Comprehensive Transcriptomic and Proteomic Analysis of Severe Pressure Ulcer Patients Identifies Molecular Signatures Associated with Impaired T Cell Function
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA Sequencing Data Download and Analysis
2.2. Proteomics Data Download and Analysis
2.3. Ingenuity Pathway Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gaspar, S.; Peralta, M.; Marques, A.; Budri, A.; Gaspar de Matos, M. Effectiveness on hospital-acquired pressure ulcers prevention: A systematic review. Int. Wound J. 2019, 16, 1087–1102. [Google Scholar] [CrossRef] [PubMed]
- Chou, R.; Dana, T.; Bougatsos, C.; Blazina, I.; Starmer, A.J.; Reitel, K.; Buckley, D.I. Pressure ulcer risk assessment and prevention: A systematic comparative effectiveness review. Ann. Intern. Med. 2013, 159, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Mervis, J.S.; Phillips, T.J. Pressure ulcers: Pathophysiology, epidemiology, risk factors, and presentation. J. Am. Acad. Dermatol. 2019, 81, 881–890. [Google Scholar] [CrossRef]
- Padula, W.V.; Delarmente, B.A. The national cost of hospital-acquired pressure injuries in the United States. Int. Wound J. 2019, 16, 634–640. [Google Scholar] [CrossRef]
- Bansal, C.; Scott, R.; Stewart, D.; Cockerell, C.J. Decubitus ulcers: A review of the literature. Int. J. Dermatol. 2005, 44, 805–810. [Google Scholar] [CrossRef]
- Li, Z.; Lin, F.; Thalib, L.; Chaboyer, W. Global prevalence and incidence of pressure injuries in hospitalised adult patients: A systematic review and meta-analysis. Int. J. Nurs. Stud. 2020, 105, 103546. [Google Scholar] [CrossRef]
- Kottner, J.; Cuddigan, J.; Carville, K.; Balzer, K.; Berlowitz, D.; Law, S.; Litchford, M.; Mitchell, P.; Moore, Z.; Pittman, J.; et al. Prevention and treatment of pressure ulcers/injuries: The protocol for the second update of the international Clinical Practice Guideline 2019. J. Tissue Viability 2019, 28, 51–58. [Google Scholar] [CrossRef]
- 8. European Pressure Ulcer Advisory Panel, National Pressure Injury Advisory Panel and Pan Pacific Pressure Injury Alliance. Prevention and Treatment of Pressure Ulcers/Injuries: Clinical Practice Guideline. The International Guideline 2019. 2019. [Third edition]. Available online: https://internationalguideline.com/2019 (accessed on 11 May 2025).
- Kottner, J.; Cuddigan, J.; Carville, K.; Balzer, K.; Berlowitz, D.; Law, S.; Litchford, M.; Mitchell, P.; Moore, Z.; Pittman, J.; et al. Pressure ulcer/injury classification today: An international perspective. J. Tissue Viability 2020, 29, 197–203. [Google Scholar] [CrossRef]
- Song, Y.P.; Shen, H.W.; Cai, J.Y.; Zha, M.L.; Chen, H.L. The relationship between pressure injury complication and mortality risk of older patients in follow-up: A systematic review and meta-analysis. Int. Wound J. 2019, 16, 1533–1544. [Google Scholar] [CrossRef]
- Anders, J.; Heinemann, A.; Leffmann, C.; Leutenegger, M.; Profener, F.; von Renteln-Kruse, W. Decubitus ulcers: Pathophysiology and primary prevention. Dtsch. Arztebl. Int. 2010, 107, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Bhamidipati, T.; Hajj, J.P.; Ghoneim, N.I.; Pradhan, A.M.; Mishra, S.; Gummalla, B.; Abouhashem, A.S.; Parmar, A.S.; Khanna, S.; Roy, S.; et al. Diabetic Wound Vasculopathy and Neuropathy: Spotlight on Wound Lipid Signaling. Adv. Wound Care 2025. Available online: https://www.liebertpub.com/doi/abs/10.1177/21621918251366681 (accessed on 7 November 2017).
- Hajj, J.; Sizemore, B.; Singh, K. Impact of Epigenetics, Diet, and Nutrition-Related Pathologies on Wound Healing. Int. J. Mol. Sci. 2024, 25, 10474. [Google Scholar] [CrossRef]
- Bhamidipati, T.; Kumar, M.; Verma, S.S.; Mohanty, S.K.; Kacar, S.; Reese, D.; Martinez, M.M.; Kamocka, M.M.; Dunn, K.W.; Sen, C.K.; et al. Epigenetic basis of diabetic vasculopathy. Front. Endocrinol. 2022, 13, 989844. [Google Scholar] [CrossRef]
- Rustagi, Y.; Abouhashem, A.S.; Verma, P.; Verma, S.S.; Hernandez, E.; Liu, S.; Kumar, M.; Guda, P.R.; Srivastava, R.; Mohanty, S.K.; et al. Endothelial Phospholipase Cgamma2 Improves Outcomes of Diabetic Ischemic Limb Rescue Following VEGF Therapy. Diabetes 2022, 71, 1149–1165. [Google Scholar] [CrossRef]
- Bhamidipati, T.; Sinha, M.; Sen, C.K.; Singh, K. Laser Capture Microdissection in the Spatial Analysis of Epigenetic Modifications in Skin: A Comprehensive Review. Oxid. Med. Cell Longev. 2022, 2022, 4127238. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Sinha, M.; Pal, D.; Tabasum, S.; Gnyawali, S.C.; Khona, D.; Sarkar, S.; Mohanty, S.K.; Soto-Gonzalez, F.; Khanna, S.; et al. Cutaneous Epithelial to Mesenchymal Transition Activator ZEB1 Regulates Wound Angiogenesis and Closure in a Glycemic Status-Dependent Manner. Diabetes 2019, 68, 2175–2190. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Pal, D.; Sinha, M.; Ghatak, S.; Gnyawali, S.C.; Khanna, S.; Roy, S.; Sen, C.K. Epigenetic Modification of MicroRNA-200b Contributes to Diabetic Vasculopathy. Mol. Ther. 2017, 25, 2689–2704. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Chen, Q.; Zhang, Y.; He, L.; Wang, J.; Cai, Y.; Li, L. Impact of diabetes on the risk of bedsore in patients undergoing surgery: An updated quantitative analysis of cohort studies. Oncotarget 2017, 8, 14516–14524. [Google Scholar] [CrossRef]
- Nasiri, E.; Mollaei, A.; Birami, M.; Lotfi, M.; Rafiei, M.H. The risk of surgery-related pressure ulcer in diabetics: A systematic review and meta-analysis. Ann. Med. Surg. 2021, 65, 102336. [Google Scholar] [CrossRef]
- Jairam, A.; Song, P.; Patel, N.B.; Wong, M.S. Pressure Sores and Systemic Inflammatory Response Syndrome: UC Davis Quality Improvement Initiative. Ann. Plast. Surg. 2018, 80 (Suppl. S5), S308–S310. [Google Scholar] [CrossRef]
- Lazaro, J.L.; Izzo, V.; Meaume, S.; Davies, A.H.; Lobmann, R.; Uccioli, L. Elevated levels of matrix metalloproteinases and chronic wound healing: An updated review of clinical evidence. J. Wound Care 2016, 25, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Santos, E.M.; Farias, L.C.; Santos, S.H.S.; de Paula, A.M.B.; Oliveira, E.S.C.S.; Guimaraes, A.L.S. Molecular finds of pressure ulcer: A bioinformatics approach in pressure ulcer. J. Tissue Viability 2017, 26, 119–124. [Google Scholar] [CrossRef]
- Li, D.; Cheng, S.; Pei, Y.; Sommar, P.; Karner, J.; Herter, E.K.; Toma, M.A.; Zhang, L.; Pham, K.; Cheung, Y.T.; et al. Single-Cell Analysis Reveals Major Histocompatibility Complex II-Expressing Keratinocytes in Pressure Ulcers with Worse Healing Outcomes. J. Investig. Dermatol. 2022, 142 (Pt A), 705–716. [Google Scholar] [CrossRef]
- Diegelmann, R.F. Excessive neutrophils characterize chronic pressure ulcers. Wound Repair. Regen. 2003, 11, 490–495. [Google Scholar] [CrossRef]
- Pena, L.T.; Escolar-Pena, A.; Solera, R.A.; Martinez, L.S.Z.; Castro, O.G.; Cerezo, C.T.; Escribese, M.M.; Lopez, J.B.; Perez, T.C.; Heredero, X.S.; et al. Systemic immune response alteration in patients with severe pressure ulcers. Sci. Rep. 2025, 15, 19579. [Google Scholar] [CrossRef] [PubMed]
- Baldan-Martin, M.; Martin-Rojas, T.; Corbacho-Alonso, N.; Lopez, J.A.; Sastre-Oliva, T.; Gil-Dones, F.; Vazquez, J.; Arevalo, J.M.; Mourino-Alvarez, L.; Barderas, M.G. Comprehensive Proteomic Profiling of Pressure Ulcers in Patients with Spinal Cord Injury Identifies a Specific Protein Pattern of Pathology. Adv. Wound Care 2020, 9, 277–294. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.M.; Omar, E.; Pai, D.R.; Sood, S. Cellular events and biomarkers of wound healing. Indian. J. Plast. Surg. 2012, 45, 220–228. [Google Scholar] [CrossRef]
- Baldan-Martin, M.; Lopez, J.A.; Corbacho-Alonso, N.; Martinez, P.J.; Rodriguez-Sanchez, E.; Mourino-Alvarez, L.; Sastre-Oliva, T.; Martin-Rojas, T.; Rincon, R.; Calvo, E.; et al. Potential role of new molecular plasma signatures on cardiovascular risk stratification in asymptomatic individuals. Sci. Rep. 2018, 8, 4802. [Google Scholar] [CrossRef]
- Cardona, M.; Lopez, J.A.; Serafin, A.; Rongvaux, A.; Inserte, J.; Garcia-Dorado, D.; Flavell, R.; Llovera, M.; Canas, X.; Vazquez, J.; et al. Executioner Caspase-3 and 7 Deficiency Reduces Myocyte Number in the Developing Mouse Heart. PLoS ONE 2015, 10, e0131411. [Google Scholar] [CrossRef]
- Navarro, P.; Trevisan-Herraz, M.; Bonzon-Kulichenko, E.; Nunez, E.; Martinez-Acedo, P.; Perez-Hernandez, D.; Jorge, I.; Mesa, R.; Calvo, E.; Carrascal, M.; et al. General statistical framework for quantitative proteomics by stable isotope labeling. J. Proteome Res. 2014, 13, 1234–1247. [Google Scholar] [CrossRef]
- Ghosh, N.; Das, A.; Biswas, N.; Gnyawali, S.; Singh, K.; Gorain, M.; Polcyn, C.; Khanna, S.; Roy, S.; Sen, C.K. Urolithin A augments angiogenic pathways in skeletal muscle by bolstering NAD(+) and SIRT1. Sci. Rep. 2020, 10, 20184. [Google Scholar] [CrossRef]
- Banerjee, P.; Das, A.; Singh, K.; Khanna, S.; Sen, C.K.; Roy, S. Collagenase-based wound debridement agent induces extracellular matrix supporting phenotype in macrophages. Sci. Rep. 2024, 14, 3257. [Google Scholar] [CrossRef]
- Das, A.; El Masry, M.S.; Gnyawali, S.C.; Ghatak, S.; Singh, K.; Stewart, R.; Lewis, M.; Saha, A.; Gordillo, G.; Khanna, S. Skin Transcriptome of Middle-Aged Women Supplemented with Natural Herbo-mineral Shilajit Shows Induction of Microvascular and Extracellular Matrix Mechanisms. J. Am. Coll. Nutr. 2019, 38, 526–536. [Google Scholar] [CrossRef]
- Trujillo-Ochoa, J.L.; Kazemian, M.; Afzali, B. The role of transcription factors in shaping regulatory T cell identity. Nat. Rev. Immunol. 2023, 23, 842–856. [Google Scholar] [CrossRef]
- Drashansky, T.T.; Helm, E.; Huo, Z.; Curkovic, N.; Kumar, P.; Luo, X.; Parthasarathy, U.; Zuniga, A.; Cho, J.J.; Lorentsen, K.J.; et al. Bcl11b prevents fatal autoimmunity by promoting T(reg) cell program and constraining innate lineages in T(reg) cells. Sci. Adv. 2019, 5, eaaw0480. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Hu, S.; Zhu, Q.; Li, C.; Kang, T.; Xie, W.; Wang, Y.; Li, Y.; Lu, Y.; Qi, J.; et al. RANKL/RANK signaling recruits Tregs via the CCL20-CCR6 pathway and promotes stemness and metastasis in colorectal cancer. Cell Death Dis. 2024, 15, 437. [Google Scholar] [CrossRef] [PubMed]
- Kersh, A.E.; Sati, S.; Huang, J.; Murphy, C.; Ahart, O.; Leung, T.H. CXCL9, CXCL10, and CCL19 synergistically recruit T lymphocytes to skin in lichen planus. JCI Insight 2024, 9, e179899. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Liang, Q.; Zhang, Q.; Zhao, S.; Lin, Y.; Liu, B.; Ma, Y.; Mai, X.; Fu, Q.; Bao, X.; et al. Ccl2-Induced Regulatory T Cells Balance Inflammation Through Macrophage Polarization During Liver Reconstitution. Adv. Sci. 2024, 11, e2403849. [Google Scholar] [CrossRef]
- Himmel, M.E.; Crome, S.Q.; Ivison, S.; Piccirillo, C.; Steiner, T.S.; Levings, M.K. Human CD4+ FOXP3+ regulatory T cells produce CXCL8 and recruit neutrophils. Eur. J. Immunol. 2011, 41, 306–312. [Google Scholar] [CrossRef]
- Marin, J.; Nixon, J.; Gorecki, C. A systematic review of risk factors for the development and recurrence of pressure ulcers in people with spinal cord injuries. Spinal Cord. 2013, 51, 522–527. [Google Scholar] [CrossRef]
- Chattopadhyay, D.; Sinha, M.; Kapoor, A.; Kumar, M.; Singh, K.; Mathew-Steiner, S.S.; Sen, C.K. Deficient functional wound closure as measured by elevated trans-epidermal water loss predicts chronic wound recurrence: An exploratory observational study. Sci. Rep. 2024, 14, 23593. [Google Scholar] [CrossRef]
- Theocharidis, G.; Thomas, B.E.; Sarkar, D.; Mumme, H.L.; Pilcher, W.J.R.; Dwivedi, B.; Sandoval-Schaefer, T.; Sirbulescu, R.F.; Kafanas, A.; Mezghani, I.; et al. Single cell transcriptomic landscape of diabetic foot ulcers. Nat. Commun. 2022, 13, 181. [Google Scholar] [CrossRef] [PubMed]
- Rani, M.; Schwacha, M.G. The composition of T-cell subsets are altered in the burn wound early after injury. PLoS ONE 2017, 12, e0179015. [Google Scholar] [CrossRef] [PubMed]
- Mulder, P.P.G.; Vlig, M.; Elgersma, A.; Rozemeijer, L.; Mastenbroek, L.S.; Middelkoop, E.; Joosten, I.; Koenen, H.; Boekema, B. Monocytes and T cells incorporated in full skin equivalents to study innate or adaptive immune reactions after burn injury. Front. Immunol. 2023, 14, 1264716. [Google Scholar] [CrossRef]
- Shi, H.; Yuan, X.; Liu, G.; Fan, W. Identifying and Validating GSTM5 as an Immunogenic Gene in Diabetic Foot Ulcer Using Bioinformatics and Machine Learning. J. Inflamm. Res. 2023, 16, 6241–6256. [Google Scholar] [CrossRef]
- Kuang, S.; He, F.; Liu, G.; Sun, X.; Dai, J.; Chi, A.; Tang, Y.; Li, Z.; Gao, Y.; Deng, C.; et al. CCR2-engineered mesenchymal stromal cells accelerate diabetic wound healing by restoring immunological homeostasis. Biomaterials 2021, 275, 120963. [Google Scholar] [CrossRef]
- Moura, J.; Rodrigues, J.; Goncalves, M.; Amaral, C.; Lima, M.; Carvalho, E. Impaired T-cell differentiation in diabetic foot ulceration. Cell Mol. Immunol. 2017, 14, 758–769. [Google Scholar] [CrossRef]
- Shan, H.; Wang, X.; Zhang, J. Dendritic epidermal T cell hydrogel induces the polarization of M2 macrophages to promote the healing of deep tissue pressure injury. J. Tissue Viability 2024, 33, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Rehak, L.; Giurato, L.; Meloni, M.; Panunzi, A.; Manti, G.M.; Uccioli, L. The Immune-Centric Revolution in the Diabetic Foot: Monocytes and Lymphocytes Role in Wound Healing and Tissue Regeneration-A Narrative Review. J. Clin. Med. 2022, 11, 889. [Google Scholar] [CrossRef]
- Leung, O.M.; Li, J.; Li, X.; Chan, V.W.; Yang, K.Y.; Ku, M.; Ji, L.; Sun, H.; Waldmann, H.; Tian, X.Y.; et al. Regulatory T Cells Promote Apelin-Mediated Sprouting Angiogenesis in Type 2 Diabetes. Cell Rep. 2018, 24, 1610–1626. [Google Scholar] [CrossRef]
- Nayer, B.; Tan, J.L.; Alshoubaki, Y.K.; Lu, Y.Z.; Legrand, J.M.D.; Lau, S.; Hu, N.; Park, A.J.; Wang, X.N.; Amann-Zalcenstein, D.; et al. Local administration of regulatory T cells promotes tissue healing. Nat. Commun. 2024, 15, 7863. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, A.S.; Mansbridge, J.N. The Innate Immune System in Acute and Chronic Wounds. Adv. Wound Care 2016, 5, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Toulon, A.; Breton, L.; Taylor, K.R.; Tenenhaus, M.; Bhavsar, D.; Lanigan, C.; Rudolph, R.; Jameson, J.; Havran, W.L. A role for human skin-resident T cells in wound healing. J. Exp. Med. 2009, 206, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Gaspari, A.A.; Katz, S.I. Induction of in vivo hyporesponsiveness to contact allergens by hapten-modified Ia+ keratinocytes. J. Immunol. 1991, 147, 4155–4161. [Google Scholar] [CrossRef]
- Bal, V.; McIndoe, A.; Denton, G.; Hudson, D.; Lombardi, G.; Lamb, J.; Lechler, R. Antigen presentation by keratinocytes induces tolerance in human T cells. Eur. J. Immunol. 1990, 20, 1893–1897. [Google Scholar] [CrossRef]
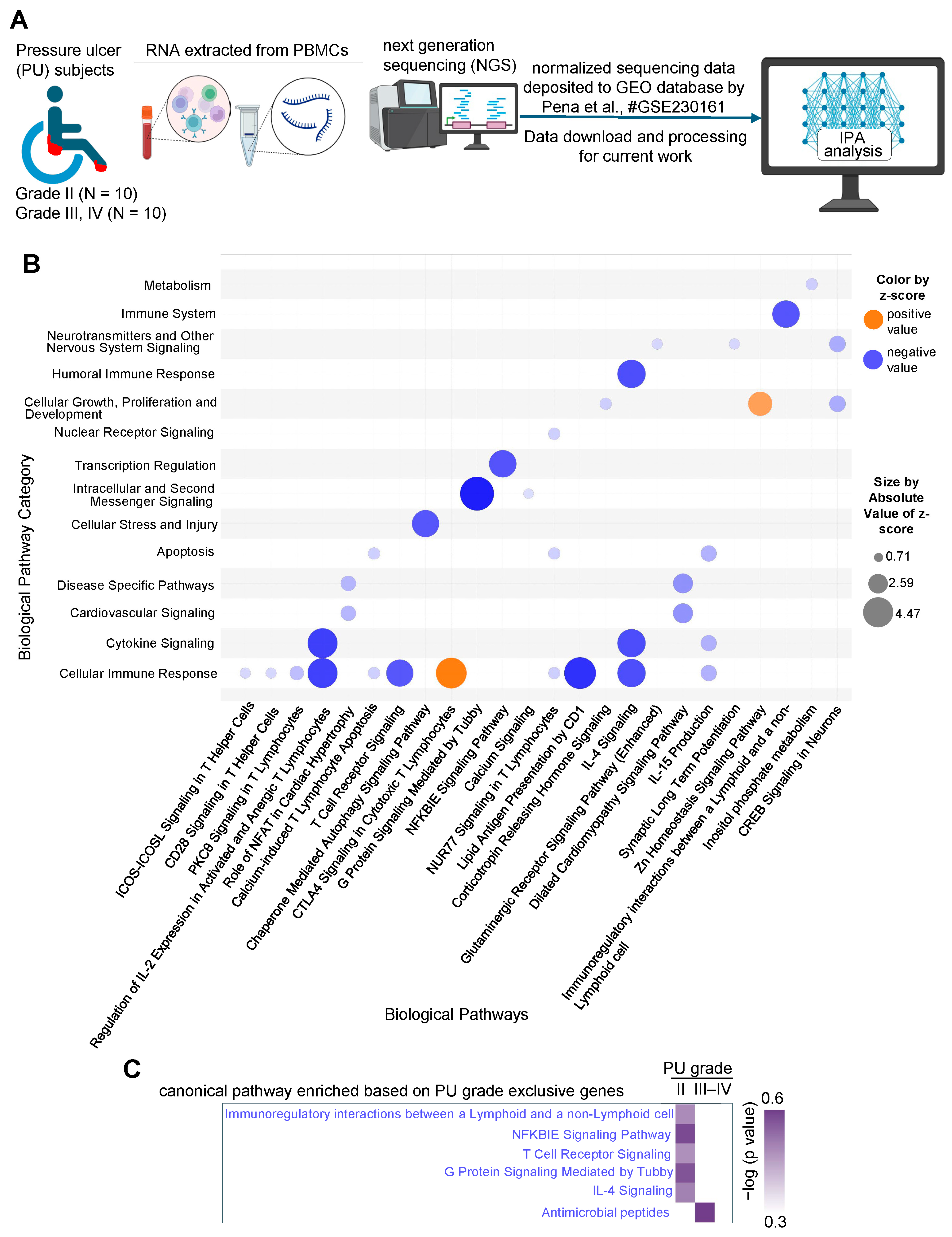
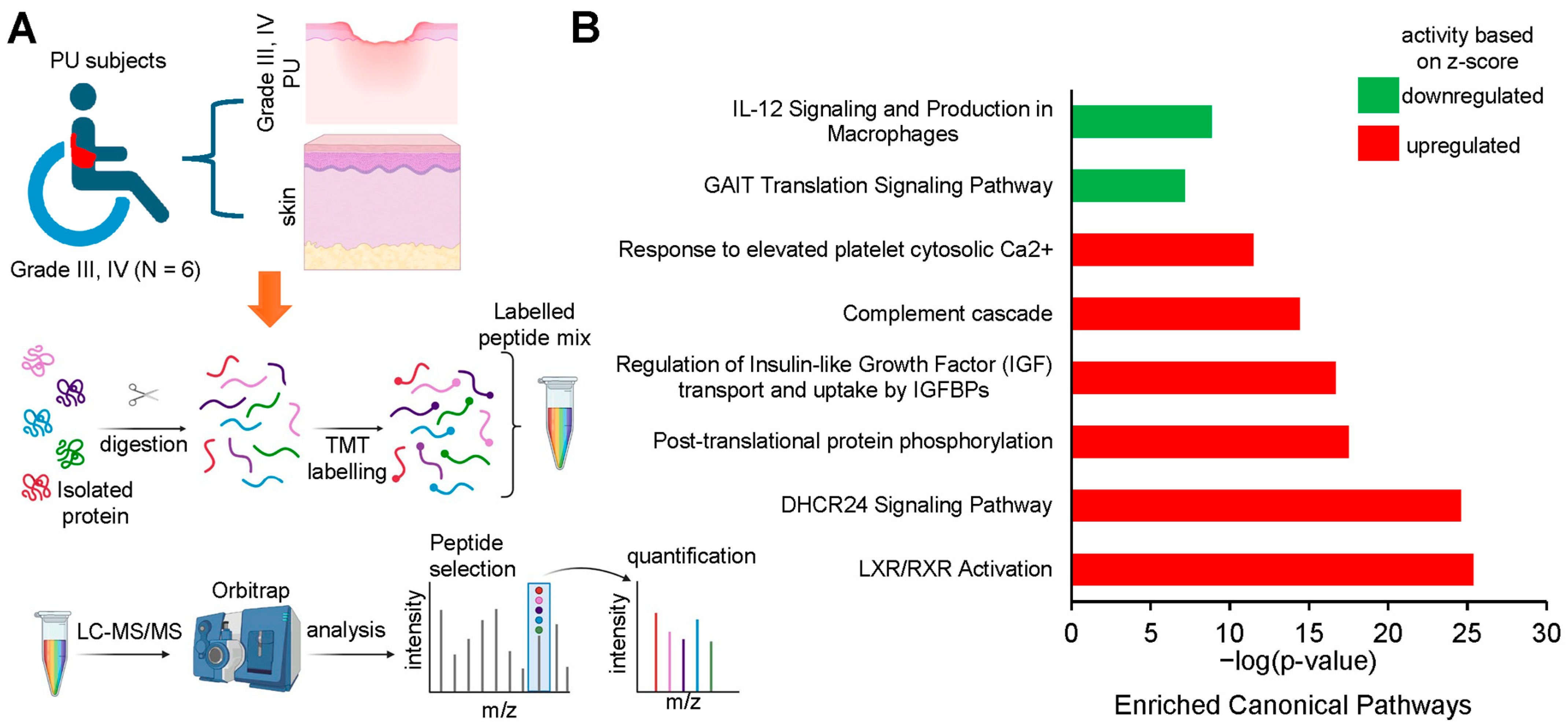
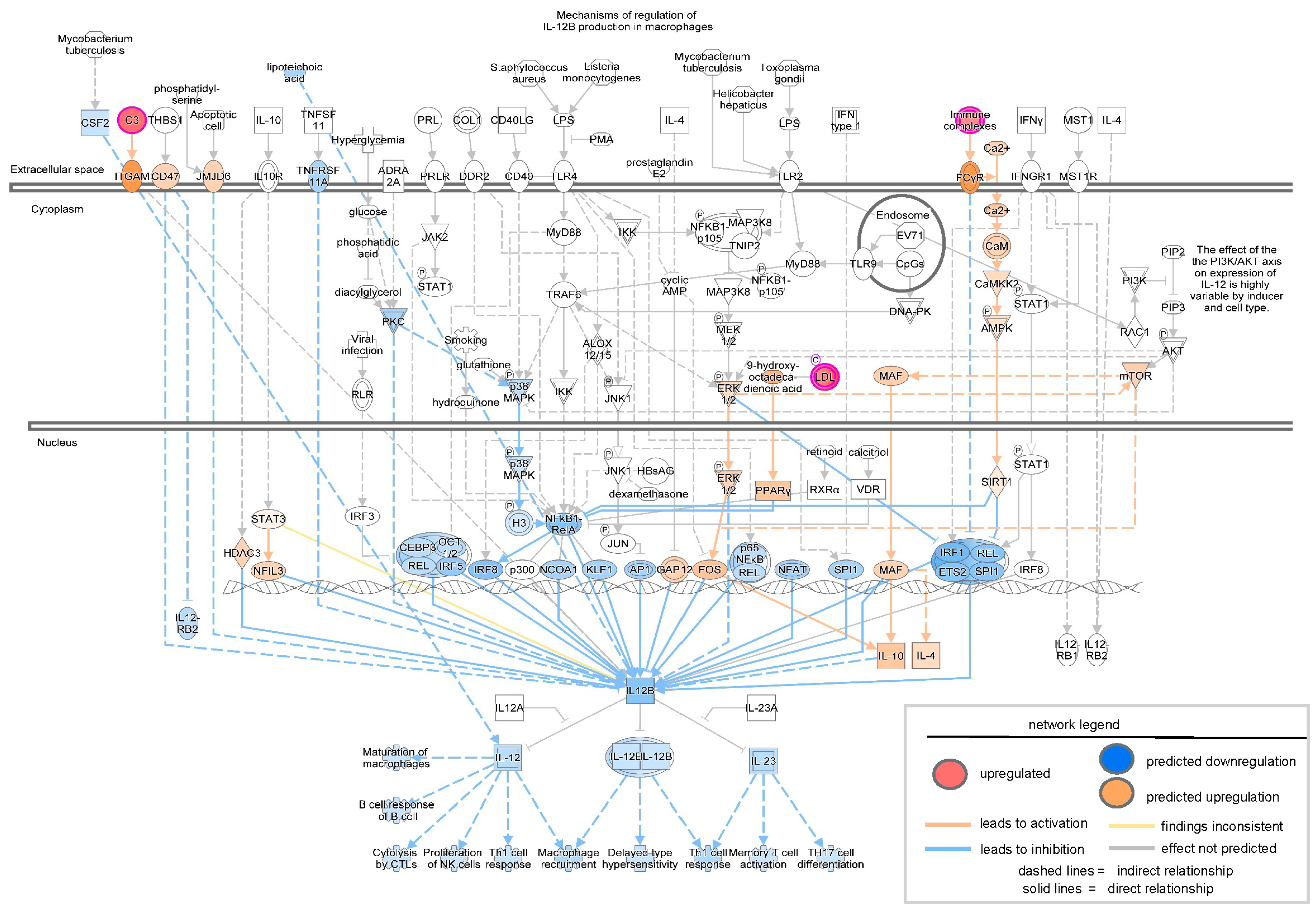
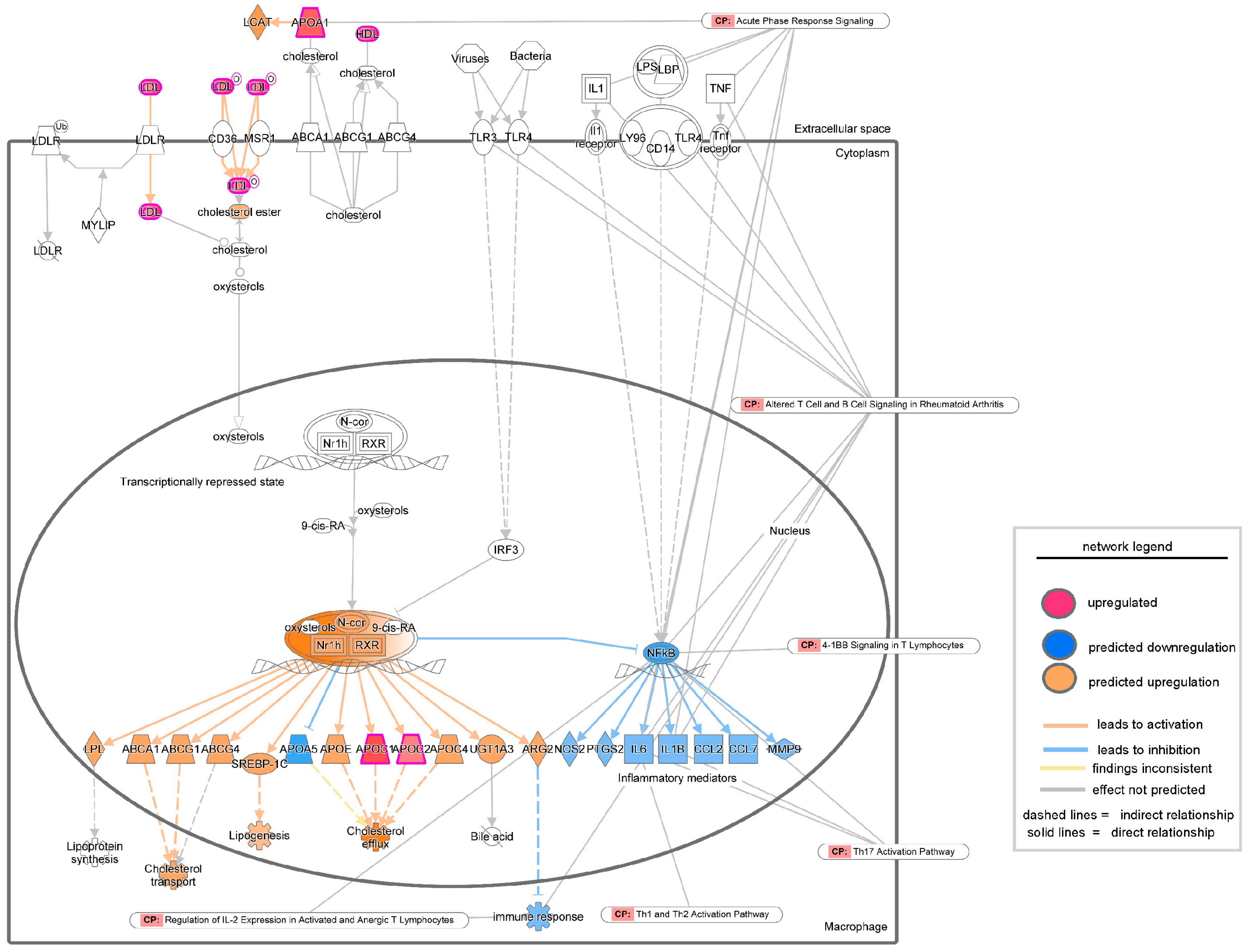
| Ingenuity Canonical Pathways | −log (p-Value) | z-Score | Molecules |
|---|---|---|---|
| ICOS-ICOSL Signaling in T Helper Cells | 5.54 | −0.816 | CD3E, CD40, ITK, LAT, PLCG1, PLEKHA2, PPP3CA, PRKCQ, REL, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| CD28 Signaling in T Helper Cells | 4.9 | −0.816 | CD3E, CTLA4, ITK, LAT, PLCG1, PPP3CA, PRKCQ, REL, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| PKCθ Signaling in T Lymphocytes | 4.87 | −1.342 | CACNA1F, CACNA1H, CACNB3, CD3E, LAT, PLCG1, PPP3CA, PRKCQ, REL, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| Regulation of IL-2 Expression in Activated and Anergic T Lymphocytes | 4.72 | −3.838 | CD3E, LAT, PLCG1, PPP3CA, PRKCQ, REL, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| Role of NFAT in Cardiac Hypertrophy | 4.38 | −1.508 | CACNA1F, CACNA1H, CACNB3, CAMK4, CSNK1A1, HDAC2, IL11, MEF2B, NOTUM, PLCG1, PPP3CA, PRKCQ, RCAN2, SHF |
| Calcium-induced T Lymphocyte Apoptosis | 4.25 | −1 | CD3E, HDAC2, PLCG1, PPP3CA, PRKCQ, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| T Cell Receptor Signaling | 4.12 | −3.4 | ATF2, CD3E, CTLA4, ITK, LAT, PLCG1, PPP3CA, PRKCQ, REL, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| Chaperone-Mediated Autophagy Signaling Pathway | 3.96 | −3.4 | CD3E, EIF2AK3, HDAC2, MMP27, NOS3, PPP3CA, PSMB3, RILP, TBK1, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| CTLA4 Signaling in Cytotoxic T Lymphocytes | 3.83 | 3.962 | CD3E, CTLA4, FOXP3, ITK, LAT, PLCG1, PPP2R2A, PRKCQ, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| G Protein Signaling Mediated by Tubby | 3.74 | −4.472 | CD3E, ITK, LAT, PLCG1, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| NFKBIE Signaling Pathway | 3.54 | −3.441 | CD3E, CD40, REL, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| Calcium Signaling | 3.42 | −0.707 | ATF2, CACNA1F, CACNA1H, CACNB3, CAMK4, GRIN2A, HDAC2, MEF2B, MYL3, PPP3CA, RCAN2, TPM2 |
| NUR77 Signaling in T Lymphocytes | 3.18 | −1 | CD3E, HDAC2, PPP3CA, PRKCQ, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| Lipid Antigen Presentation by CD1 | 3.08 | −4.123 | CD3E, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| Corticotropin Releasing Hormone Signaling | 2.86 | −1 | ATF2, CACNA1F, CACNA1H, CACNB3, CAMK4, MEF2B, NOS3, PLCG1, PRKCQ |
| IL-4 Signaling | 2.65 | −3.578 | ATF2, CD3E, COL6A1, GAB1, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| Glutaminergic Receptor Signaling Pathway (Enhanced) | 2.39 | −0.832 | ATF2, CACNA1F, CACNA1H, CACNB3, CAMK4, DGKA, FMR1, GRIN2A, NOTUM, PLCG1, PPP3CA, PRKCQ, SLC38A2 |
| Dilated Cardiomyopathy Signaling Pathway | 2.36 | −2.236 | BAG3, CACNA1F, CACNA1H, CACNB3, CAMK4, GAB1, MYL3, SGCD |
| IL-15 Production | 2.31 | −1.633 | DYRK4, ERBB3, FLT3LG, FLT4, ITK, LMTK3, REL |
| Synaptic Long-Term Potentiation | 2.27 | −0.816 | ATF2, CAMK4, GRIN2A, NOTUM, PLCG1, PPP3CA, PRKCQ |
| Zn Homeostasis Signaling Pathway | 2.17 | 2.921 | ATF2, CD3E, CYSLTR2, GPR18, GPRC5C, GRIN2A, HTR6, MMP27, PPP3CA, PRKCQ, RHO, SELENOW, SLC52A1, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30, VIPR1 |
| Immunoregulatory Interactions between a Lymphoid and a Non-Lymphoid Cell | 2.08 | −3.441 | CD3E, CD40, FCGR2B, TRAJ11, TRAJ2, TRAJ20, TRAJ22, TRAJ39, TRAJ44, TRAJ49, TRAJ54, TRAJ9, TRAV12-3, TRAV2, TRAV22, TRAV26-2, TRAV38-1, TRBV10-3, TRBV30 |
| Inositol Phosphate Metabolism | 2.05 | −1 | INPP4B, MIOX, PLCG1, PPIP5K2 |
| CREB Signaling in Neurons | 2.01 | −1.698 | ATF2, CACNA1F, CACNA1H, CACNB3, CAMK4, CYSLTR2, FLT4, GPR18, GPRC5C, GRIN2A, HTR6, NOTUM, PLCG1, POLR2F, PRKCQ, RHO, SHF, SLC52A1, VIPR1 |
| Symbol | Entrez Gene Name | log2 Fold Change | Type of Molecule |
|---|---|---|---|
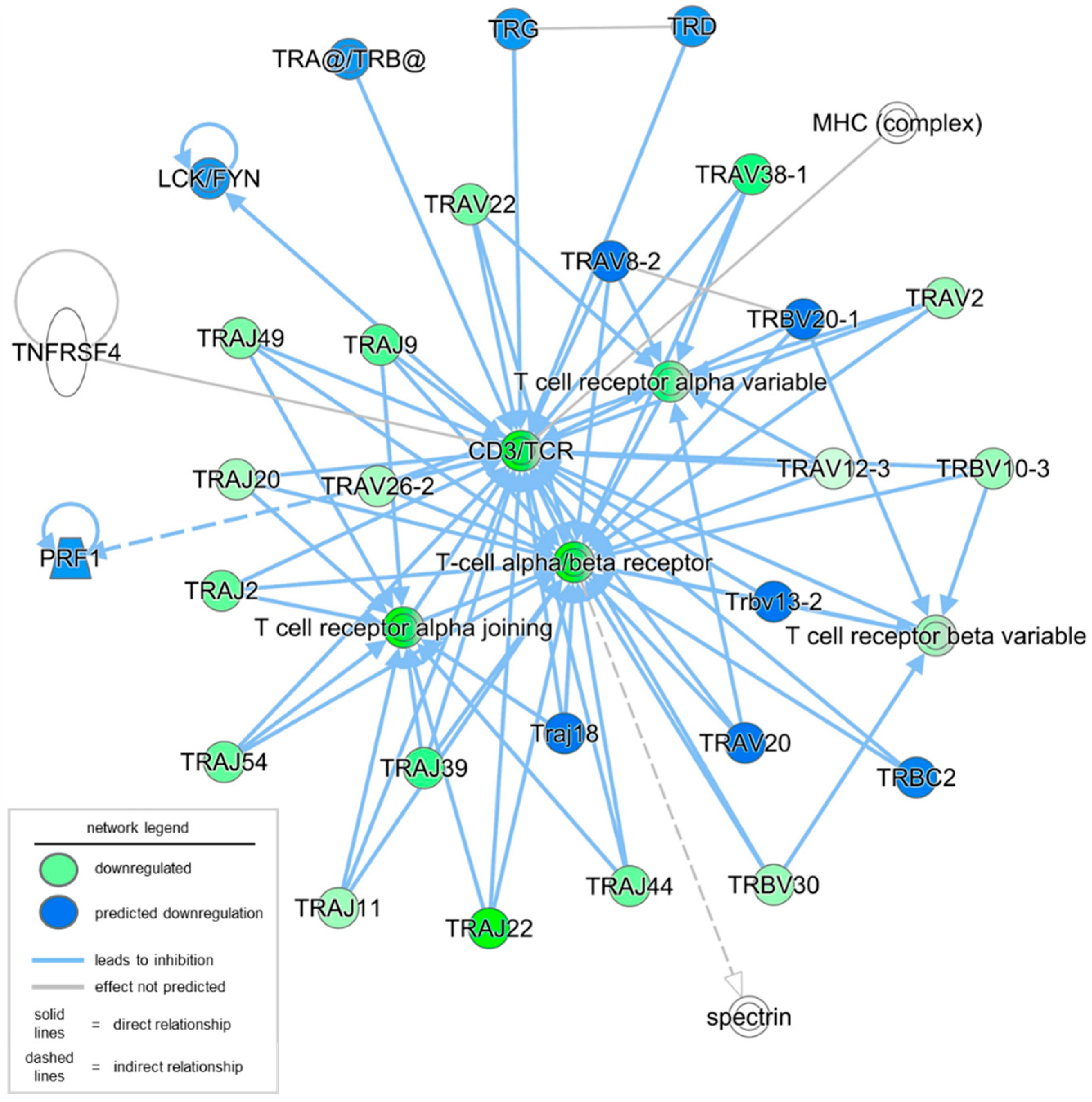
| A. Genes, groups, and complexes downregulated in the dataset | |||
| TRAJ22 | T cell receptor alpha joining 22 | −3.628619755 | Gene |
| TRAV38-1 | T cell receptor alpha variable 38-1 | −2.072440415 | Gene |
| TRAJ39 | T cell receptor alpha joining 39 | −1.823949277 | Gene |
| TRAJ9 | T cell receptor alpha joining 9 | −1.709212257 | Gene |
| TRAJ2 | T cell receptor alpha joining 2 (non-functional) | −1.588144944 | Gene |
| TRAJ54 | T cell receptor alpha joining 54 | −1.582453079 | Gene |
| TRAJ44 | T cell receptor alpha joining 44 | −1.567723326 | Gene |
| TRAV22 | T cell receptor alpha variable 22 | −1.497050695 | Gene |
| TRAJ49 | T cell receptor alpha joining 49 | −1.378685736 | Gene |
| TRAV2 | T cell receptor alpha variable 2 | −1.156064019 | Gene |
| TRBV30 | T cell receptor beta variable 30 | −1.150917618 | Gene |
| TRBV10-3 | T cell receptor beta variable 10-3 | −1.131684815 | Gene |
| TRAJ11 | T cell receptor alpha joining 11 | −1.095500661 | Gene |
| TRAJ20 | T cell receptor alpha joining 20 | −1.054293781 | Gene |
| TRAV26-2 | T cell receptor alpha variable 26-2 | −1.021698834 | Gene |
| TRAV12-3 | T cell receptor alpha variable 12-3 | −0.605153066 | Gene |
| T cell receptor alpha joining | N/A | Group | |
| T cell receptor alpha variable | N/A | Group | |
| T cell receptor beta variable | N/A | Group | |
| CD3/TCR | N/A | Complex | |
| T cell alpha/beta receptor | N/A | Complex | |
| B. Genes, groups, and complexes predicted to be downregulated in the dataset | |||
| PRF1 | perforin 1 | predicted downregulated | Gene |
| Traj18 | T cell receptor alpha joining 18 | predicted downregulated | Gene |
| TRAV20 | T cell receptor alpha variable 20 | predicted downregulated | Gene |
| TRAV8-2 | T cell receptor alpha variable 8-2 | predicted downregulated | Gene |
| TRBC2 | T cell receptor beta constant 2 | predicted downregulated | Gene |
| Trbv13-2 | T cell receptor beta, variable 13-2 | predicted downregulated | Gene |
| TRBV20-1 | T cell receptor beta variable 20-1 | predicted downregulated | Gene |
| TRD | T cell receptor delta locus | predicted downregulated | Gene |
| TRG | T cell receptor gamma locus | predicted downregulated | Gene |
| LCK/FYN | predicted downregulated | Group | |
| TRA@/TRB@ | predicted downregulated | Complex | |
| C. Genes, groups, and complexes with no activity pattern available | |||
| TNFRSF4 | TNF receptor superfamily member 4 | no activity reported | Gene |
| Spectrin | no activity reported | Group | |
| MHC (complex) | no activity reported | Complex | |
| Diseases or Functions Annotation | p-Value | Molecules |
|---|---|---|
| Quantity of T lymphocytes | 0.0298 | PRF1, TNFRSF4, Traj18, Trbv13-2, TRG |
| Abnormal morphology of T lymphocytes | 0.00233 | TNFRSF4, Traj18, TRG |
| Deletion of T lymphocytes | 0.0000256 | PRF1, TNFRSF4, Trbv13-2 |
| Quantity of activated T lymphocytes | 0.000862 | PRF1, Traj18 |
| Lack of T lymphocytes | 0.000973 | Traj18, TRG |
| Activation of CD8+ T lymphocytes | 0.0119 | PRF1, TRG |
| Activation-induced cell death of T lymphocytes | 0.00611 | PRF1, TNFRSF4 |
| Lack of natural killer T lymphocytes | 0.00795 | Traj18 |
| Lack of gamma-delta T lymphocytes | 0.0118 | TRG |
| Deletion of naive T lymphocytes | 0.00136 | PRF1 |
| Number of activated T lymphocytes | 0.0412 | PRF1 |
| Expansion of effector T lymphocytes | 0.0229 | TNFRSF4 |
| Elimination of T lymphocytes | 0.0147 | PRF1 |
| Conversion of T lymphocytes | 0.0488 | TNFRSF4 |
| Cytotoxicity of CD8+ T lymphocytes | 0.0269 | PRF1 |
| Apoptosis of naive T lymphocytes | 0.0122 | PRF1 |
| Suppression of effector T lymphocytes | 0.0105 | TNFRSF4 |
| Killing by effector T lymphocytes | 0.00136 | PRF1 |
| Differentiation of natural killer T lymphocytes | 0.0303 | Traj18 |
| Deletion of memory T lymphocytes | 0.00136 | PRF1 |
| Malignancy of natural killer T lymphocytes | 0.00136 | PRF1 |
| Activation-induced cell death of CD8+ T lymphocytes | 0.0118 | PRF1 |
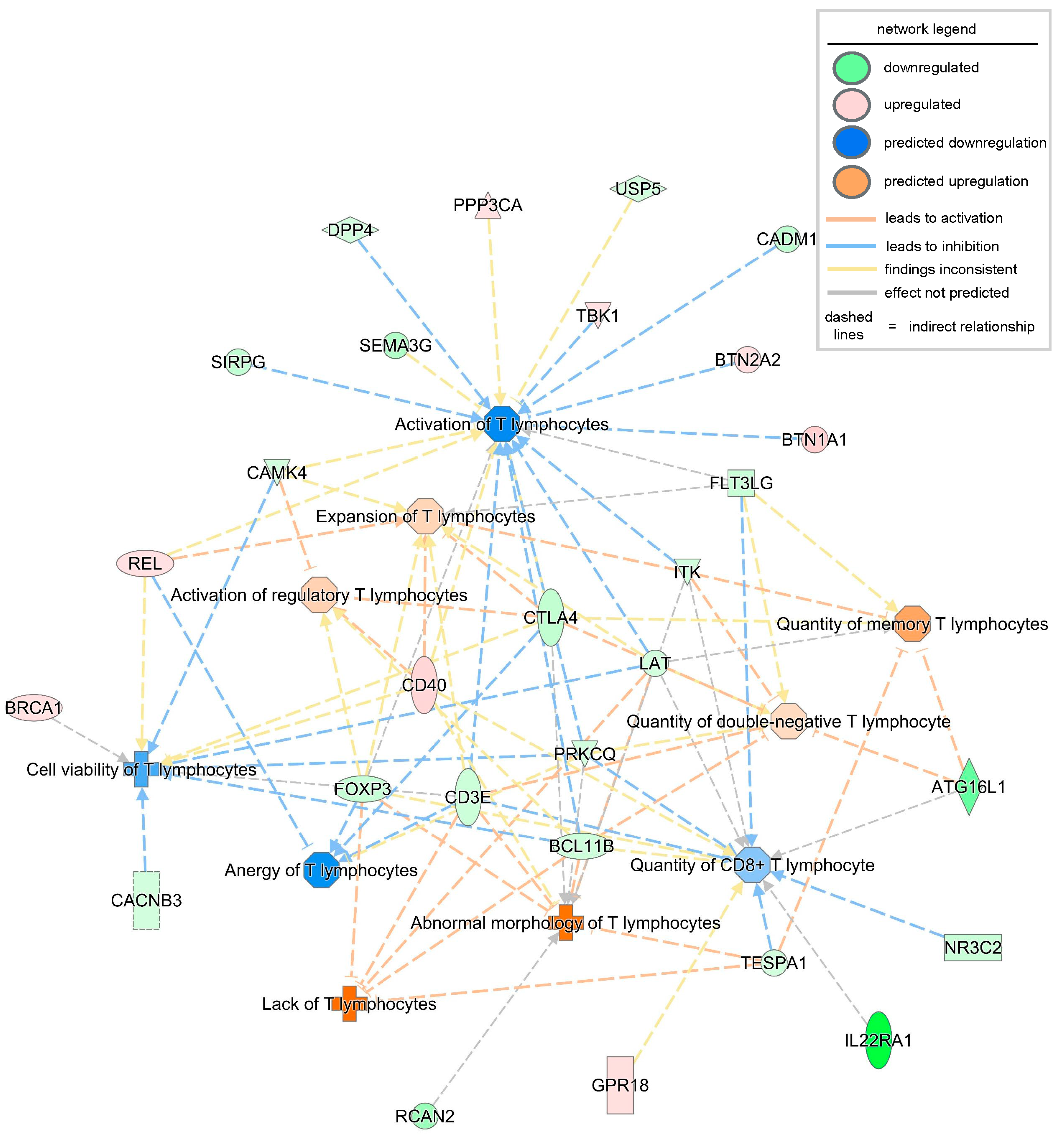
| Diseases or Functions Annotation | p-Value | Predicted Activation State | Activation z-Score | Molecules | #Molecules |
|---|---|---|---|---|---|
| Lack of T lymphocytes | 0.0000984 | Increased | 2.183 | BCL11B, CD3E, FOXP3, LAT, TESPA1 | 5 |
| Abnormal morphology of T lymphocytes | 0.000229 | 1.823 | BCL11B, CD3E, CD40, CTLA4, FOXP3, ITK, LAT, PRKCQ, RCAN2, TESPA1 | 10 | |
| Quantity of memory T lymphocytes | 0.0162 | 0.853 | ATG16L1, CTLA4, FLT3LG, ITK, LAT, TESPA1 | 6 | |
| Activation of regulatory T lymphocytes | 0.000782 | 0.391 | BCL11B, CAMK4, CD40, CTLA4, FOXP3 | 5 | |
| Expansion of T lymphocytes | 0.0101 | 0.355 | CAMK4, CD3E, CD40, CTLA4, FLT3LG, FOXP3, ITK, LAT, REL | 9 | |
| Quantity of double-negative T lymphocytes | 0.000234 | 0.314 | ATG16L1, BCL11B, CD3E, CTLA4, FLT3LG, ITK, LAT, PRKCQ | 8 | |
| Quantity of CD8+ T lymphocytes | 0.00378 | −0.513 | ATG16L1, BCL11B, CD3E, CD40, FLT3LG, FOXP3, GPR18, IL22RA1, ITK, LAT, NR3C2, PRKCQ, TESPA1 | 13 | |
| Cell viability of T lymphocytes | 0.00027 | −0.787 | BCL11B, BRCA1, CACNB3, CAMK4, CD3E, CD40, CTLA4, LAT, PRKCQ, REL | 10 | |
| Anergy of T lymphocytes | 0.000222 | −1.109 | CD3E, CTLA4, FOXP3, PRKCQ, REL | 5 | |
| Activation of T lymphocytes | 0.00502 | −1.23 | BCL11B, BTN1A1, BTN2A2, CADM1, CAMK4, CD3E, CD40, CTLA4, DPP4, FLT3LG, FOXP3, ITK, LAT, PPP3CA, PRKCQ, REL, SEMA3G, SIRPG, TBK1, USP5 | 20 |
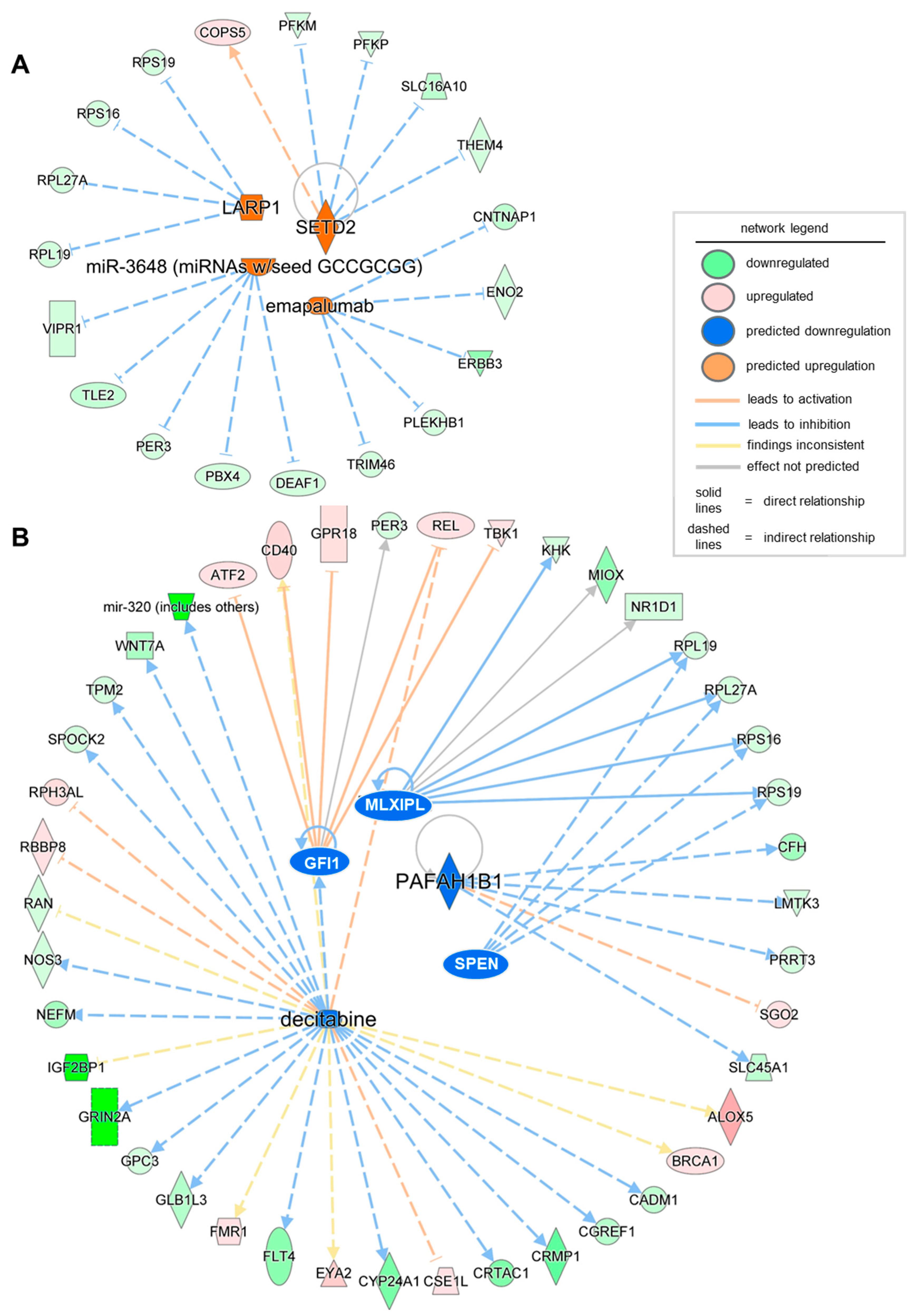
| Upstream Regulator | Molecule Type | Predicted Activation State | Activation z-Score | p-Value of Overlap | Target Molecules in Dataset |
|---|---|---|---|---|---|
| LARP1 | translation regulator | Activated | 2 | 0.045 | RPL19, RPL27A, RPS16, RPS19 |
| SETD2 | enzyme | Activated | 2.236 | 0.0178 | COPS5, PFKM, PFKP, SLC16A10, THEM4 |
| miR-3648 (miRNAs w/seed GCCGCGG) | mature microRNA | Activated | 2.236 | 0.0299 | DEAF1, PBX4, PER3, TLE2, VIPR1 |
| emapalumab | biologic drug | Activated | 2.236 | 0.000132 | CNTNAP1, ENO2, ERBB3, PLEKHB1, TRIM46 |
| GFI1 | transcription regulator | Inhibited | −2.236 | 0.041 | ATF2, CD40, GPR18, PER3, REL, TBK1 |
| PAFAH1B1 | enzyme | Inhibited | −2.236 | 0.0144 | CFH, LMTK3, PRRT3, SGO2, SLC45A1 |
| MLXIPL | transcription regulator | Inhibited | −2.177 | 0.0122 | KHK, MIOX, NR1D1, RPL19, RPL27A, RPS16, RPS19 |
| decitabine | chemical drug | Inhibited | −2.162 | 0.0157 | ALOX5, BRCA1, CADM1, CD40, CGREF1, CRMP1, CRTAC1, CSE1L, CYP24A1, EYA2 |
| SPEN | transcription regulator | Inhibited | −2 | 0.0172 | RPL19, RPL27A, RPS16, RPS19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, K. Comprehensive Transcriptomic and Proteomic Analysis of Severe Pressure Ulcer Patients Identifies Molecular Signatures Associated with Impaired T Cell Function. Biomolecules 2025, 15, 1682. https://doi.org/10.3390/biom15121682
Singh K. Comprehensive Transcriptomic and Proteomic Analysis of Severe Pressure Ulcer Patients Identifies Molecular Signatures Associated with Impaired T Cell Function. Biomolecules. 2025; 15(12):1682. https://doi.org/10.3390/biom15121682
Chicago/Turabian StyleSingh, Kanhaiya. 2025. "Comprehensive Transcriptomic and Proteomic Analysis of Severe Pressure Ulcer Patients Identifies Molecular Signatures Associated with Impaired T Cell Function" Biomolecules 15, no. 12: 1682. https://doi.org/10.3390/biom15121682
APA StyleSingh, K. (2025). Comprehensive Transcriptomic and Proteomic Analysis of Severe Pressure Ulcer Patients Identifies Molecular Signatures Associated with Impaired T Cell Function. Biomolecules, 15(12), 1682. https://doi.org/10.3390/biom15121682







