Abstract
Metastasis is the leading cause of cancer-related mortality, representing a highly coordinated, multistep process in which malignant cells gain the ability to invade, survive in the circulation, and establish secondary tumors at distant sites. While genetic mutations initiate oncogenesis, accumulating evidence shows that epigenetic and epitranscriptomic regulators, encompassing DNA methylation, RNA modifications, and noncoding RNAs (ncRNAs), reshape metastatic phenotypes. This review integrates current insights into these mechanisms and their crosstalk, with a primary focus on their methylation modification. Given their plasticity and potential reversibility, these regulators are attractive targets for therapeutic intervention. Defining the dynamic interplay between DNA and RNA modifications and ncRNAs provides a coherent framework for controlling metastasis and guides the development of precision epigenetic strategies and biomarkers. Future research that integrates multi-omics approaches and spatial transcriptomics will be essential for revealing the epigenetic and epitranscriptomic layers of the metastatic landscape.
1. Introduction
Metastasis is the spread of cancer cells from a primary tumor to distant tissues and is the leading cause of cancer-related mortality [1,2]. It unfolds as a multistep invasion–metastasis cascade comprising local invasion of surrounding tissue, intravasation into the vasculature or lymphatics, survival in circulation, extravasation at distant sites, seeding of micrometastases, and outgrowth to form macroscopic secondary tumors [1,3,4,5]. Each step requires transient phenotypic plasticity (for example, epithelial–mesenchymal transition, EMT) and adaptation to diverse microenvironments. As a result, metastasis is biologically complex and inefficient: only a minority of disseminated cells successfully colonize new niches [4,5]. Nevertheless, once established, metastatic disease is frequently incurable, underscoring the urgency of defining mechanisms that could be targeted therapeutically [1,3].
Beyond the well-characterized genetic alterations that initiate and drive tumor progression, converging evidence identifies epigenetic mechanisms as key regulators of metastasis [6]. Epigenetics refers to heritable changes in gene expression that occur without alterations of the DNA sequence. These includes covalent DNA modifications (for examples, cytosine methylation), RNA modifications, histone modifications and chromatin remodeling, and regulation by noncoding RNAs (ncRNAs) such as microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs) [6,7,8,9]. These epigenetic marks are dynamic and reversible, allowing for rapid cellular adaptation in various environments. In many cancers, global epigenomic reprogramming accompanies the acquisition of metastatic competence; comparative analyses of primary and metastatic lesions often reveal aberrant DNA methylation and dysregulated ncRNAs networks [6,8,9,10]. Such alterations confer pro-metastatic traits, including loss of adhesion, increased motility, stem-like features, and immune evasion, without requiring additional genetic change [1,3,10].
This review summarizes current knowledge on three major classes of epigenetic and epitranscriptomic regulators that shape metastasis in solid tumors: DNA methylations, RNA modifications, and ncRNAs (Figure 1).
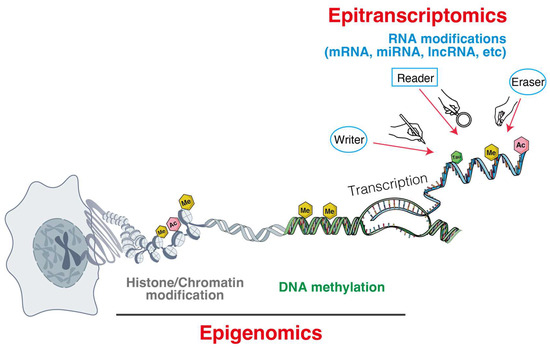
Figure 1.
Epigenomic and epitranscriptomic layers orchestrate gene regulation from chromatin to RNA.
We describe how these mechanisms reprogram gene expression and cellular state to drive metastatic progression, using examples from breast, lung, colorectal, prostate, and other cancers. We also discuss translational implications, including the development of epigenetic marks and ncRNAs as biomarkers of metastasis and as therapeutic targets. By integrating recent advances, we provide an up-to-date perspective on how epigenetic plasticity fuels metastatic evolution and how it may be leveraged to improve clinical outcomes.
2. DNA Methylation in Metastasis
DNA methylation is the addition of a methyl group to the 5-carbon of cytosine (5-methylcytosine, 5mC) within CpG dinucleotides. It is a foundational epigenetic mechanism that is frequently altered in cancer [11]. Aberrant DNA methylation patterns, including locus-specific hypermethylation (often in gene promoter CpG islands) and global hypomethylation, are hallmarks of cancer and contribute directly to metastasis [12]. Hypermethylation at gene promoters typically induces chromatin condensation and gene silencing; in cancer, this often inactivates genes that restrain tumor progression [13] (Table 1).

Table 1.
DNA methylation-driven modulators of metastasis across cancer types.
For example, the cell–cell adhesion molecule E-cadherin (encoded by CDH1) is a well-known tumor suppressor whose expression is frequently lost in metastatic carcinomas due to promoter CpG island hypermethylation [14,16,27]. Silencing of E-cadherin through DNA methylation diminishes intercellular adhesion, facilitating EMT and invasion, thereby promoting metastasis and correlating with a poor prognosis in multiple cancers [14,28]. Many other metastasis-suppressor genes such as TIMP3, BRMS1, SEMA3E, are likewise reported to undergo promoter hypermethylation during cancer progression, contributing to a more invasive phenotype [20,22]. Accordingly, increased expression or activity of DNA methyltransferases (DNMTs), the enzymes that catalyze 5mC formation, is often observed in advanced tumors [29]. Clinical studies have shown that the overexpression of DNMT1, DNMT3A, or DNMT3B in primary tumors is associated with increased risk of metastasis and poor outcomes [15,26]. In prostate cancer, upregulation of DNMT1 and DNMT3 is associated with a higher incidence of lymph node metastases [17]. In hepatocellular carcinoma, the upregulation of DNMT1 is associated with the repression of E-cadherin and the development of larger, more invasive tumors [18]. These findings suggest that abnormal DNA methylation is not just a consequence of tumorigenesis but actively contributes to the metastatic potential of tumors.
Conversely, hypomethylation of genomic DNA, such as the demethylation of normally methylated repetitive elements and heterochromatin regions, is another common alteration seen in metastatic cancers [12]. A widespread reduction in 5mC can result in chromosomal instability and the reactivation of transposable elements, which promotes genetic diversity and adaptability in tumor cells [30]. Hypomethylation at specific gene loci can also cause aberrant overexpression of pro-metastatic genes such as PRAME and S100A4, which are kept repressed in normal cells [23,31,32]. Thus, the shifting DNA methylation landscape in metastasis involves both targeted hypermethylation (shutting down inhibitors of metastasis) and diffuse hypomethylation (facilitating the permissive expression of genes that promote metastasis and instability). This dual remodeling of the methylome drives acquisition of metastatic traits, a paradoxical state of promoter hypermethylation alongside global hypomethylation. DNA methylation changes in metastasis can also affect noncoding genomic regions that control gene expression. For instance, enhancer or insulator methylation changes may activate pro-metastatic pathways or turn off genomic imprinting/tumor suppressive circuits [33]. In fact, hypermethylation can silence certain miRNA genes that normally suppress metastasis. The miR-34 family of tumor-suppressor miRNAs is frequently downregulated in metastatic disease; this downregulation has been attributed, in part, to CpG methylation of the MIR34A/B/C promoters in cancers such as colon, breast, and lung [34]. Loss of miR-34 permits increased expression of targets such as c-MET and SNAIL, which drive invasion and metastasis [35]. More broadly, a genome-wide study identified a DNA methylation signature that involves the silencing of multiple metastasis-suppressing miRNAs in metastatic tumors [36]. These findings reinforce that DNA methylation interacts with other epigenetic regulators, such as miRNAs, to orchestrate metastatic behavior.
Dynamic regulation of 5mC by DNA demethylases can also influence metastasis. The Ten-eleven translocation (TET) family of enzymes plays a crucial role in the oxidation of 5mC to 5-hydroxymethylcytosine (5hmC), a key step in the process of DNA demethylation [37]. A decrease in 5hmC is often found in advanced cancers and metastases, indicating reduced TET activity [38]. In some cases, TET-mediated demethylation can inhibit metastasis by reactivating genes that have been suppressed [39]. In addition, aberrant demethylation might activate genes that drive metastasis. An example is miR-29, which directly targets TET transcripts. In hepatocellular carcinoma, miR-29a induces the loss of 5hmC and promotes metastasis via a TET-SOCS1-MMP9 axis [19]. Here, miR-29a downregulates TET expression, leading to hypermethylation and silencing of the SOCS1 gene (a negative regulator of STAT3/MMP9 signaling), which in turn permits upregulation of MMP9 and aggressive, invasive behavior. This example highlights a convergence of epigenetic mechanisms, including DNA methylation and miRNAs, in driving metastasis.
On the other hand, excessive TET activity may facilitate metastasis under specific contexts, as demethylation can increase the expression of pro-metastatic factors. The specific outcome, however, depends on the targeted genes and the tissue environment. Translationally, insights into the role of DNA methylation in metastasis have spurred efforts to exploit these marks as biomarkers and therapeutic targets. Hypermethylated gene promoters can serve as blood-based DNA methylation biomarkers signaling occult metastatic disease or recurrence (for example, CDH1 or RARB methylation in circulating tumor DNA) [40,41,42]. DNA methylation patterns in primary tumors may also predict metastatic proclivity; for example, a “metastatic methylation signature” has been proposed for specific cancers [43]. In terms of therapy, hypomethylating agents (DNA methyltransferase inhibitors, such as 5-azacytidine and decitabine) can reactivate metastasis-suppressor genes that are silenced by methylation [44]. These drugs are already used in hematologic malignancies and are being investigated in solid tumors [45,46]; there is interest in whether they could reverse EMT or prevent metastatic outgrowth by restoring expression of adhesion molecules and other suppressors [47,48]. However, current systemic DNMT inhibitors induce broad CpG hypomethylation and can inadvertently activate oncogenes. By contrast, precise targeting of locus- or cell-restricted epigenome editing can modulate methylation at a defined regulatory element while sparing the rest of the genome. At present, CRISPR/dCas9-TET1 to demethylate a silenced metastasis suppressor or dCas9-DNMT3A to methylate a single enhancer that drives invasion; these approaches are compelling at the bench but remain preclinical [49,50]. Another approach is to target upstream regulators of aberrant methylation, such as inhibiting overexpressed DNMTs or modulating TET activity, to rebalance the methylome. Overall, DNA methylation is a double-edged sword in metastasis: it can serve as an early-warning biomarker of aggressive behavior and as a potentially malleable point of intervention to restrain metastatic progression.
3. RNA Epigenetic Modifications in Metastasis
DNA methylation influences the encoding of genomic information, but a comparable regulatory framework at the RNA level has also recently emerged as the epitranscriptome. The epitranscriptome comprises RNA modifications applied to RNA nucleotides after transcription, which can significantly impact RNA stability, processing [51], and translation. Thus, gene functions can also be modified without changing of the DNA sequence by the epitranscriptome. More than 170 distinct RNA modifications, including pseudouridine (Ψ), m1A, ac4C, m7G, and A-to-I editing, have been identified in cells. Among these, N6-methyladenosine (m6A) and 5-methylcytidine (m5C) are two of the most intensively studied marks in cancer biology [7,52]. This review focuses on m6A and m5C among the diverse RNA modifications. Dysregulation of these RNA modifications, particularly m6A and m5C, has been identified as a crucial contributor to the development of oncogenesis and metastasis [53,54]. In this section, we focus on how m6A and m5C RNA modifications, along with their writer/eraser/reader proteins, drive metastatic progression (Table 2).

Table 2.
The summary of epitranscriptomic regulators.
3.1. m6A RNA Methylation and Metastasis
N6-methyladenosine, m6A, is the most abundant internal modification in eukaryotic messenger RNAs and ncRNAs. The dynamic m6A methylome on RNA transcripts has been comprehensively revealed by m6A-specific sequencing techniques [55]. This mark, installed at the N6 position of adenine, affects RNA metabolism at multiple levels, including splicing, nuclear export, decay, and translation, depending on the context and the “reader” proteins that recognize a specific m6A [56]. The m6A landscape is dynamically regulated by “writer” enzymes (the METTL3/METTL14/WTAP methyltransferase complex and related factors) that add the mark, and “eraser” enzymes (FTO and ALKBH5 demethylases) that remove it [57]. Distinct m6A-binding reader proteins, such as the YTH domain family and IGF2BP family, then translate the presence of m6A into functional outcomes, such as altered mRNA stability or translation efficiency [58]. Aberrant regulation of m6A has been implicated in many aspects of tumor biology, including metastasis. Global m6A levels in mRNA often change during cancer progression, although findings are sometimes context-dependent [59]. Many aggressive tumors show increased m6A modification on transcripts, partly due to overexpression of m6A writers. For example, METTL3, the catalytic core component of the m6A methyltransferase complex, is frequently upregulated in advanced cancers such as lung adenocarcinoma, hepatocellular carcinoma, pancreatic cancer, and colorectal carcinoma [60,61,62,63]. High METTL3 levels have been correlated with higher metastatic capacity and poor survival (Table 3).

Table 3.
m6A epitranscriptomics-driven modulators of metastasis across cancer types.
In colorectal cancer, METTL3 expression is elevated in metastatic tumors, and METTL3 drives cell migration, invasion, and metastasis [63]. Mechanistically, METTL3 can promote metastasis by enhancing the maturation or translation of pro-metastatic transcripts. In a colorectal study, METTL3-mediated m6A on pri-miR-1246 was shown to increase processing of this miRNA, which in turn downregulated SPRED2 (a negative regulator of MAPK signaling), thereby unleashing MAPK activity and metastasis [64]. Similarly, in oral squamous cell carcinoma and pancreatic cancer, METTL3-catalyzed m6A installation on specific mRNAs such as BMI1 or E2F family transcripts leads to their increased translation and confers invasive, stem-like traits to tumor cells [76,79]. These studies illustrate that METTL3 has pro-metastatic functions by reprogramming the epitranscriptome to favor cancer cell motility and survival.
While m6A is often pro-metastatic, its role can vary and may be metastasis-suppressive in specific contexts (Figure 2).
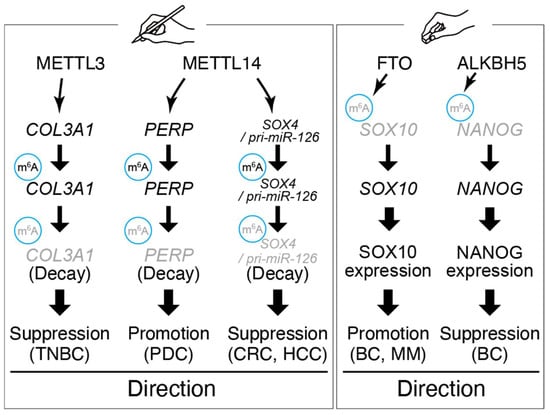
Figure 2.
Representative roles and target genes of m6A writer and eraser.
In triple-negative breast cancer (TNBC), METTL3 expression was relatively low; restoring METTL3 increased m6A on the COL3A1 mRNA, which led to reduced COL3A1 expression and impaired invasion [70]. In this case, reduced METTL3 allowed COL3A1 (a collagen gene involved in extracellular matrix remodeling) to be aberrantly upregulated, consequently enhancing metastatic spread. Thus, METTL3 can have tumor-suppressive effects in certain cellular contexts by modulating metastasis-relevant genes. Likewise, the other m6A writer enzyme METTL14 exhibits tumor-type specific roles. Although METTL14 promotes metastasis by increasing m6A on PERP mRNA in pancreatic cancer metastasis, it often acts as a metastasis suppressor in colorectal and hepatocellular carcinoma, where loss of METTL14 leads to EMT and stemness through reduced m6A abundance on key transcripts [71,77,82]. Thus, METTL14 exhibits tumor-type-dependent roles in metastasis, underscoring that m6A machinery must be interpreted within disease context and cellular wiring. For example, METTL14-mediated m6A on the SOX4 transcript in colorectal cancer inhibits SOX4 expression and metastasis; low METTL14 permits SOX4 protein upregulation and is associated with metastatic relapse [66]. These divergent observations underscore that the impact of m6A modification on metastasis depends on which RNAs are targeted and the cellular environment [59].
In addition to writers, m6A erasers and readers contribute to metastasis. The demethylase FTO is overexpressed in cancers such as breast and melanoma and can promote metastasis by erasing m6A marks on mRNAs that encode pro-metastatic proteins, thereby stabilizing those transcripts [67,80]. Conversely, the other eraser ALKBH5 has been reported to suppress metastasis in some contexts, such as breast cancer, by demethylating and thereby stabilizing transcripts of tumor-suppressive genes or by interfering with pre-mRNA splicing of EMT drivers [69]. m6A reader proteins of the YTHDF family influence metastasis by binding m6A-marked transcripts and altering their fate [68,72]. YTHDF1, which promotes translation, is often upregulated in metastatic cancers and linked to worse outcomes [72,83]. YTHDF1 enhances protein abundance encoded from pro-metastatic mRNAs and is being studied as a potential prognostic marker in various cancers [72,84]. By contrast, YTHDF2 accelerates mRNA decay and may inhibit metastasis by degrading mRNAs that encode metastasis-promoting factors, although it can also destabilize mRNAs of metastasis suppressors [81,85].
Another class of m6A readers, the IGF2BP1/2/3 proteins, bind m6A-modified transcripts and greatly stabilize them. IGF2BP3, in particular, is highly expressed in aggressive tumors and was shown to enhance the stability of m6A-modified mRNAs such as MYC or SLUG, thereby promoting invasion and metastatic colonization [58,73,74]. High IGF2BP3 level expression correlate with poor survival of cancer patients, like colorectal and pancreatic cancer, consistent with a pro-metastatic function [65,78].
Taken together, these findings paint a picture in which m6A dysregulation is a pervasive feature of metastatic progression. By rewiring post-transcriptional gene expression programs, changes in m6A deposition or recognition can tip the balance at epithelial–mesenchymal states, or between dormancy and proliferation of disseminated cells. Importantly, components of the m6A machinery are emerging as drug targets for cancer therapeutics. Several small-molecule inhibitors of METTL3 have been developed, and their effects on solid tumor metastasis are under investigation. Inhibiting METTL3 or an m6A reader could, in principle, reduce metastatic seeding in cancers where m6A drives metastasis. Conversely, restoring the levels of m6A or inhibiting erasers would be beneficial in patients with contexts where m6A has tumor-suppressive roles. The therapeutic window and safety of these strategies are active areas of cancer research. Nevertheless, targeting the “epitranscriptome” holds promise because, like other epigenetic alterations, RNA modifications are reversible and do not require that underlying genetic mutations be fixed. Precision therapies, such as modified antisense oligonucleotides targeting m6A on specific transcripts, may significantly reduce a tumor’s metastatic potential with high specificity. In summary, m6A dysregulation reshapes RNA fate to tip the balance between dormancy and invasion, underscoring its therapeutic potential.
3.2. m5C RNA Methylation and Metastasis
5-methylcytidine on RNA (m5C), analogous to DNA 5-methylcytosine, is an epitranscriptomic mark increasingly recognized for its role in cancer biology. m5C was long known to occur in ncRNAs, including rRNA and tRNAs, but recent high-resolution techniques in high-throughput mapping have shown that m5C also exists on mRNAs and lncRNAs [86,87,88]. Although m5C is a comparatively rare modification, it can exert significant influence on RNA fate, affecting RNA stability, translation, localization, and forming binding sites for specific proteins [89]. The enzymatic writers of RNA m5C include the NSUN family (NSUN1 through NSUN7) and DNMT2, which methylate cytosine residues in different RNA contexts [90]. The erasers of m5C remain unclear, but the TET enzymes and ALKBH1 have been suggested to generate 5-hydroxymethylcytidine (hm5C) or otherwise demethylate m5C [91,92]. Importantly, several reader proteins recognize m5C marks, notably ALYREF (an mRNA export adaptor) and Y-box binding protein 1 (YBX1) [93,94]. These readers bind methylated cytosines and commonly promote export or stabilization of the modified transcripts. Emerging evidence indicates that dysregulation of m5C and its associated proteins contributes to tumor progression and metastasis [95]. NSUN2, a cytosine methyltransferase, is a well-studied m5C writer in cancer. NSUN2 targets a range of RNAs, including tRNAs, mRNAs, and ncRNAs, and is frequently overexpressed in aggressive tumors [93,94,96]. In addition, NSUN2 often promotes metastasis by methylating specific mRNAs to enhance their stability and translation through m5C reader proteins (Table 4).

Table 4.
A m5C writer NSUN2–driven modulators of metastasis across cancer types.
In bladder cancer, NSUN2 methylates the 3′ untranslated region (3′-UTR) of the HDGF (hepatoma-derived growth factor) mRNA, installing m5C marks that are bound by YBX1 [94]. YBX1 binding protects the transcript from degradation, leading to higher HDGF protein levels and increased tumor cell growth, invasion, and metastasis [94,97]. NSUN2 and YBX1 thus act in concert as writer and reader to post-transcriptionally upregulate pro-metastatic factors like HDGF. Similarly, in breast cancer and melanoma, NSUN2 overexpression correlates with advanced stage and metastasis; NSUN2 can methylate mRNAs or even tRNA-derived fragments that regulate cell motility and cytoskeletal dynamics, thereby facilitating metastatic spread [107,108]. The oncogenic function of NSUN2 is underscored by studies showing that NSUN2 knockout or knockdown impairs migration and metastasis in models of esophageal squamous carcinoma, glioma, and others [98,101,109]. In esophageal cancer, NSUN2-mediated m5C on a specific lncRNA was found to promote that lncRNA’s association with a chromatin modifier (BPTF), increasing levels of matrix metalloproteinases (MMPs), which leads to degradation of the extracellular matrix, promoting invasion [99,100]. This highlights that RNA m5C can interface with chromatin regulation indirectly through lncRNAs. Other NSUN family members also contribute to metastasis. NSUN5 and NSUN6 have been linked to metastasis in certain cancers. For example, in breast cancer, NSUN6 expression is elevated in bone metastases, and NSUN6-mediated m5C on specific transcripts is associated with activation of YAP signaling, promoting osteotropic metastasis [110,111,112]. NSUN5, on the other hand, has been implicated in metastasis of head and neck squamous cell carcinoma and gliomas, although its precise targets in those contexts are still being elucidated [113,114].
Beyond the NSUN enzymes that target mRNA, the DNMT2 enzyme primarily methylates tRNA but may indirectly influence metastasis by affecting protein synthesis and cell stress responses [115,116]. The overall picture is that m5C “writer” enzymes commonly act as metastasis promoters by reprogramming the transcriptome of cancer cells at the RNA level. Crucially, the reader proteins of m5C mediate the downstream effects of these methylation events. ALYREF, a nuclear RNA-binding protein that shuttles mRNAs to the cytoplasm, binds preferentially to m5C-modified transcripts and enhances their export from the nucleus [93]. Overexpression of ALYREF in cancers has been associated with increased translation of m5C-marked oncogenic mRNAs. ALYREF was also found to bind and export transcripts modified with m5C of MYC, YAP, and PKM2, leading to their increased expression and contributing to tumor progression and resistance to therapy [93,117,118]. Meanwhile, YBX1, a cytoplasmic mRNA-binding protein, is another critical m5C reader in metastasis. YBX1 has high affinity for m5C sites in the 3′-UTRs or coding sequences of target mRNAs, and upon binding, YBX1 can recruit stabilizing factors like PABP (poly(A)-binding protein) to protect the transcript [94,119,120]. YBX1 also often enhances translation of its target mRNAs. Of note, YBX1 recognizes m5C sites on FOXC2, HDGF, and AR encoding pro-metastatic proteins, resulting in the enhancement of translation efficiencies and functions [94,102,104]. FOXC2 is a transcription factor that drives EMT and metastasis. As HDGF facilitates tumor angiogenesis and growth, YBX1’s stabilization of these mRNAs directly contributes to the metastatic phenotype. In cholangiocarcinoma (bile duct cancer), YBX1 binding to m5C-modified NKILA lncRNA was reported to stabilize that lncRNA and promote cancer cell migration [121]. Intriguingly, NKILA is known to interact with NF-κB signaling, suggesting an epigenetic feed-forward loop where m5C, lncRNAs, and inflammatory signaling intersect in metastasis [121,122].
Overall, the aberrant upregulation of m5C readers like YBX1 in cancers creates a scenario in which any mRNA bearing m5C might be aberrantly stabilized, tilting the proteome toward a metastatic state. On the other hand, disruption of the m5C machinery can impair metastasis. Loss-of-function of NSUN2 in cancer cells leads to reduced methylation of many RNAs, often resulting in decreased stability of pro-migratory transcripts and a subsequent decline in invasion [94,105]. Depletion of YBX1 or ALYREF has similarly been shown to sensitize m5C-marked transcripts to degradation and to reduce metastasis in experimental models [106,123]. These findings suggest that targeting m5C writer or reader proteins could be a viable therapeutic strategy. Indeed, NSUN2 is being explored as a potential drug target or prognostic marker. The levels of NSUN2 expression in tumors have prognostic value in certain cancers, such as high NSUN2 portends poor outcome and greater metastatic risk [103,105,124]. YBX1 has also been investigated as a therapeutic target in aggressive cancers, although being a DNA/RNA-binding protein makes it challenging to target directly. An alternative approach might be to disrupt the interaction between m5C marks and their readers. Small molecules or peptides that mask m5C sites could prevent readers like YBX1 from binding, thereby destabilizing those transcripts [125]. Such approaches remain speculative but exemplify the new avenues of intervention that epitranscriptomic research is just beginning to open. In summary, RNA m5C modification is now recognized as an important regulator of metastasis. Through m5C modification on coding and ncRNAs, cancer cells can post-transcriptionally upregulate entire cohorts of genes that endow metastatic capabilities, including invasion, anoikis resistance, and colonization. The NSUN methyltransferases and YBX1/ALYREF reader proteins form an axis that frequently drives metastasis in diverse tumors by synergistically methylating and stabilizing metastasis-related RNAs. RNA modifications can quickly alter protein expression without new transcription. This ability is particularly useful for cancer cells during metastasis. As detection methods for m5C improve, such as high-resolution m5C RNA sequencing, we are likely to discover specific “m5C-regulated RNA networks” that control metastatic colonization and perhaps metastatic latency. Investigating components of the m5C regulatory network and utilizing m5C markers as potential biomarkers, such as altered tRNA methylation patterns in circulating vesicles, may provide new approaches to combat metastatic cancer. Overall, aberrant m5C regulation establishes a post-transcriptional program that supports invasion and colonization, highlighting the NSUN–YBX1/ALYREF axis as a therapeutic opportunity.
4. Noncoding RNAs in Metastasis
ncRNAs are RNA molecules that are not translated into proteins. In this review, we explicitly separate housekeeping ncRNAs (rRNA, tRNA, snRNA, snoRNA) from regulatory ncRNAs (miRNAs, lncRNAs, circRNAs). Regulatory ncRNAs have emerged as central players in post-transcriptional and epigenetic regulation of gene expression and are particularly implicated in cancer phenotypes such as metastasis. In various human cancers, multiple pathways by which ncRNAs regulate metastasis through RNA modifications and epigenetic mechanisms have been elucidated. miRNAs suppress or promote metastasis by altering histone modifications and DNA methylation states through regulating the expression levels of target epigenetic factors. Meanwhile, lncRNAs and circRNAs can comprehensively regulate metastasis-related gene expression by recruiting epigenetic factors as scaffold molecules or by acquiring functions through RNA modifications applied to themselves. These findings suggest that ncRNAs occupy a central position within the epigenome and epitranscriptome of the regulatory network governing the complex process of metastasis. Here, we focus on representative ncRNAs, miRNA, and lncRNA, to discuss the relationship between cancer metastasis and RNA modifications and epigenetics.
4.1. microRNAs and Metastasis
miRNAs are ~20–24 nucleotide small RNAs that typically bind to complementary sequences in the 3′-UTRs of target mRNAs, inducing mRNA degradation or translational repression [126,127]. A single miRNA can regulate dozens of genes mainly by the seed-region of miRNA, and conversely, a given mRNA may be targeted by multiple miRNAs, making them potent modulators of gene networks [128]. In cancer, numerous miRNAs are aberrantly expressed, functioning either as oncogenic miRNAs or tumor-suppressor miRNAs [129] (Table 5).

Table 5.
Key factors in miRNA functions.
4.1.1. RNA Modification of miRNAs
RNA modifications are known to impact miRNA functions in various ways, affecting cancer phenotypes. N6-methyladenosine (m6A) marks on primary miRNAs (pri-miRNAs) modulate the activity of Microprocessor, which consists of Drosha and DGCR8 [130]. The nuclear RNA-binding protein HNRNPA2B1 acts as an m6A “reader” on a subset of pri-miRNAs, physically associates with DGCR8, and thereby promotes pri-miRNA processing [131]. Thus, loss of HNRNPA2B1 phenocopies METTL3 depletion for those substrates. Therefore, the METTL3/METTL14–m6A–HNRNPA2B1–DGCR8 axis affects the abundance of mature miRNA. In hepatocellular carcinoma (HCC), METTL14 binds DGCR8 and facilitates m6A-dependent processing of pri-miR-126 [71]. Reduced METTL14 expression represses the mature miR-126 level, enhancing migratory and invasive activity. In orthotopic and tail-vein models, downregulation of METTL14 increased metastatic burden, while restoring METTL14 or miR-126 suppressed dissemination, linking an m6A-programmed miRNA defect to metastasis in vivo [71,132]. Additionally, m6A-sensitive pri-miRNAs in HCC reinforce the notion that m6A-guided processing is a metastasis checkpoint [133,134].
METTL1 installs 7-methylguanosine (m7G) into specific pri-/pre-miRNAs. In the let-7 family, m7G disrupts a form of G-quadruplex, enhancing the stability of let-7 maturation and reduction in migration activity [135]. By a mass spectrometry approach, m7G modification was identified in a single guanosine in let-7e-5p, and genetic or catalytic inactivation of METTL1 reduces mature let-7 form and increases migratory activity [135]. While these data are limited to migration/invasion rather than overt metastasis, they pin a defined chemical mark on a defined miRNA to a pro-metastatic phenotype in human cancer cells.
At the 3′ ends of miRNA, terminal uridylyl transferases TUT4/ZCCHC11 and TUT7/ZCCHC6, recruited by LIN28A/B to the conserved GGAG element in pre-let-7, catalyze oligo-uridylation [136]. The uridylation on pre-let-7 prevents Dicer cleavage and suppresses the maturation of let-7. In this process, LIN28 switches TUT4/7 from mono- to oligo-uridylation to enforce this block [137]. In contrast, in somatic cells lacking LIN28, mono-uridylation facilitates the processing of group II pre-let-7 [138]. Functionally, the LIN28–TUT4/7 uridylation gate sits directly on migratory and invasive activity through the let-7 network.
Adenosine-to-inosine (A-to-I) editing, changing a specific nucleotide base within the seed region of miRNAs, affects the recognition of target genes by miRNA [139]. In melanoma, a decrease in the editing of miR-455-5p changes its role from a suppressor of growth and metastasis (the edited form) to a regulator that promotes metastasis (the unedited form) [140]. Either restoring ADAR1 or expressing the edited form of miR-455-5p reduces tumor growth and metastases in the lung metastasis model [141]. Conversely, the unedited isoform of miR-455-5p suppresses CPEB1 and encourages tumor dissemination [140,141]. Additionally, attenuated editing of miR-376a* enhances migration and invasion in vitro and increases aggressiveness in orthotopic glioma models [142]. Although brain tumors do not conventionally “metastasize” outside the CNS, this study demonstrates that editing state dictates invasive behavior in vivo. These experiments provide a direct causal chain from miRNA editing status to metastatic outcome.
4.1.2. Epigenetics of miRNAs
miRNAs influence cancer cell functions in various ways, both directly and indirectly, through epigenetic mechanisms. Loss of miR-101 relieves repression of EZH2, the catalytic subunit of PRC2, expanding H3K27me3 at anti-metastatic loci and shifting chromatin toward a pro-invasive program [143]. In breast cancer patients, progression to advanced and metastatic disease is accompanied by decreased miR-101 and reciprocal EZH2 upregulation, and functional studies show that restoring miR-101 restrains invasion by lowering EZH2 and its repressive mark [143,144]. Collectively, these data position the miR-101-PRC2/EZH2 axis as a core epigenetic route to metastatic competence in various tumors.
In gastric cancer, miR-29b/c and DNMT3A form a methylation circuit that converges on CDH1. Loss of miR-29b/c with gain of DNMT3A drives CDH1 promoter hypermethylation, represses E-cadherin, and increases migration/invasion, an epigenetic route to EMT and metastatic competence [21]. Thus, direct targeting of DNMT3A by miR-29b/c is associated with methylation-dependent silencing of miR-29b/c itself.
miR-148a directly suppresses DNMT1, leading to promoter demethylation and re-expression of metastasis-suppressor genes in pancreatic cancers [24,25]. Further, restoring miR-148a (or inhibiting DNMT1) reduces migration and invasion, indicating that the DNMT1–miR-148a axis functions as an epigenetic brake on metastatic traits [25]. In clinical samples, reduced miR-148a expression correlated with increased expression of DNMT1, consistent with pathway disruption in aggressive disease [24,25].
As in breast cancer, miR-101 loss in prostate tumors unleashes EZH2 and H3K27me3-mediated silencing across metastasis-suppressive programs in prostate cancer [143,145]. Genomic analyses reveal frequent deletions of miR-101 loci in metastatic samples. In addition, functional studies indicate that restoring miR-101 reduces EZH2 expression and invasive behavior, making this miR-101–PRC2 axis one of the most precise miRNA-driven epigenetic mechanisms linked to metastasis [143,146,147].
Taken together, the functional regulation of cancer metastasis through complex feedback loops and networks involving miRNAs, epigenetics, and RNA modifications is only beginning to be elucidated.
4.2. Long Noncoding RNAs and Metastasis
lncRNAs are a broad class of ncRNAs of over 200 nucleotides in length that do not encode proteins. lncRNAs are involved in gene regulation through diverse mechanisms. They can act as scaffolds for protein complexes, guides to recruit chromatin modifiers to specific genomic loci, decoys that sequester proteins or miRNAs, or even as precursors for small RNAs. The human genome encodes thousands of lncRNAs, and aberrant lncRNA expression is a common feature of cancer. Many lncRNAs have been identified as key drivers or suppressors of metastasis, often functioning in a cell-type-specific manner (Table 6).

Table 6.
Representative lncRNAs that remodel the epigenome and epitranscriptome in metastasis.
4.2.1. RNA Modification of lncRNAs
HOTAIR is a well-established lncRNA that rewires chromatin in favor of metastatic activity in multiple cancer types [148,149,167]. HOTAIR carries a conserved m6A site (A783) that is required for its chromatin repression program [164,168]. Mutating this single site weakens HOTAIR’s interaction with the nuclear m6A reader YTHDC1, blunts H3K27me-linked silencing, and reduces malignant growth and invasion of breast cancer cells [148,164,168]. In patient datasets, tumors with high HOTAIR and intact A783-dependent signaling show stronger Polycomb target repression and worse outcomes, consistent with an epitranscriptomic “licensing” of HOTAIR function [148,169].
One of the classical lncRNAs, H19, is a direct substrate of NSUN2 in hepatocellular carcinoma. m5C on H19 increases transcript stability and promotes the recruitment of the oncoprotein G3BP1, supporting proliferation and tumor progression; higher H19 m5C and expression levels are associated with poor differentiation in patients with liver cancer [96,98].
METTL14, in part, promotes m6A-dependent decay of oncogenic XIST transcripts through YTHDF2. Dampening XIST through this pathway reduces proliferation and metastatic potential in models and correlates with improved prognosis of the patients with colorectal cancer [82,165].
The lncRNA, RNA Component of Mitochondrial RNA Processing Endoribonuclease (RMRP), carries abundant m6A, which increases its stability. RMRP recruits DNA methyltransferases to the SCARA5 promoter, induces promoter methylation, and suppresses SCARA5 expression. Silencing RMRP reduces proliferation, migration, and invasion of bladder cancer [170]. LINC01833 is m6A-modified by METTL3 and engages hnRNPA2B1 [171]. This module promotes growth and invasive traits of NSCLC, which are associated with adverse clinicopathologic features [172]. PCAT6 is m6A-modified and bound by the reader IGF2BP2, which stabilizes IGF1R mRNA. This lncRNA–reader circuit fuels proliferation and bone metastasis of prostate cancer in mouse models and is enriched in high-risk clinical situations [58].
YTHDF3, an m6A reader, recognizes m6A-modified GAS5 and accelerates its decay in colorectal cancer [173]. Although the binding of GAS5 to YAP promotes its phosphorylation and ubiquitylation for degradation, loss of GAS5 lifts the restraint on YAP signaling, resulting in enhancement of cell proliferation, migration, and further in vivo tumor progression [173,174]. In a clinical study, a low GAS5 level is inversely correlated with YTHDF3 and YAP protein levels in colorectal tumors [173].
The host gene of the miR-100, MIR100HG, binds the m6A reader/adapter hnRNPA2B1 to stabilize m6A-modified TCF7L2 mRNA, amplifying Wnt target expression and driving colorectal tumor growth and invasion [131,161,162]. Disruption of METTL3-installed m6A or of the MIR100HG–hnRNPA2B1 interaction destabilizes TCF7L2 and curtails malignant behavior [162,163].
The m6A eraser ALKBH5 binds and demethylates NEAT1 transcript in gastric cancer, increasing NEAT1 activity [154]. Elevated ALKBH5/NEAT1 enhances invasion and metastasis, in part by modulating EZH2 expression and downstream EMT programs [155]. Suppression of NEAT1 reverses these effects in vitro and in mouse models [156]. In addition, NEAT1 is m6A-modified in aggressive prostate cancer [157]. m6A enhances NEAT1’s nuclear functions that foster chromatin remodeling and transcriptional programs conducive to osteotropism, increasing bone colonization in vivo; genetic interference with NEAT1 or m6A recognition suppresses bone metastasis [158]. Furthermore, reduced level of ALKBH5 provides m6A accumulation on the metastasis-restraining lncRNA KCNK15-AS1 in pancreatic ductal adenocarcinoma. Restoration of ALKBH5 expression recovers demethylation and stabilization of KCNK15-AS1, suppressing migration and invasion [159]. KCNK15-AS1, in turn, modulates PTEN–AKT signaling and downstream motility circuits [160].
Collectively, the relations between lncRNA and RNA modification hard-wire EMT, motility, survival under stress, and metastatic colonization into the cancer transcriptome.
4.2.2. Epigenetics of lncRNAs
Overexpressed HOTAIR binds PRC2 through EZH2 and the LSD1/CoREST complex and retargets them across the genome, increasing H3K27me3 and reducing H3K4 methylation at anti-metastatic loci [148,149,150]. These changes enhance invasion in vitro and promote distant colonization in mouse models, and high HOTAIR levels in primary tumors correlate with subsequent metastasis and poorer survival.
GClnc1 illustrates a chromatin-scaffolding lncRNA in gastric cancer [151]. It assembles WDR5 (to promote H3K4 methylation) and KAT2A/GCN5 (to promote histone acetylation) at specific promoters, establishing modification patterns that support EMT, migration, and invasion; expression rises with tumor size, depth of invasion, and nodal spread [152].
Aggressive prostate cancers often activate SChLAP1, which directly antagonizes the SWI/SNF (BAF) chromatin-remodeling complex [175]. Gain- and loss-of-function studies show that SChLAP1 impairs SWI/SNF occupancy genome-wide, shifting chromatin states toward oncogenic, pro-metastatic transcriptional programs; this remodeling increases invasion and distant spread and is enriched in high-risk clinical disease [153,176].
Across organs, a recurring theme is that lncRNAs reshape the epigenome by recruiting or opposing core chromatin regulators (PRC2, LSD1/CoREST, SWI/SNF) or, in some cases, by exploiting RNA modifications (for example, m6A installed by METTL3) to stabilize scaffolds that then alter chromatin-linked signaling. The downstream effect is a durable transcriptional program that supports EMT, invasion, survival in circulation, and colonization at distant sites.
4.3. circRNAs and Metastasis
Circular RNAs (circRNAs) arise from back-splicing of pre-mRNAs, yielding covalently closed loops that are typically exon-derived, lack 5′/3′ ends, and are relatively resistant to exonucleases, enabling stable accumulation in cancer cells and extracellular vesicles [177]. Their subcellular localization (nuclear vs. cytoplasmic) and binding partners dictate function.
Epitranscriptomic modification of circular RNAs, particularly m6A, regulates biogenesis, nuclear export, subcellular localization, translation, and decay, which together shape post-transcriptional programs relevant to EMT, invasion, survival in circulation, and colonization [178]. m6A marks on circRNAs are recognized by nuclear and cytoplasmic readers that direct RNAs toward export, degradation, or cap-independent translation, establishing routing decisions that can influence metastatic behavior in specific contexts [178].
A metastasis-focused exemplar is circNSUN2 in colorectal cancer, where m6A on circNSUN2 is recognized by YTHDC1 and promotes nuclear export to the cytoplasm [179]. In the cytoplasm, circNSUN2 scaffolds IGF2BP2 on HMGA2 mRNA, which stabilizes HMGA2 and enhances liver metastatic colonization, and loss-of-function for circNSUN2, YTHDC1, or IGF2BP2 reduces HMGA2 stability and metastatic traits with enrichment of cytoplasmic circNSUN2 in liver metastases relative to primary tumors [179].
An additional circuit relevant to epithelial cancers involves circITGB6 and IGF2BP3, where TGFβ induction of circITGB6 promotes formation of a complex with IGF2BP3 that stabilizes PDPN mRNA and increases PDPN protein, driving EMT features and metastatic dissemination in mouse models [75]. The biochemical plausibility of this stabilization step is supported by the capacity of IGF2BP proteins to bind m6A-modified transcripts and enhance their stability and translation under defined conditions [58].
m6A also licenses translation of circRNAs in a cap-independent manner, with a single m6A site sufficient to initiate translation and with YTHDF3 and eIF4G2 promoting ribosome engagement while METTL3 and METTL14 enhance this output [178]. In colorectal cancer, circYAP encodes the YAP-220aa micro-protein in a translation program that requires YTHDF3 and eIF4G2 and that supports invasion and liver metastasis, providing a metastasis-proximal example of m6A-dependent circRNA translation [180].
Conversely, m6A can designate circRNAs for targeted decay because YTHDF2 together with the adaptor HRSP12 recruits the RNase P or RNase MRP complex to cleave m6A-bearing substrates, reducing circRNA abundance and counterbalancing export or translation programs [181].
Hepatocellular carcinoma provides an additional illustration in which circHPS5 accumulates with increased m6A; METTL3 facilitates circHPS5 production, YTHDC1 supports cytoplasmic export, and circHPS5 functions as a miRNA sponge that elevates HMGA2 expression and promotes malignant phenotypes aligned with invasion and colonization [182].
Taken together, YTHDC1-guided export, YTHDF3 and eIF4G2-assisted translation, YTHDF2–HRSP12–RNase P or RNase MRP-mediated decay, and reader-dependent stabilization exemplified by the circITGB6–IGF2BP3–PDPN and circNSUN2–IGF2BP2–HMGA2 axes outline a coherent framework for how circRNA modifications intersect with metastatic biology; continued work that quantifies per-site modification stoichiometry, reader competition, and in vivo causality is likely to refine these connections across tumor types.
5. Conclusions and Future Directions
Metastasis remains the most lethal aspect of cancer, responsible for the majority of deaths in cancer patients. Metastasis is understood to be not only be caused by serial genetic mutations, but also epigenetic plasticity that allows cancer cells to adapt and thrive in new environments. In this review, we highlighted three major epigenetic mechanisms—DNA methylation, RNA modifications, and ncRNAs—and described how their dysregulation drives the multi-step metastatic cascade across various solid tumors. DNA methylation alterations in metastasis can silence key suppressors of invasion while activating prometastatic pathways, and they provide potential biomarkers and targets for intervention. RNA modifications like m6A and m5C on mRNAs add a post-transcriptional layer of control that cancer cells exploit to fine-tune gene expression for metastasis; targeting the enzymes or readers of these marks is a burgeoning therapeutic concept. ncRNAs, miRNAs, and lncRNAs form extensive regulatory networks that integrate with canonical signaling and epigenetic machinery to promote or restrain metastasis. They not only serve as mechanistic drivers through their effects on gene and protein networks, but also as convenient biomarkers detectable in blood.
A unifying theme is the reversibility and context-dependence of epigenetic modifications. This plasticity is what enables metastatic cells to switch phenotypes like toggling between epithelial and mesenchymal states, and to remain latent or reactivate after years. Importantly, these changes are, in principle, therapeutically reversed.
Epigenetic and RNA-based therapies are still in development, but they offer opportunities for precision medicine approaches to metastasis [183]. For instance, if a patient’s metastasis is found to be driven by overactive m6A writer, METTL3, and low miR-200, a combination of a METTL3 inhibitor and a miR-200 mimic could be considered to tackle the disease on two epigenetic fronts. This kind of rationale may soon be testable as our toolkit of epigenetic drugs expands. Looking ahead, continued research is needed to address several challenges and unanswered questions. We must decipher the global epigenomic landscapes of metastases: How do they differ from primary tumors? Single-cell multi-omics and spatial transcriptomics are powerful new approaches to chart heterogeneity within metastases and their surrounding microenvironments. These technologies can reveal how cancer cells at the invasive edge vs. the core of a metastatic lesion differ in chromatin state or ncRNA expression, or how cancer-associated fibroblasts and immune cells in the metastatic niche are epigenetically modulated to support tumor growth. Integrating these layers will provide a holistic view of metastasis and identify critical “epigenetic crosstalk” between tumor cells and stroma.
Another priority is understanding temporal dynamics: which epigenetic changes are early drivers of metastasis, and which are later adaptations or consequences? Discriminating between drivers and passengers enables prioritization of targets. Longitudinal studies can elucidate the sequence of epigenetic events in metastatic progression by analyzing patient samples at the point of initial diagnosis, during remission, and metastatic recurrence.
Epigenetic drugs can have broad effects given that they target fundamental cellular processes. Combining epigenetic therapy with standard treatments (chemotherapy, targeted therapy, immunotherapy) in rational ways will be critical to prevent escape. Epigenetic priming (using a drug to alter tumor cell state to make them more susceptible to another therapy) is an active strategy. Such sequential or combinatorial approaches represent the forefront of translational epigenetics.
Interactions between DNA and RNA modifications and ncRNAs forms an “epigenetic blueprint” that drives metastasis. By encoding this blueprint, we not only gain fundamental insights into how cancer spreads but also open new avenues to intercept the spread of cancer. In recent years, various technologies, such as long-read sequencers and nanopore sequencers, have been developed and have achieved significant results as tools for deciphering these blueprints. While this review does not discuss them in detail, it has become clear that various modifications, such as acetylation and lactylation, influence epigenetic changes, affecting metabolic reprogramming of cancer cells [184,185]. Furthermore, advances in mass spectrometry continue to identify novel RNA modifications. While technological advances are welcome, several caveats temper interpretation. For instance, as antibody-based m6A profiling and RNA bisulfite sequencing provide relative enrichment, not absolute stoichiometry, these techniques may misread the sites. A reader-CLIP confirms binding but not causality. Bulk assays confound signals from tumor, stromal, and immune compartments, and many functional studies rely on overexpression or deletion outside physiological ranges. More work using orthogonal chemistries, quantitative standards, and endogenous perturbations is required to build causal maps linking specific marks on defined transcripts or loci to metastatic phenotype in vivo [186].
Epigenetic mechanisms are architects of cellular identity and behavior. However, in cancer, they are hijacked to construct the phenotype of metastasis. Importantly, we are beginning to elucidate these regulatory blueprints, and we can design therapies to rewrite the narrative of metastasis. Multidisciplinary efforts that bridge molecular biology, bioinformatics, and clinical oncology, will be essential to translate epigenetic and ncRNA-targeted interventions from bench to bedside. The ultimate vision is that, in the future, a diagnosis of metastatic cancer will come not with resignation, but with a well-informed, personalized plan to epigenetically disarm the metastases and improve patient outcomes.
Author Contributions
Conceptualization, T.H.; writing—original draft preparation, T.H. and M.S.; writing—review and editing, T.H. and M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the Takeda Science Foundation.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We sincerely thank Anisimov Sergei for his proofreading and constructive feedback.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ncRNA circRNA | noncoding RNA circular RNA |
| lncRNA | long noncoding RNA |
| miRNA | microRNA |
| m6A | N6-methyladenosine; RNA |
| m5C | 5-methylcytidine; RNA |
| hm5C | 5-hydroxymethylcytidine; RNA |
| 5mC | 5-methylcytosine; DNA |
| 5hmC | 5-hydroxymethylcytosine; DNA |
| TNBC | Triple-negative breast cancer |
| HCC | Hepatocellular carcinoma |
| BC | breast cancer |
| PDC | Pancreatic ductal adenocarcinoma |
| MM | Malignant Melanoma |
| CRC | Colorectal cancer |
| NSCLC | Non-small cell lung cancer |
References
- Valastyan, S.; Weinberg, R.A. Tumor Metastasis: Molecular Insights and Evolving Paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef]
- Dillekås, H.; Rogers, M.S.; Straume, O. Are 90% of Deaths from Cancer Caused by Metastases? Cancer Med. 2019, 8, 5574–5576. [Google Scholar] [CrossRef]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef]
- Luzzi, K.J.; MacDonald, I.C.; Schmidt, E.E.; Kerkvliet, N.; Morris, V.L.; Chambers, A.F.; Groom, A.C. Multistep Nature of Metastatic Inefficiency: Dormancy of Solitary Cells after Successful Extravasation and Limited Survival of Early Micrometastases. Am. J. Pathol. 1998, 153, 865–873. [Google Scholar] [CrossRef]
- Fidler, I.J. The Pathogenesis of Cancer Metastasis: The “seed and Soil” Hypothesis Revisited. Nat. Rev. Cancer 2003, 3, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The Fundamental Role of Epigenetic Events in Cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef]
- Nombela, P.; Miguel-López, B.; Blanco, S. The Role of M6A, M5C and Ψ RNA Modifications in Cancer: Novel Therapeutic Opportunities. Mol. Cancer 2021, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M. The Emerging Role of LncRNAs in Cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The Biogenesis, Biology and Characterization of Circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Garcia-Recio, S.; Hinoue, T.; Wheeler, G.L.; Kelly, B.J.; Garrido-Castro, A.C.; Pascual, T.; De Cubas, A.A.; Xia, Y.; Felsheim, B.M.; McClure, M.B.; et al. Multiomics in Primary and Metastatic Breast Tumors from the AURORA US Network Finds Microenvironment and Epigenetic Drivers of Metastasis. Nat. Cancer 2023, 4, 128–147. [Google Scholar] [CrossRef]
- Robertson, K.D. DNA Methylation, Methyltransferases, and Cancer. Oncogene 2001, 20, 3139–3155. [Google Scholar] [CrossRef]
- Feinberg, A.P.; Vogelstein, B. Hypomethylation Distinguishes Genes of Some Human Cancers from Their Normal Counterparts. Nature 1983, 301, 89–92. [Google Scholar] [CrossRef]
- Yang, J.; Xu, J.; Wang, W.; Zhang, B.; Yu, X.; Shi, S. Epigenetic Regulation in the Tumor Microenvironment: Molecular Mechanisms and Therapeutic Targets. Signal Transduct. Target. Ther. 2023, 8, 210. [Google Scholar] [CrossRef]
- Caldeira, J.R.F.; Prando, E.C.; Quevedo, F.C.; Neto, F.A.M.; Rainho, C.A.; Rogatto, S.R. CDH1 Promoter Hypermethylation and E-Cadherin Protein Expression in Infiltrating Breast Cancer. BMC Cancer 2006, 6, 48. [Google Scholar] [CrossRef]
- Jahangiri, R.; Jamialahmadi, K.; Gharib, M.; Emami Razavi, A.; Mosaffa, F. Expression and Clinicopathological Significance of DNA Methyltransferase 1, 3A and 3B in Tamoxifen-Treated Breast Cancer Patients. Gene 2019, 685, 24–31. [Google Scholar] [CrossRef]
- Graff, J.R.; Herman, J.G.; Lapidus, R.G.; Chopra, H.; Xu, R.; Jarrard, D.F.; Isaacs, W.B.; Pitha, P.M.; Davidson, N.E.; Baylin, S.B. E-Cadherin Expression Is Silenced by DNA Hypermethylation in Human Breast and Prostate Carcinomas. Cancer Res. 1995, 55, 5195–5199. [Google Scholar]
- Tzelepi, V.; Logotheti, S.; Efstathiou, E.; Troncoso, P.; Aparicio, A.; Sakellakis, M.; Hoang, A.; Perimenis, P.; Melachrinou, M.; Logothetis, C.; et al. Epigenetics and Prostate Cancer: Defining the Timing of DNA Methyltransferase Deregulation during Prostate Cancer Progression. Pathology 2020, 52, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.-K.; Kim, H.; Park, H.-J.; Shim, Y.-H.; Choi, J.; Park, C.; Park, Y.N. DNA Methyltransferase Expression and DNA Methylation in Human Hepatocellular Carcinoma and Their Clinicopathological Correlation. Int. J. Mol. Med. 2007, 20, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yin, D.; Zhang, Y.; Yu, L.; Li, X.-D.; Zhou, Z.-J.; Zhou, S.-L.; Gao, D.-M.; Hu, J.; Jin, C.; et al. MicroRNA-29a Induces Loss of 5-Hydroxymethylcytosine and Promotes Metastasis of Hepatocellular Carcinoma through a TET-SOCS1-MMP9 Signaling Axis. Cell Death Dis. 2017, 8, e2906. [Google Scholar] [CrossRef] [PubMed]
- Guan, Z.; Zhang, J.; Song, S.; Dai, D. Promoter Methylation and Expression of TIMP3 Gene in Gastric Cancer. Diagn. Pathol. 2013, 8, 110. [Google Scholar] [CrossRef]
- Cui, H.; Wang, L.; Gong, P.; Zhao, C.; Zhang, S.; Zhang, K.; Zhou, R.; Zhao, Z.; Fan, H. Deregulation between MiR-29b/c and DNMT3A Is Associated with Epigenetic Silencing of the CDH1 Gene, Affecting Cell Migration and Invasion in Gastric Cancer. PLoS ONE 2015, 10, e0123926. [Google Scholar] [CrossRef]
- Yang, J.; Shen, Y.; Liu, B.; Tong, Y. Promoter Methylation of BRMS1 Correlates with Smoking History and Poor Survival in Non-Small Cell Lung Cancer Patients. Lung Cancer 2011, 74, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Maitra, A.; Fukushima, N.; van Heek, N.T.; Matsubayashi, H.; Iacobuzio-Donahue, C.A.; Rosty, C.; Goggins, M. Frequent Hypomethylation of Multiple Genes Overexpressed in Pancreatic Ductal Adenocarcinoma. Cancer Res. 2003, 63, 4158–4166. [Google Scholar]
- Zhan, Q.; Fang, Y.; Deng, X.; Chen, H.; Jin, J.; Lu, X.; Peng, C.; Li, H.; Shen, B. The Interplay Between MiR-148a and DNMT1 Might Be Exploited for Pancreatic Cancer Therapy. Cancer Investig. 2015, 33, 267–275. [Google Scholar] [CrossRef]
- Hong, L.; Sun, G.; Peng, L.; Tu, Y.; Wan, Z.; Xiong, H.; Li, Y.; Xiao, W. The Interaction between MiR 148a and DNMT1 Suppresses Cell Migration and Invasion by Reactivating Tumor Suppressor Genes in Pancreatic Cancer. Oncol. Rep. 2018, 40, 2916–2925. [Google Scholar] [CrossRef]
- Bai, X.; Song, Z.; Fu, Y.; Yu, Z.; Zhao, L.; Zhao, H.; Yao, W.; Huang, D.; Mi, X.; Wang, E.; et al. Clinicopathological Significance and Prognostic Value of DNA Methyltransferase 1, 3a, and 3b Expressions in Sporadic Epithelial Ovarian Cancer. PLoS ONE 2012, 7, e40024. [Google Scholar] [CrossRef]
- Graff, J.R.; Gabrielson, E.; Fujii, H.; Baylin, S.B.; Herman, J.G. Methylation Patterns of the E-Cadherin 5′ CpG Island Are Unstable and Reflect the Dynamic, Heterogeneous Loss of E-Cadherin Expression during Metastatic Progression. J. Biol. Chem. 2000, 275, 2727–2732. [Google Scholar] [CrossRef]
- Kanai, Y.; Ushijima, S.; Hui, A.M.; Ochiai, A.; Tsuda, H.; Sakamoto, M.; Hirohashi, S. The E-Cadherin Gene Is Silenced by CpG Methylation in Human Hepatocellular Carcinomas. Int. J. Cancer 1997, 71, 355–359. [Google Scholar] [CrossRef]
- Mensah, I.K.; Norvil, A.B.; AlAbdi, L.; McGovern, S.; Petell, C.J.; He, M.; Gowher, H. Misregulation of the Expression and Activity of DNA Methyltransferases in Cancer. NAR Cancer 2021, 3, zcab045. [Google Scholar] [CrossRef]
- Jang, H.S.; Shah, N.M.; Du, A.Y.; Dailey, Z.Z.; Pehrsson, E.C.; Godoy, P.M.; Zhang, D.; Li, D.; Xing, X.; Kim, S.; et al. Transposable Elements Drive Widespread Expression of Oncogenes in Human Cancers. Nat. Genet. 2019, 51, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Field, M.G.; Durante, M.A.; Decatur, C.L.; Tarlan, B.; Oelschlager, K.M.; Stone, J.F.; Kuznetsov, J.; Bowcock, A.M.; Kurtenbach, S.; Harbour, J.W. Epigenetic Reprogramming and Aberrant Expression of PRAME Are Associated with Increased Metastatic Risk in Class 1 and Class 2 Uveal Melanomas. Oncotarget 2016, 7, 59209–59219. [Google Scholar] [CrossRef]
- Nakamura, N.; Takenaga, K. Hypomethylation of the Metastasis-Associated S100A4 Gene Correlates with Gene Activation in Human Colon Adenocarcinoma Cell Lines. Clin. Exp. Metastasis 1998, 16, 471–479. [Google Scholar] [CrossRef]
- Kim, J.; Bretz, C.L.; Lee, S. Epigenetic Instability of Imprinted Genes in Human Cancers. Nucleic Acids Res. 2015, 43, 10689–10699. [Google Scholar] [CrossRef] [PubMed]
- Lodygin, D.; Tarasov, V.; Epanchintsev, A.; Berking, C.; Knyazeva, T.; Körner, H.; Knyazev, P.; Diebold, J.; Hermeking, H. Inactivation of MiR-34a by Aberrant CpG Methylation in Multiple Types of Cancer. Cell Cycle 2008, 7, 2591–2600. [Google Scholar] [CrossRef]
- He, L.; He, X.; Lim, L.P.; de Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A MicroRNA Component of the P53 Tumour Suppressor Network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef]
- Lujambio, A.; Calin, G.A.; Villanueva, A.; Ropero, S.; Sánchez-Céspedes, M.; Blanco, D.; Montuenga, L.M.; Rossi, S.; Nicoloso, M.S.; Faller, W.J.; et al. A MicroRNA DNA Methylation Signature for Human Cancer Metastasis. Proc. Natl. Acad. Sci. USA 2008, 105, 13556–13561. [Google Scholar] [CrossRef]
- Kohli, R.M.; Zhang, Y. TET Enzymes, TDG and the Dynamics of DNA Demethylation. Nature 2013, 502, 472–479. [Google Scholar] [CrossRef]
- An, J.; González-Avalos, E.; Chawla, A.; Jeong, M.; López-Moyado, I.F.; Li, W.; Goodell, M.A.; Chavez, L.; Ko, M.; Rao, A. Acute Loss of TET Function Results in Aggressive Myeloid Cancer in Mice. Nat. Commun. 2015, 6, 10071. [Google Scholar] [CrossRef] [PubMed]
- Pastor, W.A.; Aravind, L.; Rao, A. TETonic Shift: Biological Roles of TET Proteins in DNA Demethylation and Transcription. Nat. Rev. Mol. Cell Biol. 2013, 14, 341–356. [Google Scholar] [CrossRef] [PubMed]
- Fackler, M.J.; Lopez Bujanda, Z.; Umbricht, C.; Teo, W.W.; Cho, S.; Zhang, Z.; Visvanathan, K.; Jeter, S.; Argani, P.; Wang, C.; et al. Novel Methylated Biomarkers and a Robust Assay to Detect Circulating Tumor DNA in Metastatic Breast Cancer. Cancer Res. 2014, 74, 2160–2170. [Google Scholar] [CrossRef]
- Li, L.; Sun, Y. Circulating Tumor DNA Methylation Detection as Biomarker and Its Application in Tumor Liquid Biopsy: Advances and Challenges. MedComm 2024, 5, e766. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Rong, H.; Xu, J.; Cao, R.; Li, S.; Gao, Y.; Cheng, B.; Zhou, T. DNA Methylation: An Important Biomarker and Therapeutic Target for Gastric Cancer. Front. Genet. 2022, 13, 823905. [Google Scholar] [CrossRef]
- Zuccato, J.A.; Mamatjan, Y.; Nassiri, F.; Ajisebutu, A.; Liu, J.C.; Muazzam, A.; Singh, O.; Zhang, W.; Voisin, M.; Mirhadi, S.; et al. Prediction of Brain Metastasis Development with DNA Methylation Signatures. Nat. Med. 2025, 31, 116–125. [Google Scholar] [CrossRef]
- Pfeifer, G.P.; Kadam, S.; Jin, S.-G. 5-Hydroxymethylcytosine and Its Potential Roles in Development and Cancer. Epigenet. Chromatin 2013, 6, 10. [Google Scholar] [CrossRef]
- Cheishvili, D.; Boureau, L.; Szyf, M. DNA Demethylation and Invasive Cancer: Implications for Therapeutics. Br. J. Pharmacol. 2015, 172, 2705–2715. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Issa, J.-P.J.; Kropf, P. DNA Hypomethylating Drugs in Cancer Therapy. Cold Spring Harb. Perspect. Med. 2017, 7, a026948. [Google Scholar] [CrossRef]
- Dong, B.; Qiu, Z.; Wu, Y. Tackle Epithelial-Mesenchymal Transition with Epigenetic Drugs in Cancer. Front. Pharmacol. 2020, 11, 596239. [Google Scholar] [CrossRef]
- Hu, C.; Liu, X.; Zeng, Y.; Liu, J.; Wu, F. DNA Methyltransferase Inhibitors Combination Therapy for the Treatment of Solid Tumor: Mechanism and Clinical Application. Clin. Epigenet. 2021, 13, 166. [Google Scholar] [CrossRef]
- Morita, S.; Noguchi, H.; Horii, T.; Nakabayashi, K.; Kimura, M.; Okamura, K.; Sakai, A.; Nakashima, H.; Hata, K.; Nakashima, K.; et al. Targeted DNA Demethylation in Vivo Using DCas9-Peptide Repeat and ScFv-TET1 Catalytic Domain Fusions. Nat. Biotechnol. 2016, 34, 1060–1065. [Google Scholar] [CrossRef]
- Vojta, A.; Dobrinić, P.; Tadić, V.; Bočkor, L.; Korać, P.; Julg, B.; Klasić, M.; Zoldoš, V. Repurposing the CRISPR-Cas9 System for Targeted DNA Methylation. Nucleic Acids Res. 2016, 44, 5615–5628. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Li, L.; Wang, Y.; Wu, P.; Hou, X.; Ouyang, J.; Fan, C.; Li, Z.; Wang, F.; Guo, C.; et al. RNA Modifications in Cancer. Br. J. Cancer 2023, 129, 204–221. [Google Scholar] [CrossRef]
- Dang, Q.; Shao, B.; Zhou, Q.; Chen, C.; Guo, Y.; Wang, G.; Liu, J.; Kan, Q.; Yuan, W.; Sun, Z. RNA N 6-Methyladenosine in Cancer Metastasis: Roles, Mechanisms, and Applications. Front. Oncol. 2021, 11, 681781. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, F.; Chen, W.; Miao, H.; Liang, H.; Liao, Z.; Zhang, Z.; Zhang, B. The Role of RNA M5C Modification in Cancer Metastasis. Int. J. Biol. Sci. 2021, 17, 3369–3380. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the Human and Mouse M6A RNA Methylomes Revealed by M6A-Seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Ćorović, M.; Hoch-Kraft, P.; Zhou, Y.; Hallstein, S.; König, J.; Zarnack, K. M6A in the Coding Sequence: Linking Deposition, Translation, and Decay. Trends Genet. 2025, 41, 963–973. [Google Scholar] [CrossRef]
- Fang, Z.; Mei, W.; Qu, C.; Lu, J.; Shang, L.; Cao, F.; Li, F. Role of M6A Writers, Erasers and Readers in Cancer. Exp. Hematol. Oncol. 2022, 11, 45. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-Methyladenosine by IGF2BP Proteins Enhances MRNA Stability and Translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Deng, X.; Qing, Y.; Horne, D.; Huang, H.; Chen, J. The Roles and Implications of RNA M6A Modification in Cancer. Nat. Rev. Clin. Oncol. 2023, 20, 507–526. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef]
- Chen, M.; Wei, L.; Law, C.-T.; Tsang, F.H.-C.; Shen, J.; Cheng, C.L.-H.; Tsang, L.-H.; Ho, D.W.-H.; Chiu, D.K.-C.; Lee, J.M.-F.; et al. RNA N6-Methyladenosine Methyltransferase-like 3 Promotes Liver Cancer Progression through YTHDF2-Dependent Posttranscriptional Silencing of SOCS2. Hepatology 2018, 67, 2254–2270. [Google Scholar] [CrossRef]
- Taketo, K.; Konno, M.; Asai, A.; Koseki, J.; Toratani, M.; Satoh, T.; Doki, Y.; Mori, M.; Ishii, H.; Ogawa, K. The Epitranscriptome M6A Writer METTL3 Promotes Chemo- and Radioresistance in Pancreatic Cancer Cells. Int. J. Oncol. 2018, 52, 621–629. [Google Scholar] [CrossRef]
- Zhang, L.; Mao, Z.; Yin, K.; Wang, S. Review of METTL3 in Colorectal Cancer: From Mechanisms to the Therapeutic Potential. Int. J. Biol. Macromol. 2024, 277, 134212. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Li, J.; Chen, R.; Gu, Q.; Yang, P.; Qian, W.; Ji, D.; Wang, Q.; Zhang, Z.; Tang, J.; et al. Upregulated METTL3 Promotes Metastasis of Colorectal Cancer via MiR-1246/SPRED2/MAPK Signaling Pathway. J. Exp. Clin. Cancer Res. 2019, 38, 393. [Google Scholar] [CrossRef] [PubMed]
- Lochhead, P.; Imamura, Y.; Morikawa, T.; Kuchiba, A.; Yamauchi, M.; Liao, X.; Qian, Z.R.; Nishihara, R.; Wu, K.; Meyerhardt, J.A.; et al. Insulin-like Growth Factor 2 Messenger RNA Binding Protein 3 (IGF2BP3) Is a Marker of Unfavourable Prognosis in Colorectal Cancer. Eur. J. Cancer 2012, 48, 3405–3413. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, M.; Xu, X.; Zeng, K.; Liu, X.; Pan, B.; Li, C.; Sun, L.; Qin, J.; Xu, T.; et al. METTL14-Mediated N6-Methyladenosine Modification of SOX4 MRNA Inhibits Tumor Metastasis in Colorectal Cancer. Mol. Cancer 2020, 19, 106. [Google Scholar] [CrossRef]
- Niu, Y.; Lin, Z.; Wan, A.; Chen, H.; Liang, H.; Sun, L.; Wang, Y.; Li, X.; Xiong, X.-F.; Wei, B.; et al. RNA N6-Methyladenosine Demethylase FTO Promotes Breast Tumor Progression through Inhibiting BNIP3. Mol. Cancer 2019, 18, 46. [Google Scholar] [CrossRef]
- Chang, G.; Shi, L.; Ye, Y.; Shi, H.; Zeng, L.; Tiwary, S.; Huse, J.T.; Huo, L.; Ma, L.; Ma, Y.; et al. YTHDF3 Induces the Translation of M6A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 2020, 38, 857–871.e7. [Google Scholar] [CrossRef]
- Woodcock, C.L.; Alsaleem, M.; Toss, M.S.; Lothion-Roy, J.; Harris, A.E.; Jeyapalan, J.N.; Blatt, N.; Rizvanov, A.A.; Miftakhova, R.R.; Kariri, Y.A.; et al. The Role of the ALKBH5 RNA Demethylase in Invasive Breast Cancer. Discov. Oncol. 2024, 15, 343. [Google Scholar] [CrossRef]
- Shi, Y.; Zheng, C.; Jin, Y.; Bao, B.; Wang, D.; Hou, K.; Feng, J.; Tang, S.; Qu, X.; Liu, Y.; et al. Reduced Expression of METTL3 Promotes Metastasis of Triple-Negative Breast Cancer by M6A Methylation-Mediated COL3A1 Up-Regulation. Front. Oncol. 2020, 10, 1126. [Google Scholar] [CrossRef]
- Ma, J.-Z.; Yang, F.; Zhou, C.-C.; Liu, F.; Yuan, J.-H.; Wang, F.; Wang, T.-T.; Xu, Q.-G.; Zhou, W.-P.; Sun, S.-H. METTL14 Suppresses the Metastatic Potential of Hepatocellular Carcinoma by Modulating N6 -Methyladenosine-Dependent Primary MicroRNA Processing. Hepatology 2017, 65, 529–543. [Google Scholar] [CrossRef]
- Pi, J.; Wang, W.; Ji, M.; Wang, X.; Wei, X.; Jin, J.; Liu, T.; Qiang, J.; Qi, Z.; Li, F.; et al. YTHDF1 Promotes Gastric Carcinogenesis by Controlling Translation of FZD7. Cancer Res. 2021, 81, 2651–2665. [Google Scholar] [CrossRef]
- Du, M.; Peng, Y.; Li, Y.; Sun, W.; Zhu, H.; Wu, J.; Zong, D.; Wu, L.; He, X. MYC-Activated RNA N6-Methyladenosine Reader IGF2BP3 Promotes Cell Proliferation and Metastasis in Nasopharyngeal Carcinoma. Cell Death Discov. 2022, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Huang, R.; Xia, T.; Wang, C.; Xiao, X.; Lu, S.; Chen, X.; Ouyang, Y.; Deng, X.; Miao, J.; et al. The M6A Reader IGF2BP3 Preserves NOTCH3 MRNA Stability to Sustain Notch3 Signaling and Promote Tumor Metastasis in Nasopharyngeal Carcinoma. Oncogene 2023, 42, 3564–3574. [Google Scholar] [CrossRef]
- Li, K.; Guo, J.; Ming, Y.; Chen, S.; Zhang, T.; Ma, H.; Fu, X.; Wang, J.; Liu, W.; Peng, Y. A Circular RNA Activated by TGFβ Promotes Tumor Metastasis through Enhancing IGF2BP3-Mediated PDPN MRNA Stability. Nat. Commun. 2023, 14, 6876. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Gao, G.; Xia, W.-W.; Wang, J.-B. METTL3 Promotes the Growth and Metastasis of Pancreatic Cancer by Regulating the M6A Modification and Stability of E2F5. Cell. Signal. 2022, 99, 110440. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, J.; Zhao, Y.; He, R.; Xu, X.; Guo, X.; Li, X.; Xu, S.; Miao, J.; Guo, J.; et al. Upregulation of METTL14 Mediates the Elevation of PERP MRNA N6 Adenosine Methylation Promoting the Growth and Metastasis of Pancreatic Cancer. Mol. Cancer 2020, 19, 130. [Google Scholar] [CrossRef]
- Schaeffer, D.F.; Owen, D.R.; Lim, H.J.; Buczkowski, A.K.; Chung, S.W.; Scudamore, C.H.; Huntsman, D.G.; Ng, S.S.W.; Owen, D.A. Insulin-like Growth Factor 2 MRNA Binding Protein 3 (IGF2BP3) Overexpression in Pancreatic Ductal Adenocarcinoma Correlates with Poor Survival. BMC Cancer 2010, 10, 59. [Google Scholar] [CrossRef]
- Liu, L.; Wu, Y.; Li, Q.; Liang, J.; He, Q.; Zhao, L.; Chen, J.; Cheng, M.; Huang, Z.; Ren, H.; et al. METTL3 Promotes Tumorigenesis and Metastasis through BMI1 M6A Methylation in Oral Squamous Cell Carcinoma. Mol. Ther. 2020, 28, 2177–2190. [Google Scholar] [CrossRef]
- Yang, S.; Wei, J.; Cui, Y.-H.; Park, G.; Shah, P.; Deng, Y.; Aplin, A.E.; Lu, Z.; Hwang, S.; He, C.; et al. M6A MRNA Demethylase FTO Regulates Melanoma Tumorigenicity and Response to Anti-PD-1 Blockade. Nat. Commun. 2019, 10, 2782. [Google Scholar] [CrossRef]
- Li, J.; Xie, H.; Ying, Y.; Chen, H.; Yan, H.; He, L.; Xu, M.; Xu, X.; Liang, Z.; Liu, B.; et al. YTHDF2 Mediates the MRNA Degradation of the Tumor Suppressors to Induce AKT Phosphorylation in N6-Methyladenosine-Dependent Way in Prostate Cancer. Mol. Cancer 2020, 19, 152. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, S.; He, C.; Xue, P.; Zhang, L.; He, Z.; Zang, L.; Feng, B.; Sun, J.; Zheng, M. METTL14 Suppresses Proliferation and Metastasis of Colorectal Cancer by Down-Regulating Oncogenic Long Non-Coding RNA XIST. Mol. Cancer 2020, 19, 46. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-Methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Hu, J.; Qiu, D.; Yu, A.; Hu, J.; Deng, H.; Li, H.; Yi, Z.; Chen, J.; Zu, X. YTHDF1 Is a Potential Pan-Cancer Biomarker for Prognosis and Immunotherapy. Front. Oncol. 2021, 11, 607224. [Google Scholar] [CrossRef]
- Hou, J.; Zhang, H.; Liu, J.; Zhao, Z.; Wang, J.; Lu, Z.; Hu, B.; Zhou, J.; Zhao, Z.; Feng, M.; et al. YTHDF2 Reduction Fuels Inflammation and Vascular Abnormalization in Hepatocellular Carcinoma. Mol. Cancer 2019, 18, 163. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread Occurrence of 5-Methylcytosine in Human Coding and Non-Coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef]
- Edelheit, S.; Schwartz, S.; Mumbach, M.R.; Wurtzel, O.; Sorek, R. Transcriptome-Wide Mapping of 5-Methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals M5C within Archaeal MRNAs. PLoS Genet. 2013, 9, e1003602. [Google Scholar] [CrossRef] [PubMed]
- Khoddami, V.; Cairns, B.R. Identification of Direct Targets and Modified Bases of RNA Cytosine Methyltransferases. Nat. Biotechnol. 2013, 31, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yuan, Y.; Zhou, F.; Huang, X.; Li, L.; Pu, J.; Zeng, Y.; Jiang, X. RNA M5C Modification: From Physiology to Pathology and Its Biological Significance. Front. Immunol. 2025, 16, 1599305. [Google Scholar] [CrossRef]
- Tuorto, F.; Liebers, R.; Musch, T.; Schaefer, M.; Hofmann, S.; Kellner, S.; Frye, M.; Helm, M.; Stoecklin, G.; Lyko, F. RNA Cytosine Methylation by Dnmt2 and NSun2 Promotes TRNA Stability and Protein Synthesis. Nat. Struct. Mol. Biol. 2012, 19, 900–905. [Google Scholar] [CrossRef]
- Fu, L.; Guerrero, C.R.; Zhong, N.; Amato, N.J.; Liu, Y.; Liu, S.; Cai, Q.; Ji, D.; Jin, S.-G.; Niedernhofer, L.J.; et al. Tet-Mediated Formation of 5-Hydroxymethylcytosine in RNA. J. Am. Chem. Soc. 2014, 136, 11582–11585. [Google Scholar] [CrossRef]
- Kawarada, L.; Suzuki, T.; Ohira, T.; Hirata, S.; Miyauchi, K.; Suzuki, T. ALKBH1 Is an RNA Dioxygenase Responsible for Cytoplasmic and Mitochondrial TRNA Modifications. Nucleic Acids Res. 2017, 45, 7401–7415. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, Y.; Sun, B.-F.; Chen, Y.-S.; Xu, J.-W.; Lai, W.-Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-Methylcytosine Promotes MRNA Export—NSUN2 as the Methyltransferase and ALYREF as an M5C Reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef]
- Chen, X.; Li, A.; Sun, B.-F.; Yang, Y.; Han, Y.-N.; Yuan, X.; Chen, R.-X.; Wei, W.-S.; Liu, Y.; Gao, C.-C.; et al. 5-Methylcytosine Promotes Pathogenesis of Bladder Cancer through Stabilizing MRNAs. Nat. Cell Biol. 2019, 21, 978–990. [Google Scholar] [CrossRef]
- Zhao, T.; Zhang, Z.; Chen, Z.; Xu, G.; Wang, Y.; Wang, F. Biological Functions of 5-Methylcytosine RNA-Binding Proteins and Their Potential Mechanisms in Human Cancers. Front. Oncol. 2025, 15, 1534948. [Google Scholar] [CrossRef]
- Hussain, S.; Sajini, A.A.; Blanco, S.; Dietmann, S.; Lombard, P.; Sugimoto, Y.; Paramor, M.; Gleeson, J.G.; Odom, D.T.; Ule, J.; et al. NSun2-Mediated Cytosine-5 Methylation of Vault Noncoding RNA Determines Its Processing into Regulatory Small RNAs. Cell Rep. 2013, 4, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Chen, R.-X.; Deng, M.-H.; Wei, W.-S.; Zhou, Z.-H.; Ning, K.; Li, Y.-H.; Li, X.-D.; Ye, Y.-L.; Wen, J.-H.; et al. M5C-Dependent Cross-Regulation between Nuclear Reader ALYREF and Writer NSUN2 Promotes Urothelial Bladder Cancer Malignancy through Facilitating RABL6/TK1 MRNAs Splicing and Stabilization. Cell Death Dis. 2023, 14, 139. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Wu, G.; Ye, Y.; Zhang, J.; Zeng, L.; Huang, X.; Zheng, Y.; Bai, R.; Zhuang, L.; Li, M.; et al. NSUN2-Mediated RNA 5-Methylcytosine Promotes Esophageal Squamous Cell Carcinoma Progression via LIN28B-Dependent GRB2 MRNA Stabilization. Oncogene 2021, 40, 5814–5828. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Luo, M.; Zhou, C.; Shi, X.; Yang, W.; Lu, Z.; Chen, Z.; Sun, N.; He, J. Novel Long Noncoding RNA NMR Promotes Tumor Progression via NSUN2 and BPTF in Esophageal Squamous Cell Carcinoma. Cancer Lett. (Lond) 2018, 430, 57–66. [Google Scholar] [CrossRef]
- Su, M.; Xiao, Y.; Ma, J.; Cao, D.; Zhou, Y.; Wang, H.; Liao, Q.; Wang, W. Long Non-Coding RNAs in Esophageal Cancer: Molecular Mechanisms, Functions, and Potential Applications. J. Hematol. Oncol. 2018, 11, 118. [Google Scholar] [CrossRef]
- Zou, S.; Huang, Y.; Yang, Z.; Zhang, J.; Meng, M.; Zhang, Y.; Feng, J.; Sun, R.; Li, W.; Wang, W.; et al. NSUN2 Promotes Colorectal Cancer Progression by Enhancing SKIL MRNA Stabilization. Clin. Transl. Med. 2024, 14, e1621. [Google Scholar] [CrossRef]
- Zhu, W.; Wan, F.; Xu, W.; Liu, Z.; Wang, J.; Zhang, H.; Huang, S.; Ye, D. Positive Epigenetic Regulation Loop between AR and NSUN2 Promotes Prostate Cancer Progression. Clin. Transl. Med. 2022, 12, e1028. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, L.; Li, J.; Chen, Y.; Wang, Y.; Zhang, Y.; Dong, Z.; Xue, W.; Sun, R.; Cui, G. NSUN2 Stimulates Tumor Progression via Enhancing TIAM2 MRNA Stability in Pancreatic Cancer. Cell Death Discov. 2023, 9, 219. [Google Scholar] [CrossRef]
- Yan, J.; Liu, J.; Huang, Z.; Huang, W.; Lv, J. FOXC2-AS1 Stabilizes FOXC2 MRNA via Association with NSUN2 in Gastric Cancer Cells. Hum. Cell 2021, 34, 1755–1764. [Google Scholar] [CrossRef]
- Fang, L.; Huang, H.; Lv, J.; Chen, Z.; Lu, C.; Jiang, T.; Xu, P.; Li, Y.; Wang, S.; Li, B.; et al. m5C-methylated lncRNA NR_033928 promotes gastric cancer proliferation by stabilizing GLS mRNA to promote glutamine metabolism reprogramming. Cell Death Dis. 2023, 14, 520. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Xu, P.; Lv, J.; Ge, H.; Yan, Z.; Huang, S.; Li, B.; Xu, H.; Yang, L.; Xu, Z.; et al. Peritoneal High-Fat Environment Promotes Peritoneal Metastasis of Gastric Cancer Cells through Activation of NSUN2-Mediated ORAI2 M5C Modification. Oncogene 2023, 42, 1980–1993. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, H.; Liu, X.; Nguyen, H.C.B.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous TRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef]
- Yi, J.; Gao, R.; Chen, Y.; Yang, Z.; Han, P.; Zhang, H.; Dou, Y.; Liu, W.; Wang, W.; Du, G.; et al. Overexpression of NSUN2 by DNA Hypomethylation Is Associated with Metastatic Progression in Human Breast Cancer. Oncotarget 2017, 8, 20751–20765. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, Y.; Zhang, J.; Zhang, X. NSun2 Promotes Cell Migration through Methylating Autotaxin MRNA. J. Biol. Chem. 2020, 295, 18134–18147. [Google Scholar] [CrossRef]
- Zhang, Y.-Y.; Li, C.-T.; Zhou, Y.-J.; Li, H.; Li, J.; Xiong, Q.-P.; Zhou, W.; Huang, W.; Zhang, Q.C.; Xiang, Y.; et al. A Cohort of MRNAs Undergo High-Stoichiometry NSUN6-Mediated Site-Specific M5C Modification. Nat. Commun. 2025, 16, 6119. [Google Scholar] [CrossRef]
- Li, C.; Wang, S.; Xing, Z.; Lin, A.; Liang, K.; Song, J.; Hu, Q.; Yao, J.; Chen, Z.; Park, P.K.; et al. A ROR1-HER3-LncRNA Signalling Axis Modulates the Hippo-YAP Pathway to Regulate Bone Metastasis. Nat. Cell Biol. 2017, 19, 106–119. [Google Scholar] [CrossRef]
- Selmi, T.; Hussain, S.; Dietmann, S.; Heiß, M.; Borland, K.; Flad, S.; Carter, J.-M.; Dennison, R.; Huang, Y.-L.; Kellner, S.; et al. Sequence- and Structure-Specific Cytosine-5 MRNA Methylation by NSUN6. Nucleic Acids Res. 2021, 49, 1006–1022. [Google Scholar] [CrossRef]
- Xue, M.; Shi, Q.; Zheng, L.; Li, Q.; Yang, L.; Zhang, Y. Gene Signatures of M5C Regulators May Predict Prognoses of Patients with Head and Neck Squamous Cell Carcinoma. Am. J. Transl. Res. 2020, 12, 6841–6852. [Google Scholar] [PubMed]
- Zhou, J.; Kong, Y.S.; Vincent, K.M.; Dieters-Castator, D.; Bukhari, A.B.; Glubrecht, D.; Liu, R.-Z.; Quilty, D.; Findlay, S.D.; Huang, X.; et al. RNA Cytosine Methyltransferase NSUN5 Promotes Protein Synthesis and Tumorigenic Phenotypes in Glioblastoma. Mol. Oncol. 2023, 17, 1763–1783. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.-L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of TRNAAsp by the DNA Methyltransferase Homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA Methylation by Dnmt2 Protects Transfer RNAs against Stress-Induced Cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef]
- Huang, S.; Shi, D.; Dai, S.; Jiang, X.; Wang, R.; Yang, M.; Chen, B.; Chen, X.; Kong, L.; He, L.; et al. RNF31 Induces Paclitaxel Resistance by Sustaining ALYREF Cytoplasmic-Nuclear Shuttling in Human Triple-Negative Breast Cancer. Clin. Transl. Med. 2025, 15, e70203. [Google Scholar] [CrossRef]
- Wang, J.-Z.; Zhu, W.; Han, J.; Yang, X.; Zhou, R.; Lu, H.-C.; Yu, H.; Yuan, W.-B.; Li, P.-C.; Tao, J.; et al. The Role of the HIF-1α/ALYREF/PKM2 Axis in Glycolysis and Tumorigenesis of Bladder Cancer. Cancer Commun. (Lond) 2021, 41, 560–575. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, L.; Han, X.; Yang, W.-L.; Zhang, M.; Ma, H.-L.; Sun, B.-F.; Li, A.; Xia, J.; Chen, J.; et al. RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal MRNA Decay. Mol. Cell 2019, 75, 1188–1202.e11. [Google Scholar] [CrossRef]
- Meng, H.; Miao, H.; Zhang, Y.; Chen, T.; Yuan, L.; Wan, Y.; Jiang, Y.; Zhang, L.; Cheng, W. YBX1 Promotes Homologous Recombination and Resistance to Platinum-Induced Stress in Ovarian Cancer by Recognizing M5C Modification. Cancer Lett. 2024, 597, 217064. [Google Scholar] [CrossRef]
- Zheng, H.; Zhu, M.; Li, W.; Zhou, Z.; Wan, X. M5 C and M6 A Modification of Long Noncoding NKILA Accelerates Cholangiocarcinoma Progression via the MiR-582-3p-YAP1 Axis. Liver Int. 2022, 42, 1144–1157. [Google Scholar] [CrossRef]
- Liu, B.; Sun, L.; Liu, Q.; Gong, C.; Yao, Y.; Lv, X.; Lin, L.; Yao, H.; Su, F.; Li, D.; et al. A Cytoplasmic NF-ΚB Interacting Long Noncoding RNA Blocks IκB Phosphorylation and Suppresses Breast Cancer Metastasis. Cancer Cell 2015, 27, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Yao, J.; Fu, J.; Huang, Q.; Luo, Y.; You, Y.; Zhang, W.; Zhong, Q.; Xia, T.; Xia, L. ALYREF Promotes the Metastasis of Nasopharyngeal Carcinoma by Increasing the Stability of NOTCH1 MRNA. Cell Death Dis. 2024, 15, 578. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Gu, X.; Liu, Y.; Xu, L.; He, Y.; Xue, C. NSUN2 Regulates Wnt Signaling Pathway Depending on the M5C RNA Modification to Promote the Progression of Hepatocellular Carcinoma. Oncogene 2024, 43, 3469–3482. [Google Scholar] [CrossRef]
- El Hage, K.; Babault, N.; Maciejak, O.; Desforges, B.; Craveur, P.; Steiner, E.; Rengifo-Gonzalez, J.C.; Henrie, H.; Clement, M.-J.; Joshi, V.; et al. Targeting RNA:Protein Interactions with an Integrative Approach Leads to the Identification of Potent YBX1 Inhibitors. Elife 2023, 12, e80387. [Google Scholar] [CrossRef]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian MicroRNAs Predominantly Act to Decrease Target MRNA Levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates That Thousands of Human Genes Are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Gregory, R.I. MicroRNA Biogenesis Pathways in Cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-Methyladenosine Marks Primary MicroRNAs for Processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhuang, Y.; Zhang, J.; Chen, M.; Wu, S. METTL14 Inhibits Hepatocellular Carcinoma Metastasis Through Regulating EGFR/PI3K/AKT Signaling Pathway in an M6A-Dependent Manner. Cancer Manag. Res. 2020, 12, 13173–13184. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, X.; Yang, F.; Shang, J.; Yang, Q. METTL3-Mediated M6A Modification of Pri-MiR-93 Promotes Hepatocellular Carcinoma Progression via CDKN1A Suppression. Cancer Genet. 2025, 296–297, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, B.; Jin, N.; Lv, J.; Zhou, H. MiRNA-1293 Promotes Hepatocellular Carcinoma Cell Proliferation and Invasion by METTL3-Mediated M6 A Modification of Pri-MiRNA-1293. Appl. Biochem. Biotechnol. 2025, 197, 4409–4422. [Google Scholar] [CrossRef] [PubMed]
- Pandolfini, L.; Barbieri, I.; Bannister, A.J.; Hendrick, A.; Andrews, B.; Webster, N.; Murat, P.; Mach, P.; Brandi, R.; Robson, S.C.; et al. METTL1 Promotes Let-7 MicroRNA Processing via M7G Methylation. Mol. Cell 2019, 74, 1278–1290.e9. [Google Scholar] [CrossRef]
- Nam, Y.; Chen, C.; Gregory, R.I.; Chou, J.J.; Sliz, P. Molecular Basis for Interaction of Let-7 MicroRNAs with Lin28. Cell 2011, 147, 1080–1091. [Google Scholar] [CrossRef]
- Heo, I.; Joo, C.; Kim, Y.-K.; Ha, M.; Yoon, M.-J.; Cho, J.; Yeom, K.-H.; Han, J.; Kim, V.N. TUT4 in Concert with Lin28 Suppresses MicroRNA Biogenesis through Pre-MicroRNA Uridylation. Cell 2009, 138, 696–708. [Google Scholar] [CrossRef]
- King, C.E.; Wang, L.; Winograd, R.; Madison, B.B.; Mongroo, P.S.; Johnstone, C.N.; Rustgi, A.K. LIN28B Fosters Colon Cancer Migration, Invasion and Transformation through Let-7-Dependent and -Independent Mechanisms. Oncogene 2011, 30, 4185–4193. [Google Scholar] [CrossRef]
- Kawahara, Y.; Zinshteyn, B.; Sethupathy, P.; Iizasa, H.; Hatzigeorgiou, A.G.; Nishikura, K. Redirection of Silencing Targets by Adenosine-to-Inosine Editing of MiRNAs. Science 2007, 315, 1137–1140. [Google Scholar] [CrossRef]
- Shoshan, E.; Mobley, A.K.; Braeuer, R.R.; Kamiya, T.; Huang, L.; Vasquez, M.E.; Salameh, A.; Lee, H.J.; Kim, S.J.; Ivan, C.; et al. Reduced Adenosine-to-Inosine MiR-455-5p Editing Promotes Melanoma Growth and Metastasis. Nat. Cell Biol. 2015, 17, 311–321. [Google Scholar] [CrossRef]
- Nemlich, Y.; Greenberg, E.; Ortenberg, R.; Besser, M.J.; Barshack, I.; Jacob-Hirsch, J.; Jacoby, E.; Eyal, E.; Rivkin, L.; Prieto, V.G.; et al. MicroRNA-Mediated Loss of ADAR1 in Metastatic Melanoma Promotes Tumor Growth. J. Clin. Investig. 2013, 123, 2703–2718. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, Y.; Tay, F.C.; Lam, D.H.; Sandanaraj, E.; Tang, C.; Ang, B.-T.; Wang, S. Attenuated Adenosine-to-Inosine Editing of MicroRNA-376a* Promotes Invasiveness of Glioblastoma Cells. J. Clin. Investig. 2012, 122, 4059–4076. [Google Scholar] [CrossRef]
- Varambally, S.; Cao, Q.; Mani, R.-S.; Shankar, S.; Wang, X.; Ateeq, B.; Laxman, B.; Cao, X.; Jing, X.; Ramnarayanan, K.; et al. Genomic Loss of MicroRNA-101 Leads to Overexpression of Histone Methyltransferase EZH2 in Cancer. Science 2008, 322, 1695–1699. [Google Scholar] [CrossRef]
- Li, J.-T.; Jia, L.-T.; Liu, N.-N.; Zhu, X.-S.; Liu, Q.-Q.; Wang, X.-L.; Yu, F.; Liu, Y.-L.; Yang, A.-G.; Gao, C.-F. MiRNA-101 Inhibits Breast Cancer Growth and Metastasis by Targeting CX Chemokine Receptor 7. Oncotarget 2015, 6, 30818–30830. [Google Scholar] [CrossRef][Green Version]
- Chen, H.; Tu, S.; Hsieh, J.-T. Down-Regulation of Human DAB2IP Gene Expression Mediated by Polycomb Ezh2 Complex and Histone Deacetylase in Prostate Cancer. J. Biol. Chem. 2005, 280, 22437–22444. [Google Scholar] [CrossRef]
- Cao, P.; Deng, Z.; Wan, M.; Huang, W.; Cramer, S.D.; Xu, J.; Lei, M.; Sui, G. MicroRNA-101 Negatively Regulates Ezh2 and Its Expression Is Modulated by Androgen Receptor and HIF-1alpha/HIF-1beta. Mol. Cancer 2010, 9, 108. [Google Scholar] [CrossRef]
- Friedman, J.M.; Liang, G.; Liu, C.-C.; Wolff, E.M.; Tsai, Y.C.; Ye, W.; Zhou, X.; Jones, P.A. The Putative Tumor Suppressor MicroRNA-101 Modulates the Cancer Epigenome by Repressing the Polycomb Group Protein EZH2. Cancer Res. 2009, 69, 2623–2629. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long Non-Coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.-T.; He, J.; Liang, Q.; Ren, L.-L.; Yan, T.-T.; Yu, T.-C.; Tang, J.-Y.; Bao, Y.-J.; Hu, Y.; Lin, Y.; et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016, 6, 784–801. [Google Scholar] [CrossRef]
- Ma, M.; Zhang, Y.; Weng, M.; Hu, Y.; Xuan, Y.; Hu, Y.; Lv, K. LncRNA GCAWKR Promotes Gastric Cancer Development by Scaffolding the Chromatin Modification Factors WDR5 and KAT2A. Mol. Ther. 2018, 26, 2658–2668. [Google Scholar] [CrossRef]
- Mehra, R.; Shi, Y.; Udager, A.M.; Prensner, J.R.; Sahu, A.; Iyer, M.K.; Siddiqui, J.; Cao, X.; Wei, J.; Jiang, H.; et al. A Novel RNA in Situ Hybridization Assay for the Long Noncoding RNA SChLAP1 Predicts Poor Clinical Outcome after Radical Prostatectomy in Clinically Localized Prostate Cancer. Neoplasia 2014, 16, 1121–1127. [Google Scholar] [CrossRef]
- Zhang, J.; Guo, S.; Piao, H.-Y.; Wang, Y.; Wu, Y.; Meng, X.-Y.; Yang, D.; Zheng, Z.-C.; Zhao, Y. ALKBH5 Promotes Invasion and Metastasis of Gastric Cancer by Decreasing Methylation of the LncRNA NEAT1. J. Physiol. Biochem. 2019, 75, 379–389. [Google Scholar] [CrossRef]
- Fu, J.-W.; Kong, Y.; Sun, X. Long Noncoding RNA NEAT1 Is an Unfavorable Prognostic Factor and Regulates Migration and Invasion in Gastric Cancer. J. Cancer Res. Clin. Oncol. 2016, 142, 1571–1579. [Google Scholar] [CrossRef]
- Chakravarty, D.; Sboner, A.; Nair, S.S.; Giannopoulou, E.; Li, R.; Hennig, S.; Mosquera, J.M.; Pauwels, J.; Park, K.; Kossai, M.; et al. The Oestrogen Receptor Alpha-Regulated LncRNA NEAT1 Is a Critical Modulator of Prostate Cancer. Nat. Commun. 2014, 5, 5383. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Wei, Y.; Zen, C.; Xiong, W.; Niu, Y.; Zhao, Y. Long Non-Coding RNA NEAT1 Promotes Bone Metastasis of Prostate Cancer through N6-Methyladenosine. Mol. Cancer 2020, 19, 171. [Google Scholar] [CrossRef] [PubMed]
- Mo, C.; Huang, B.; Zhuang, J.; Jiang, S.; Guo, S.; Mao, X. LncRNA Nuclear-Enriched Abundant Transcript 1 Shuttled by Prostate Cancer Cells-Secreted Exosomes Initiates Osteoblastic Phenotypes in the Bone Metastatic Microenvironment via MiR-205-5p/Runt-Related Transcription Factor 2/Splicing Factor Proline- and Glutamine-Rich/Polypyrimidine Tract-Binding Protein 2 Axis. Clin. Transl. Med. 2021, 11, e493. [Google Scholar] [CrossRef] [PubMed]
- Devi, K.T.R..; Karthik, D.; Mahendran, T.; Jaganathan, M.K.; Hemdev, S.P. Long noncoding RNAs: Role and contribution in pancreatic cancer. Transcription 2018, 12, 12–27. [Google Scholar] [CrossRef]
- He, Y.; Yue, H.; Cheng, Y.; Ding, Z.; Xu, Z.; Lv, C.; Wang, Z.; Wang, J.; Yin, C.; Hao, H.; et al. ALKBH5-Mediated M6A Demethylation of KCNK15-AS1 Inhibits Pancreatic Cancer Progression via Regulating KCNK15 and PTEN/AKT Signaling. Cell Death Dis. 2021, 12, 1121. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhao, X.; Liu, Q.; Li, C.; Graves-Deal, R.; Cao, Z.; Singh, B.; Franklin, J.L.; Wang, J.; Hu, H.; et al. LncRNA MIR100HG-Derived MiR-100 and MiR-125b Mediate Cetuximab Resistance via Wnt/β-Catenin Signaling. Nat. Med. 2017, 23, 1331–1341. [Google Scholar] [CrossRef]
- Liu, H.; Li, D.; Sun, L.; Qin, H.; Fan, A.; Meng, L.; Graves-Deal, R.; Glass, S.E.; Franklin, J.L.; Liu, Q.; et al. Interaction of LncRNA MIR100HG with HnRNPA2B1 Facilitates M6A-Dependent Stabilization of TCF7L2 MRNA and Colorectal Cancer Progression. Mol. Cancer 2022, 21, 74. [Google Scholar] [CrossRef]
- Chen, S.; Li, T.; Liu, D.; Liu, Y.; Long, Z.; Wu, Y.; Zhong, Y.; Zhao, J.; Wu, T.; He, W.; et al. Interaction of PHGDH with IGF2BP1 Facilitates M6A-Dependent Stabilization of TCF7L2 MRNA to Confer Multidrug Resistance in Gastric Cancer. Oncogene 2025, 44, 2064–2077. [Google Scholar] [CrossRef]
- Patil, D.P.; Chen, C.-K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. M(6)A RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Chen, D.-L.; Chen, L.-Z.; Lu, Y.-X.; Zhang, D.-S.; Zeng, Z.-L.; Pan, Z.-Z.; Huang, P.; Wang, F.-H.; Li, Y.-H.; Ju, H.-Q.; et al. Long Noncoding RNA XIST Expedites Metastasis and Modulates Epithelial-Mesenchymal Transition in Colorectal Cancer. Cell Death Dis. 2017, 8, e3011. [Google Scholar] [CrossRef]
- Lu, Z.; Li, Y.; Wang, J.; Che, Y.; Sun, S.; Huang, J.; Chen, Z.; He, J. Long Non-Coding RNA NKILA Inhibits Migration and Invasion of Non-Small Cell Lung Cancer via NF-ΚB/Snail Pathway. J. Exp. Clin. Cancer Res. 2017, 36, 54. [Google Scholar] [CrossRef] [PubMed]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long Noncoding RNA HOTAIR Regulates Polycomb-Dependent Chromatin Modification and Is Associated with Poor Prognosis in Colorectal Cancers. Cancer Res. 2011, 71, 6320–6326. [Google Scholar] [CrossRef] [PubMed]
- Porman, A.M.; Roberts, J.T.; Duncan, E.D.; Chrupcala, M.L.; Levine, A.A.; Kennedy, M.A.; Williams, M.M.; Richer, J.K.; Johnson, A.M. A Single N6-Methyladenosine Site Regulates LncRNA HOTAIR Function in Breast Cancer Cells. PLoS Biol. 2022, 20, e3001885. [Google Scholar] [CrossRef]
- Gökmen-Polar, Y.; Vladislav, I.T.; Neelamraju, Y.; Janga, S.C.; Badve, S. Prognostic Impact of HOTAIR Expression Is Restricted to ER-Negative Breast Cancers. Sci. Rep. 2015, 5, 8765. [Google Scholar] [CrossRef]
- Lu, X.; Chen, L.; Liu, S.; Cao, Y.; Huang, Z. m6A-mediated upregulation of lncRNA RMRP boosts the progression of bladder cancer via epigenetically suppressing SCARA5. Epigenomics 2023, 15, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Fu, Z.; Dong, C.; Song, Y. Methyltransferase 3, N6-Adenosine-Methyltransferase Complex Catalytic Subunit-Induced Long Intergenic Non-Protein Coding RNA 1833 N6-Methyladenosine Methylation Promotes the Non-Small Cell Lung Cancer Progression via Regulating Heterogeneous Nuclear Ribonucleoprotein A2/B1 Expression. Bioengineered 2022, 13, 10493–10503. [Google Scholar] [CrossRef]
- Liu, W.; Wan, Q.; Zhou, E.; He, P.; Tang, L. LncRNA LINC01833 Is a Prognostic Biomarker and Correlates with Immune Infiltrates in Patients with Lung Adenocarcinoma by Integrated Bioinformatics Analysis. J. Oncol. 2023, 2023, 3965198. [Google Scholar] [CrossRef]
- Ni, W.; Yao, S.; Zhou, Y.; Liu, Y.; Huang, P.; Zhou, A.; Liu, J.; Che, L.; Li, J. Long Noncoding RNA GAS5 Inhibits Progression of Colorectal Cancer by Interacting with and Triggering YAP Phosphorylation and Degradation and Is Negatively Regulated by the M6A Reader YTHDF3. Mol. Cancer 2019, 18, 143. [Google Scholar] [CrossRef]
- Zhao, B.; Li, L.; Tumaneng, K.; Wang, C.-Y.; Guan, K.-L. A Coordinated Phosphorylation by Lats and CK1 Regulates YAP Stability through SCF(Beta-TRCP). Genes Dev. 2010, 24, 72–85. [Google Scholar] [CrossRef]
- Prensner, J.R.; Iyer, M.K.; Sahu, A.; Asangani, I.A.; Cao, Q.; Patel, L.; Vergara, I.A.; Davicioni, E.; Erho, N.; Ghadessi, M.; et al. The Long Noncoding RNA SChLAP1 Promotes Aggressive Prostate Cancer and Antagonizes the SWI/SNF Complex. Nat. Genet. 2013, 45, 1392–1398. [Google Scholar] [CrossRef] [PubMed]
- Prensner, J.R.; Zhao, S.; Erho, N.; Schipper, M.; Iyer, M.K.; Dhanasekaran, S.M.; Magi-Galluzzi, C.; Mehra, R.; Sahu, A.; Siddiqui, J.; et al. RNA Biomarkers Associated with Metastatic Progression in Prostate Cancer: A Multi-Institutional High-Throughput Analysis of SChLAP1. Lancet Oncol. 2014, 15, 1469–1480. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs Are a Large Class of Animal RNAs with Regulatory Potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y.; et al. Extensive Translation of Circular RNAs Driven by N6-Methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Chen, R.-X.; Chen, X.; Xia, L.-P.; Zhang, J.-X.; Pan, Z.-Z.; Ma, X.-D.; Han, K.; Chen, J.-W.; Judde, J.-G.; Deas, O.; et al. N6-Methyladenosine Modification of CircNSUN2 Facilitates Cytoplasmic Export and Stabilizes HMGA2 to Promote Colorectal Liver Metastasis. Nat. Commun. 2019, 10, 4695. [Google Scholar] [CrossRef] [PubMed]
- Zeng, K.; Peng, J.; Xing, Y.; Zhang, L.; Zeng, P.; Li, W.; Zhang, W.; Pan, Z.; Zhou, C.; Lin, J. A Positive Feedback Circuit Driven by M6A-Modified Circular RNA Facilitates Colorectal Cancer Liver Metastasis. Mol. Cancer 2023, 22, 202. [Google Scholar] [CrossRef]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic Cleavage of M6A-Containing RNAs by RNase P/MRP Complex. Mol. Cell 2019, 74, 494–507.e8. [Google Scholar] [CrossRef]
- Rong, D.; Wu, F.; Lu, C.; Sun, G.; Shi, X.; Chen, X.; Dai, Y.; Zhong, W.; Hao, X.; Zhou, J.; et al. M6A Modification of CircHPS5 and Hepatocellular Carcinoma Progression through HMGA2 Expression. Mol. Ther. Nucleic Acids 2021, 26, 637–648. [Google Scholar] [CrossRef]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small-Molecule Inhibition of METTL3 as a Strategy against Myeloid Leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, H.; Gao, P. Metabolic Reprogramming and Epigenetic Modifications on the Path to Cancer. Protein Cell 2022, 13, 877–919. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Huang, Z.; Chen, Y.; Tian, H.; Chai, P.; Shen, Y.; Yao, Y.; Xu, S.; Ge, S.; Jia, R. Lactate and Lactylation in Cancer. Signal Transduct. Target. Ther. 2025, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Mateos, P.A.; Sethi, A.J.; Ravindran, A.; Srivastava, A.; Woodward, K.; Mahmud, S.; Kanchi, M.; Guarnacci, M.; Xu, J.; Yuen, Z.W.S.; et al. Prediction of M6A and M5C at Single-Molecule Resolution Reveals a Transcriptome-Wide Co-Occurrence of RNA Modifications. Nat. Commun. 2024, 15, 3899. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).