, E.; Matthiesen, R.; Martinez-Chantar, M.L. Integrative genomic signatures of hepatocellular carcinoma derived from nonalcoholic Fatty liver disease. PLoS ONE 2015, 10, e0124544. [Google Scholar] [CrossRef]
- Bronner, I.F.; Bochdanovits, Z.; Rizzu, P.; Kamphorst, W.; Ravid, R.; van Swieten, J.C.; Heutink, P. Comprehensive mRNA expression profiling distinguishes tauopathies and identifies shared molecular pathways. PLoS ONE 2009, 4, e6826. [Google Scholar] [CrossRef][Green Version]
- Platts, A.E.; Dix, D.J.; Chemes, H.E.; Thompson, K.E.; Goodrich, R.; Rockett, J.C.; Rawe, V.Y.; Quintana, S.; Diamond, M.P.; Strader, L.F.; et al. Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Hum. Mol. Genet. 2007, 16, 763–773. [Google Scholar] [CrossRef]
| Physiological Function | Cell, Tissue, or Organism | Source |
|---|---|---|
| Countering Oxidative Stress | Human aortic endothelial cells | [20] |
| Human myeloid leukemia U937 cell line | [59] | |
| Regulating Cytokinesis | C. elegans | [23] |
| Mediating Apoptosis | Human osteosarcoma U2OS cells Human breast cancer MCF-7 cells Human hepatocellular carcinoma tissue samples Human spinal cord astrocytes Human A549 lung adenocarcinoma cells Human bronchial epithelial cells Mouse C17.2 neural stem cells | [60] [53] [52] [58] [61] [62] |
| Mediating Ferroptosis | Human pancreatic ductal adenocarcinoma cells | [56] |
| Human SKOV3 and ES-2 ovarian cancer cell lines | [57] | |
| Mediating Autophagy | Human primary airway basal stem cells | [63] |
| Human breast cancer MCF-7 cells | [59] | |
| Human umbilical vein endothelial cells | [64] | |
| Human pancreatic tissue | [65] | |
| Human coronary artery endothelial cells | [29] |
| Physiological Function | Cell or Tissue Type | Source |
|---|---|---|
| Countering Oxidative Stress | Human liver tissue biopsy sample | [66] |
| Human soft tissue sarcoma sample | [67] | |
| Regulating Cell Proliferation | NUGC3 and HGC27 gastric cancer cells | [68] |
| Regulating Osteogenesis | Osteoporotic jawbone bone marrow mesenchymal stem cells | [69] |
| Maintaining Mitochondrial Biogenesis | XTC.UC1 and B-CPAP human thyroid cell lines | [70] |
| Mediating Autophagy | Human coronary artery endothelial cells | [29] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussey, G.; Royster, M.; Vaidy, N.; Culkin, M.; Saha, M.S. The Osgin Gene Family: Underexplored Yet Essential Mediators of Oxidative Stress. Biomolecules 2025, 15, 409. https://doi.org/10.3390/biom15030409
Hussey G, Royster M, Vaidy N, Culkin M, Saha MS. The Osgin Gene Family: Underexplored Yet Essential Mediators of Oxidative Stress. Biomolecules. 2025; 15(3):409. https://doi.org/10.3390/biom15030409
Chicago/Turabian StyleHussey, Grace, Marcus Royster, Nivedha Vaidy, Michael Culkin, and Margaret S. Saha. 2025. "The Osgin Gene Family: Underexplored Yet Essential Mediators of Oxidative Stress" Biomolecules 15, no. 3: 409. https://doi.org/10.3390/biom15030409
APA StyleHussey, G., Royster, M., Vaidy, N., Culkin, M., & Saha, M. S. (2025). The Osgin Gene Family: Underexplored Yet Essential Mediators of Oxidative Stress. Biomolecules, 15(3), 409. https://doi.org/10.3390/biom15030409






