LncRNA-Mediated miR-145 Sponging Drives FN1 and CCND1 Expression: Prognostic and Therapeutic Targets in NSCLC
Abstract
1. Introduction
2. Methods
2.1. Data Acquisition and Pre-Processing
2.2. Identification of Differentially Expressed Genes and miRNAs
2.3. Target Prediction and Intersection Analysis
2.4. Functional Enrichment Analysis
2.5. ceRNA Network Construction
2.6. RT-qPCR Validation of miR-145 in NSCLC Tissues and Cell Lines
2.7. GEPIA Survival and Correlation Analysis
2.8. Statistical Analysis
3. Results
3.1. Demographic and Clinical Characteristics of the Study Cohort
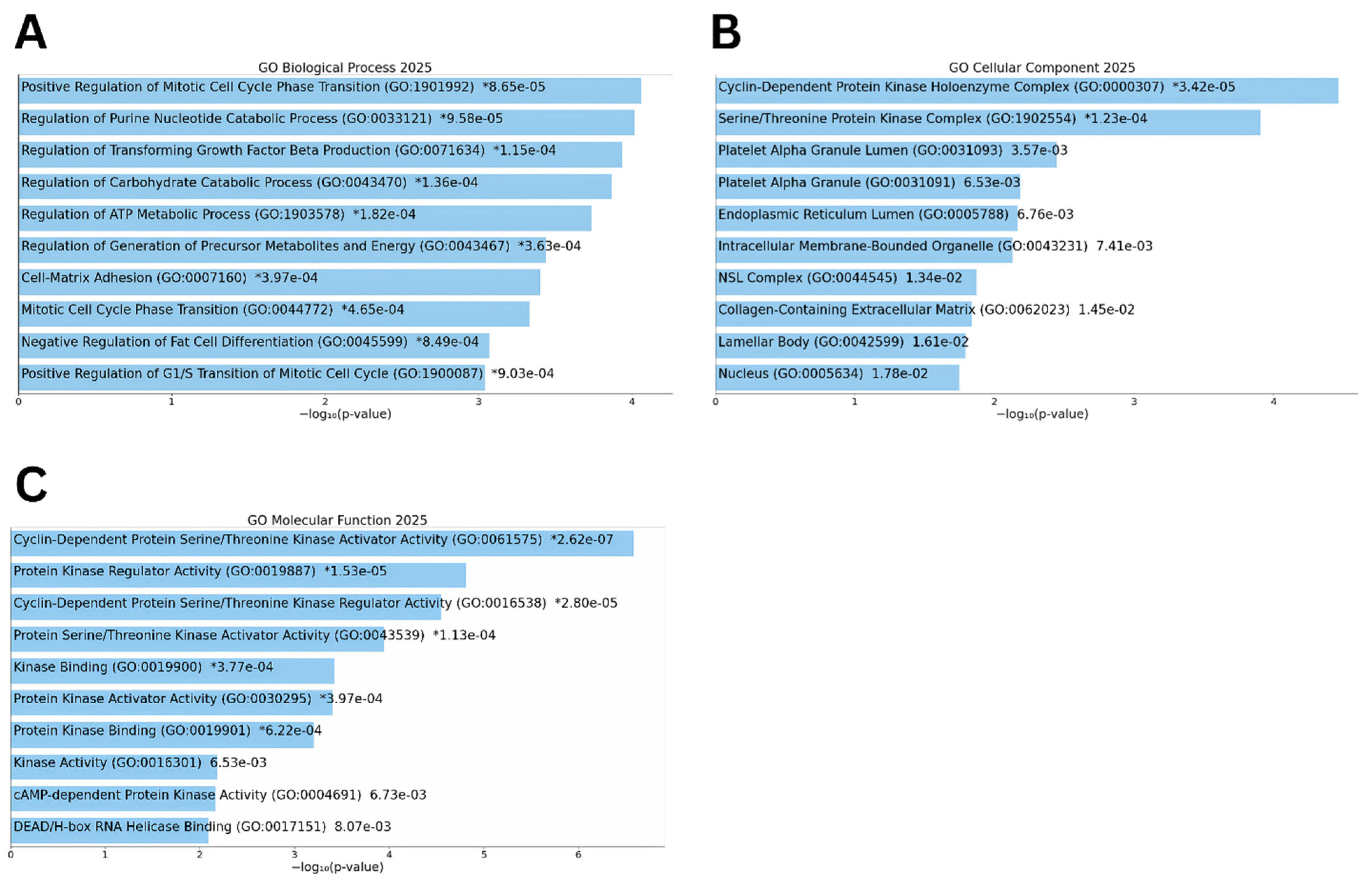
3.2. Functional Enrichment of Differentially Expressed Genes
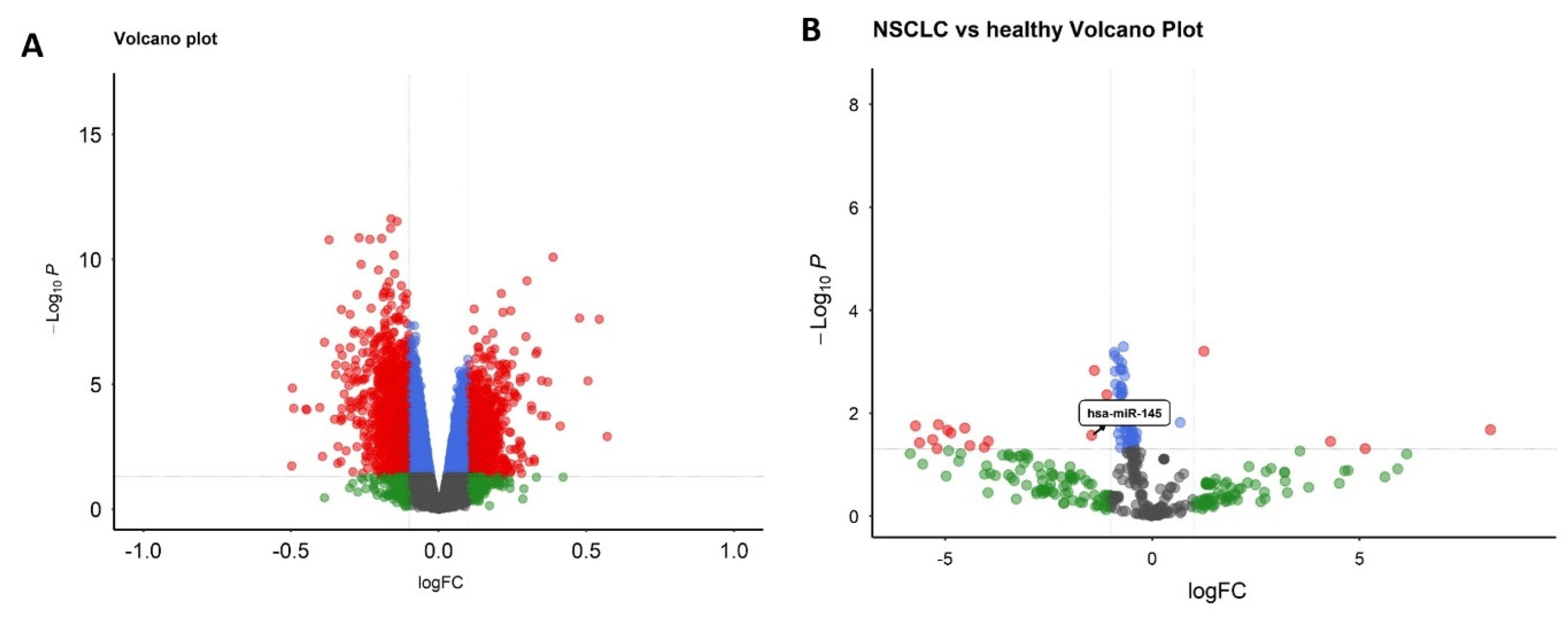
3.3. Selection of hsa-miR-145-3p and Target Identification
3.4. Pathway and Functional Enrichment
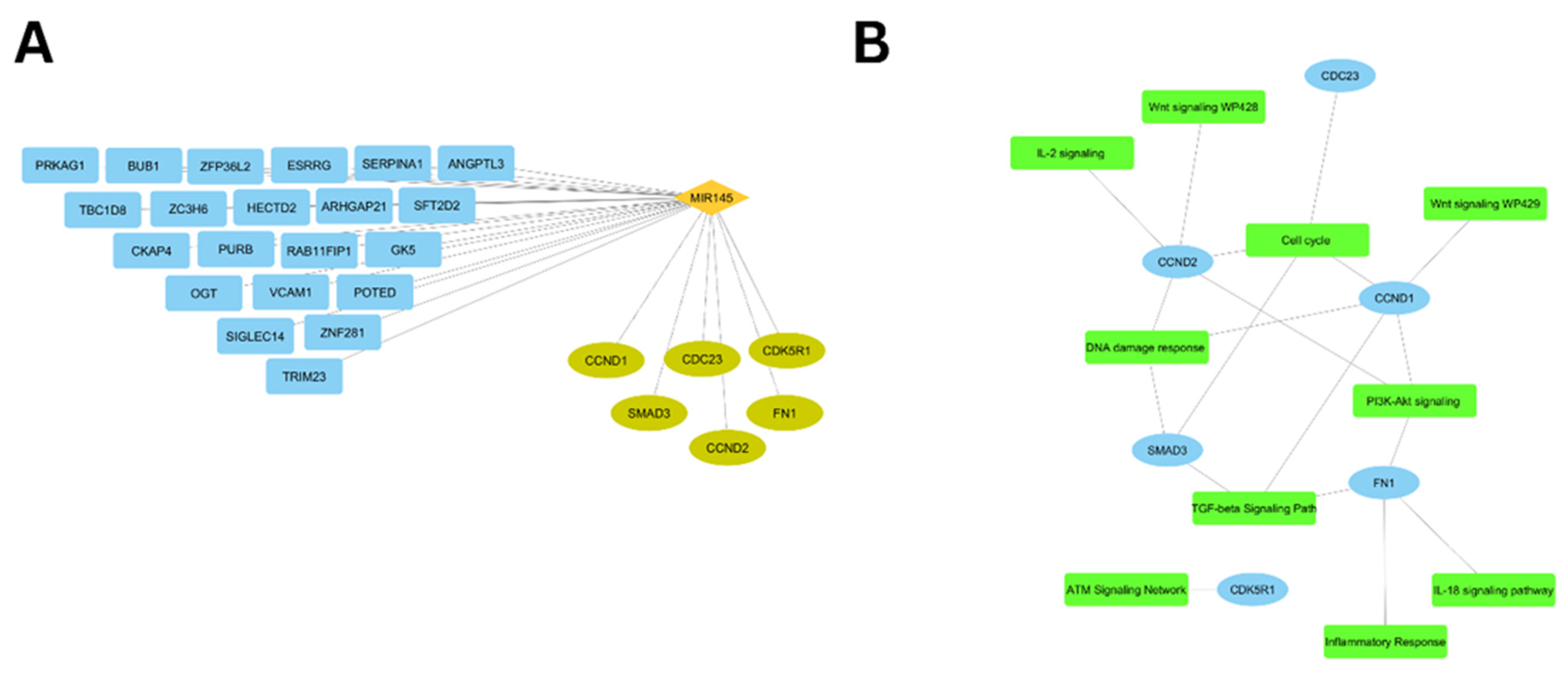
3.5. Immune-Related Network Visualization
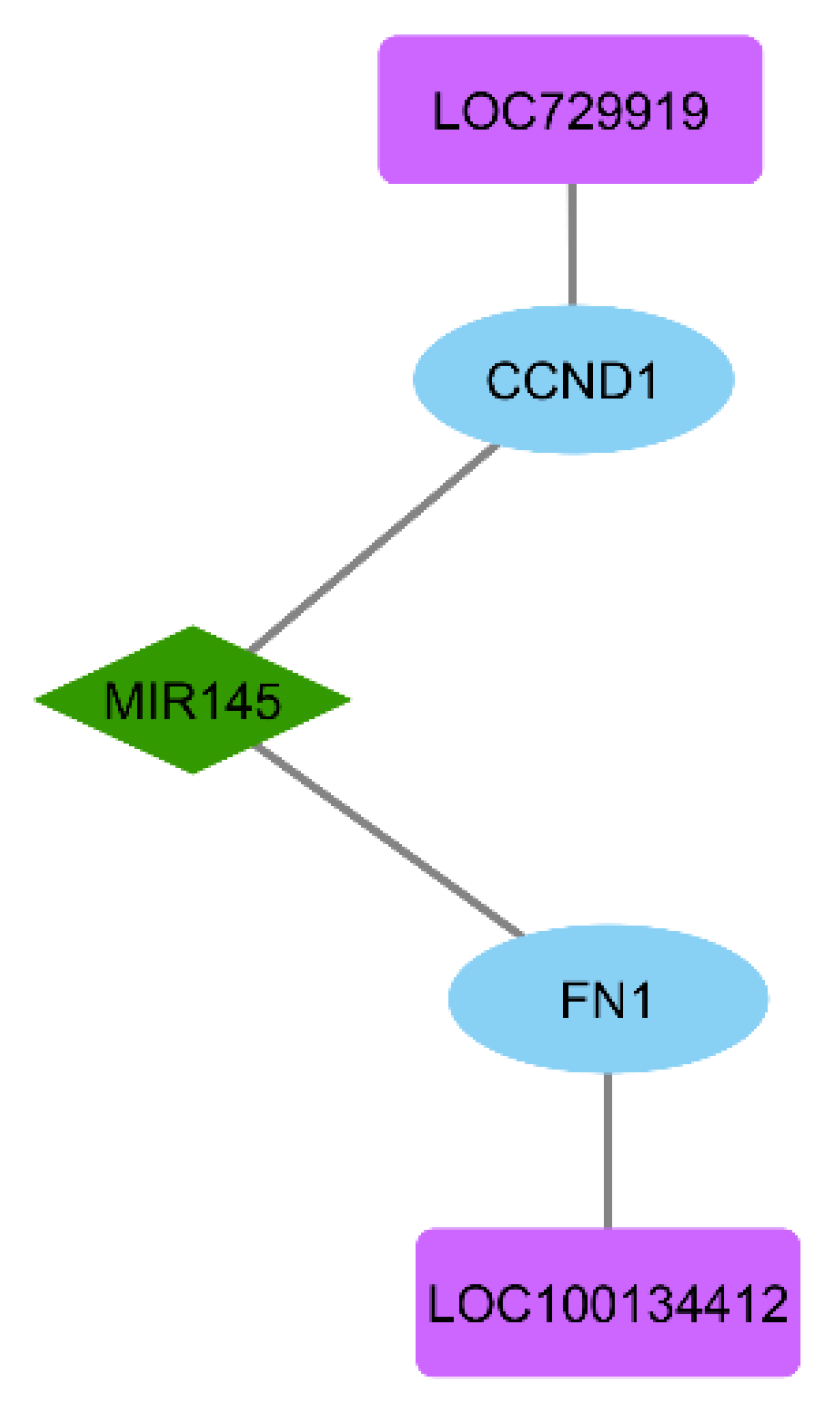
3.6. ceRNA Network Construction
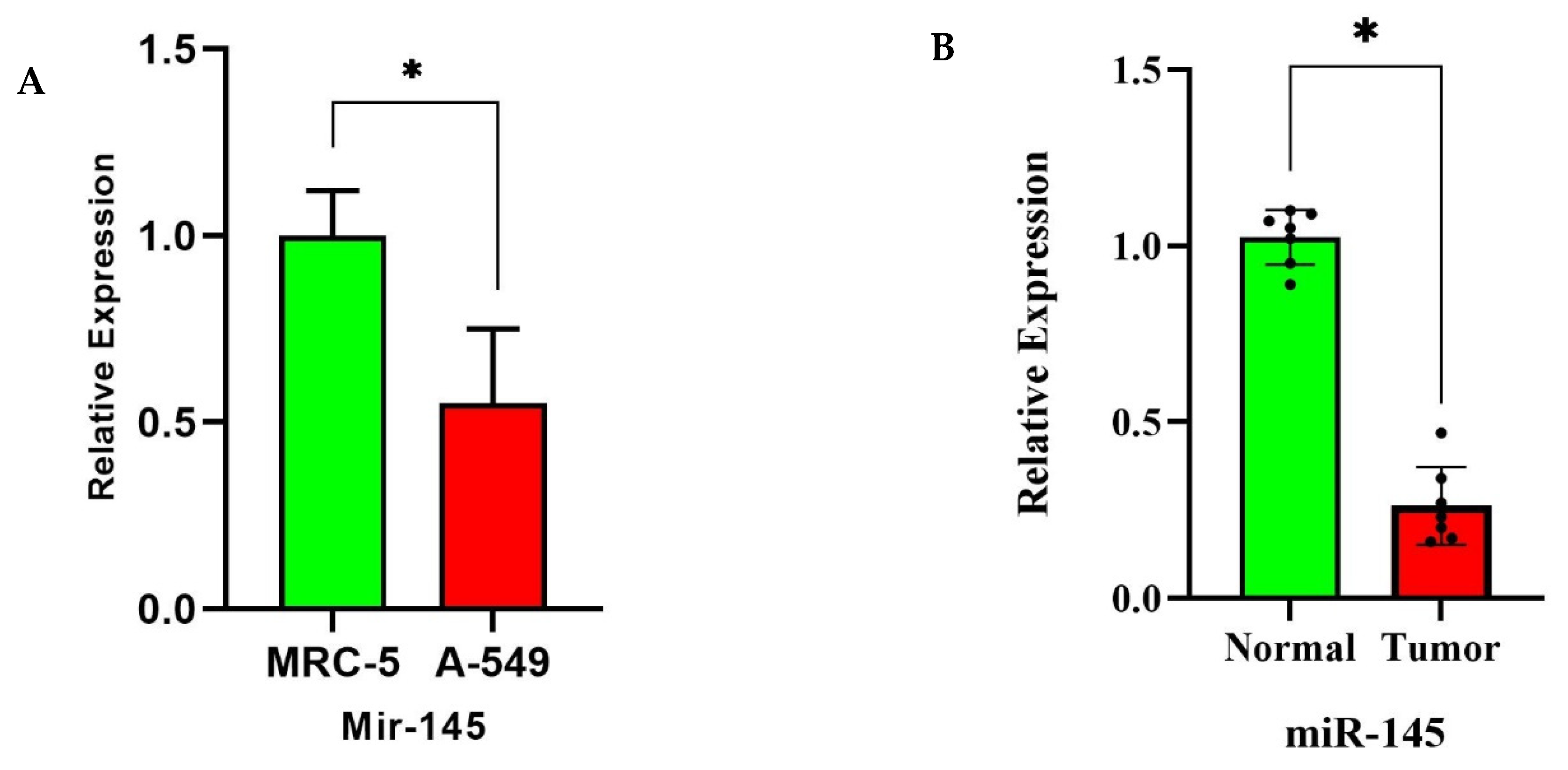
3.7. miR-145 Downregulation in NSCLC Tissues and Cell Lines via RT-qPCR
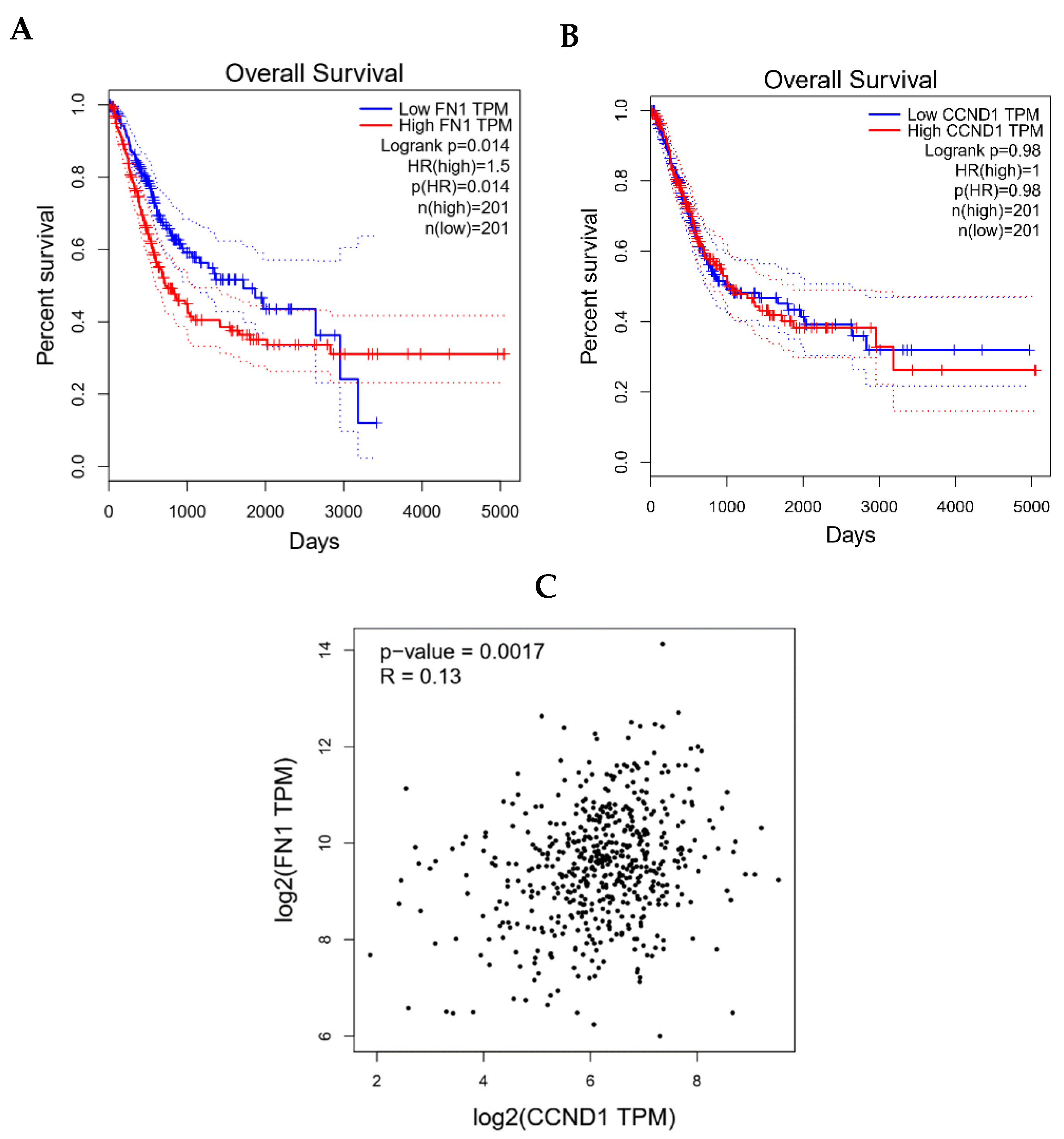
3.8. Clinical Validation of FN1/CCND1 Relevance via GEPIA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hendriks, L.E.; Remon, J.; Faivre-Finn, C.; Garassino, M.C.; Heymach, J.V.; Kerr, K.M.; Tan, D.S.; Veronesi, G.; Reck, M. Non-small-cell lung cancer. Nat. Rev. Dis. Primers 2024, 10, 71. [Google Scholar] [CrossRef]
- Gridelli, C.; Rossi, A.; Carbone, D.P.; Guarize, J.; Karachaliou, N.; Mok, T.; Petrella, F.; Spaggiari, L.; Rosell, R. Non-small-cell lung cancer. Nat. Rev. Dis. Primers 2015, 1, 15009. [Google Scholar] [CrossRef]
- Alduais, Y.; Zhang, H.; Fan, F.; Chen, J.; Chen, B. Non-small cell lung cancer (NSCLC): A review of risk factors, diagnosis, and treatment. Medicine 2023, 102, e32899. [Google Scholar] [CrossRef]
- Pellini, B.; Madison, R.W.; Childress, M.A.; Miller, S.T.; Gjoerup, O.; Cheng, J.; Huang, R.S.; Krainock, M.; Gupta, P.; Zou, W. Circulating tumor DNA monitoring on chemo-immunotherapy for risk stratification in advanced non–small cell lung cancer. Clin. Cancer Res. 2023, 29, 4596–4605. [Google Scholar] [CrossRef]
- Qi, X.; Zhang, D.-H.; Wu, N.; Xiao, J.-H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar] [CrossRef]
- Guo, Q.; Li, D.; Luo, X.; Yuan, Y.; Li, T.; Liu, H.; Wang, X. The regulatory network and potential role of LINC00973-miRNA-mRNA ceRNA in the progression of non-small-cell lung cancer. Front. Immunol. 2021, 12, 684807. [Google Scholar] [CrossRef] [PubMed]
- Sen, R.; Ghosal, S.; Das, S.; Balti, S.; Chakrabarti, J. Competing endogenous RNA: The key to posttranscriptional regulation. Sci. World J. 2014, 2014, 896206. [Google Scholar] [CrossRef] [PubMed]
- Luan, W.; Li, L.; Shi, Y.; Bu, X.; Xia, Y.; Wang, J.; Djangmah, H.S.; Liu, X.; You, Y.; Xu, B. Long non-coding RNA MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget 2016, 7, 63901. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Liu, Y.; Liu, Q.; Xiao, Y.; Liu, B.; Ren, X.; Qi, X.; Zhou, H.; Zeng, C.; Jia, L. HOTAIR/miR-326/FUT6 axis facilitates colorectal cancer progression through regulating fucosylation of CD44 via PI3K/AKT/mTOR pathway. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2019, 1866, 750–760. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Y.; Xing, L.; Zhou, Y.; Chang, N.; Xi, H.; Xu, X.; Zhang, J. LINC01559 drives osimertinib resistance in NSCLC through a ceRNA network regulating miR-320a/IGF2BP3 axis. Front. Pharmacol. 2025, 16, 1592846. [Google Scholar] [CrossRef]
- Nie, W.; Ge, H.-j.; Yang, X.-q.; Sun, X.; Huang, H.; Tao, X.; Chen, W.-s.; Li, B. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016, 371, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Drexler, H.; Dirks, W.; MacLeod, R.; Quentmeier, H.; Steube, K.; Uphoff, C. DSMZ Catalogue of Human and Animal Cell Lines; DSMZ: Braunschweig, Germany, 2001. [Google Scholar]
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2008; pp. 584–594. [Google Scholar]
- Wang, M.; Herbst, R.S.; Boshoff, C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat. Med. 2021, 27, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B. The 2015 World Health Organization classification of lung tumors: Impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef]
- Garinet, S.; Wang, P.; Mansuet-Lupo, A.; Fournel, L.; Wislez, M.; Blons, H. Updated prognostic factors in localized NSCLC. Cancers 2022, 14, 1400. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef]
- Braga, E.A.; Fridman, M.V.; Burdennyy, A.M.; Loginov, V.I.; Dmitriev, A.A.; Pronina, I.V.; Morozov, S.G. Various LncRNA mechanisms in gene regulation involving miRNAs or RNA-binding proteins in non-small-cell lung cancer: Main signaling pathways and networks. Int. J. Mol. Sci. 2023, 24, 13617. [Google Scholar] [CrossRef]
- Yu, W.; Ding, J.; He, M.; Chen, Y.; Wang, R.; Han, Z.; Xing, E.Z.; Zhang, C.; Yeh, S. Estrogen receptor β promotes the vasculogenic mimicry (VM) and cell invasion via altering the lncRNA-MALAT1/miR-145-5p/NEDD9 signals in lung cancer. Oncogene 2019, 38, 1225–1238. [Google Scholar] [CrossRef]
- Zeinali, T.; Mansoori, B.; Mohammadi, A.; Baradaran, B. Regulatory mechanisms of miR-145 expression and the importance of its function in cancer metastasis. Biomed. Pharmacother. 2019, 109, 195–207. [Google Scholar] [CrossRef]
- Sachdeva, M.; Mo, Y.-Y. miR-145-mediated suppression of cell growth, invasion and metastasis. Am. J. Transl. Res. 2010, 2, 170. [Google Scholar]
- Ren, D.; Wang, M.; Guo, W.; Zhao, X.; TU, X.A.; Huang, S.; Zou, X.; Peng, X. Wild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR-145. Int. J. Oncol. 2013, 42, 1473–1481. [Google Scholar] [CrossRef]
- Cui, Y.; Li, G.; Zhang, X.; Dai, F.; Zhang, R. Increased MALAT1 expression contributes to cisplatin resistance in non–small cell lung cancer. Oncol. Lett. 2018, 16, 4821–4828. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, L.; Zhou, J.; Chen, Y.; Xie, D.; Yao, Y.; Cui, D. Novel insights into MALAT1 function as a MicroRNA sponge in NSCLC. Front. Oncol. 2021, 11, 758653. [Google Scholar] [CrossRef]
- Huang, S.; Guo, W.; Tang, Y.; Ren, D.; Zou, X.; Peng, X. miR-143 and miR-145 inhibit stem cell characteristics of PC-3 prostate cancer cells. Oncol. Rep. 2012, 28, 1831–1837. [Google Scholar] [PubMed]
- Hughes, D.J.; Kapiris, M.; Podvez Nevajda, A.; McGrath, H.; Stavraka, C.; Ahmad, S.; Taylor, B.; Cook, G.J.R.; Ghosh, S.; Josephs, D.; et al. Non-Small Cell Lung Cancer (NSCLC) in Young Adults, Age < 50, Is Associated with Late Stage at Presentation and a Very Poor Prognosis in Patients That Do Not Have a Targeted Therapy Option: A Real-World Study. Cancers 2022, 14, 6056. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, J.; Morgensztern, D.; Goodgame, B.; Baggstrom, M.Q.; Gao, F.; Piccirillo, J.; Govindan, R. Distinctive characteristics of non-small cell lung cancer (NSCLC) in the young: A surveillance, epidemiology, and end results (SEER) analysis. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2010, 5, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Cai, X.; Yu, W.; Lv, C.; Fu, X. Clinical significance of age at diagnosis among young non-small cell lung cancer patients under 40 years old: A population-based study. Oncotarget 2015, 6, 44963–44970. [Google Scholar] [CrossRef]
- Rivera, G.A.; Wakelee, H. Lung Cancer in Never Smokers. Adv. Exp. Med. Biol. 2016, 893, 43–57. [Google Scholar] [CrossRef]
- Suda, K.; Mitsudomi, T. Role of EGFR mutations in lung cancers: Prognosis and tumor chemosensitivity. Arch. Toxicol. 2015, 89, 1227–1240. [Google Scholar] [CrossRef]
- Thomas, D.; Maloney, M.E.; Raval, G. Concomitant EGFR Mutations and ALK Rearrangements in Lung Adenocarcinoma Treated With Osimertinib. Cureus 2023, 15, e48122. [Google Scholar] [CrossRef]
- Del Vescovo, V.; Denti, M.A. microRNA and Lung Cancer. Adv. Exp. Med. Biol. 2015, 889, 153–177. [Google Scholar] [CrossRef]
- Betticher, D.C.; Heighway, J.; Hasleton, P.S.; Altermatt, H.J.; Ryder, W.D.; Cerny, T.; Thatcher, N. Prognostic significance of CCND1 (cyclin D1) overexpression in primary resected non-small-cell lung cancer. Br. J. Cancer 1996, 73, 294–300. [Google Scholar] [CrossRef]
- Ji, J.; Chen, L.; Zhuang, Y.; Han, Y.; Tang, W.; Xia, F. Fibronectin 1 inhibits the apoptosis of human trophoblasts by activating the PI3K/Akt signaling pathway. Int. J. Mol. Med. 2020, 46, 1908–1922. [Google Scholar] [CrossRef]
- Li, B.; Shen, W.; Peng, H.; Li, Y.; Chen, F.; Zheng, L.; Xu, J.; Jia, L. Fibronectin 1 promotes melanoma proliferation and metastasis by inhibiting apoptosis and regulating EMT. Oncotargets Ther. 2019, 12, 3207–3221. [Google Scholar]
- Gao, W.; Liu, Y.; Qin, R.; Liu, D.; Feng, Q. Silence of fibronectin 1 increases cisplatin sensitivity of non-small cell lung cancer cell line. Biochem. Biophys. Res. Commun. 2016, 476, 35–41. [Google Scholar] [CrossRef]
- Wu, H.; Zeng, C.; Wu, G.; Fang, F.; Xiao, C.; Li, L.; Luo, Y.; Ouyang, Z.; Zhou, C.; Qian, Y. Exosomal LRG1 promotes non-small cell lung cancer proliferation and metastasis by binding FN1 protein. Exp. Cell Res. 2024, 439, 114097. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Rao, B.; Huang, Z.; Xie, C.; Yu, Y.; Yang, B.; Wu, D.; Wang, D.; Qiu, F.; Zhou, Y. Circular RNA hsa_circ_0050386 suppresses non-small cell lung cancer progression via regulating the SRSF3/FN1 axis. J. Transl. Med. 2024, 22, 47. [Google Scholar] [CrossRef]
- Zhang, G.; Han, L.; Li, Z.; Wang, Y.; Ma, Y.; Ke, X.; Wang, Z. A newly discovered Lnc-PDZD7-3 increased metastatic and proliferative potential of lung adenocarcinoma cells via modulating FN1/fibronectin signaling. Res. Sq. 2023; preprint. [Google Scholar]
- Sun, C.-C.; Li, S.-J.; Li, D.-J. Hsa-miR-134 suppresses non-small cell lung cancer (NSCLC) development through down-regulation of CCND1. Oncotarget 2016, 7, 35960. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-L.; Wang, J.; Zhang, C.-Y.; Shen, Y.-Q.; Wang, H.-M.; Ding, L.; Gu, Y.-C.; Lou, J.-T.; Zhao, X.-T.; Ma, Z.-L. MiR-146a-5p inhibits cell proliferation and cell cycle progression in NSCLC cell lines by targeting CCND1 and CCND2. Oncotarget 2016, 7, 59287. [Google Scholar] [PubMed]
- Gautschi, O.; Hugli, B.; Ziegler, A.; Bigosch, C.; Bowers, N.L.; Ratschiller, D.; Jermann, M.; Stahel, R.A.; Heighway, J.; Betticher, D.C. Cyclin D1 (CCND1) A870G gene polymorphism modulates smoking-induced lung cancer risk and response to platinum-based chemotherapy in non-small cell lung cancer (NSCLC) patients. Lung Cancer 2006, 51, 303–311. [Google Scholar] [CrossRef]
- Singh, S.; Gouri, V.; Samant, M. TGF-β in correlation with tumor progression, immunosuppression and targeted therapy in colorectal cancer. Med. Oncol. 2023, 40, 335. [Google Scholar] [CrossRef]
- Dituri, F.; Mancarella, S.; Cigliano, A.; Giannelli, G. TGF-β as multifaceted orchestrator in HCC progression: Signaling, EMT, immune microenvironment, and novel therapeutic perspectives. Semin. Liver Dis. 2019, 39, 53–69. [Google Scholar]
- Shiloh, Y. ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Cancer 2003, 3, 155–168. [Google Scholar] [CrossRef]
- Pan, D. The hippo signaling pathway in development and cancer. Dev. Cell 2010, 19, 491–505. [Google Scholar] [CrossRef]
- Yu, F.-X.; Zhao, B.; Guan, K.-L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 2015, 163, 811–828. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef]
- Thu, K.L.; Yoon, J.Y. ATM-the gene at the moment in non-small cell lung cancer. Transl. Lung Cancer Res. 2024, 13, 699–705. [Google Scholar] [CrossRef]
- Gong, X.; Zhou, Y.; Deng, Y. Targeting DNA Damage Response-Mediated Resistance in Non-Small Cell Lung Cancer: From Mechanistic Insights to Drug Development. Curr. Oncol. 2025, 32, 367. [Google Scholar] [CrossRef]
- Deng, T.; Xie, L.; Xiaofang, C.; Zhang, Z.; Xiao, Y.; Peng, Y.; Yin, L.; Fu, Y.; Li, X. ATM-Mediated translocation of RanBPM regulates DNA damage response by stabilizing p21 in non-small cell lung cancer cells. Cell. Oncol. 2024, 47, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Baker, D.; Ten Dijke, P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int. J. Mol. Sci. 2019, 20, 2767. [Google Scholar] [CrossRef]
- Kim, B.N.; Ahn, D.H.; Kang, N.; Yeo, C.D.; Kim, Y.K.; Lee, K.Y.; Kim, T.-J.; Lee, S.H.; Park, M.S.; Yim, H.W.; et al. TGF-β induced EMT and stemness characteristics are associated with epigenetic regulation in lung cancer. Sci. Rep. 2020, 10, 10597. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Fan, T.; Xiao, C.; Tian, H.; Zheng, Y.; Li, C.; He, J. TGF-β signaling in health, disease and therapeutics. Signal Transduct. Target. Ther. 2024, 9, 61. [Google Scholar] [CrossRef]
- Jeon, H.S.; Jen, J. TGF-beta signaling and the role of inhibitory Smads in non-small cell lung cancer. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2010, 5, 417–419. [Google Scholar] [CrossRef]
- Cao, S.; Xiao, S.; Zhang, J.; Li, S. Identification of the cell cycle characteristics of non-small cell lung cancer and its relationship with tumor immune microenvironment, cell death pathways, and metabolic reprogramming. Front. Endocrinol. 2023, 14, 1147366. [Google Scholar] [CrossRef]
- Caputi, M.; Russo, G.; Esposito, V.; Mancini, A.; Giordano, A. Role of cell-cycle regulators in lung cancer. J. Cell. Physiol. 2005, 205, 319–327. [Google Scholar] [CrossRef]
- Eymin, B.; Gazzeri, S. Role of cell cycle regulators in lung carcinogenesis. Cell Adhes. Migr. 2010, 4, 114–123. [Google Scholar] [CrossRef]
- Sterlacci, W.; Fiegl, M.; Tzankov, A. Prognostic and predictive value of cell cycle deregulation in non-small-cell lung cancer. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 2012, 79, 175–194. [Google Scholar] [CrossRef]
- Feng, Y.; Gan, X.; Rong, X.; Han, Q. The components and regulation of the Hippo pathway and its relationships with the progression and treatment of Non-small cell lung cancer (NSCLC). Cancer Cell Int. 2025, 25, 309. [Google Scholar] [CrossRef]
- Liang, H.; Xu, Y.; Zhao, J.; Chen, M.; Wang, M. Hippo pathway in non-small cell lung cancer: Mechanisms, potential targets, and biomarkers. Cancer Gene Ther. 2024, 31, 652–666. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Li, X.; Liu, J.; Hao, J. The Hippo/YAP signaling pathway: The driver of cancer metastasis. Cancer Biol. Med. 2023, 20, 483–489. [Google Scholar] [CrossRef]
- Nakasuka, F.; Tabata, S.; Sakamoto, T.; Hirayama, A.; Ebi, H.; Yamada, T.; Umetsu, K.; Ohishi, M.; Ueno, A.; Goto, H.; et al. TGF-β-dependent reprogramming of amino acid metabolism induces epithelial-mesenchymal transition in non-small cell lung cancers. Commun. Biol. 2021, 4, 782. [Google Scholar] [CrossRef]
- Gordian, E.; Welsh, E.A.; Gimbrone, N.; Siegel, E.M.; Shibata, D.; Creelan, B.C.; Cress, W.D.; Eschrich, S.A.; Haura, E.B.; Muñoz-Antonia, T. Transforming growth factor β-induced epithelial-to-mesenchymal signature predicts metastasis-free survival in non-small cell lung cancer. Oncotarget 2019, 10, 810–824. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Lu, S. Transforming growth factor-β1-induced epithelial to mesenchymal transition increases mitochondrial content in the A549 non-small cell lung cancer cell line. Mol. Med. Rep. 2015, 11, 417–421. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Term | Genes | Adjusted p-Value |
|---|---|---|
| DNA damage response (only ATM dependent) WP710 | CCND2; SMAD3; CCND1; SHC1; SOD2; MAP3K7 | 5.55 × 10−5 |
| TGF-beta Signaling Pathway WP366 | SMAD3; COL1A2; CCND1; SHC1; FN1; MAP3K7 | 1.52 × 10−4 |
| Hypothesized Pathways in Pathogenesis of Cardiovascular Disease WP3668 | SMAD3; SHC1; FBN1 | 4.51 × 10−4 |
| Cell cycle WP179 | CCND2; CDC23; SMAD3; CCND1; BUB1 | 8.17 × 10−4 |
| The influence of laminopathies on Wnt signaling WP4844 | CCND1; AGO2; HMGA2 | 0.001227 |
| Mammary gland development pathway—Puberty (Stage 2 of 4) WP2814 | CCND1; FN1 | 0.002711 |
| Pathways Regulating Hippo Signaling WP4540 | NTRK2; SMAD3; PRKAG1; CDH18 | 0.002972 |
| The Overlap Between Signal Transduction Pathways that Contribute to a Range of LMNA Laminopathies WP4879 | SMAD3; AGO2; HMGA2 | 0.004507 |
| Canonical and non-canonical TGF-B signaling WP3874 | SMAD3; LOX | 0.004652 |
| TGF-B Signaling in Thyroid Cells for Epithelial–Mesenchymal Transition WP3859 | SMAD3; FN1 | 0.005213 |
| Terms | Genes | Adjusted p-Value | GO |
|---|---|---|---|
| Biological Process | |||
| Positive Regulation of Mitotic Cell Cycle Phase Transition | CDC23; CCND2; CCND1 | 0.013 | 1901992 |
| Regulation of Purine Nucleotide Catabolic Process | PRKAG1; OGT | 0.013 | 0033121 |
| Regulation of Transforming Growth Factor Beta Production | SMAD3; FN1 | 0.013 | 0071634 |
| Regulation of Carbohydrate Catabolic Process | PRKAG1; OGT | 0.013 | 0043470 |
| Regulation of ATP Metabolic Process | PRKAG1; OGT | 0.014 | 1903578 |
| Regulation of Generation of Precursor Metabolites and Energy | PRKAG1; OGT | 0.021 | 0043467 |
| Cell–Matrix Adhesion | VCAM1; FN1; ANGPTL3 | 0.021 | 0007160 |
| Mitotic Cell Cycle Phase Transition | CDC23; CCND2; CCND1 | 0.022 | 0044772 |
| Negative Regulation of Fat Cell Differentiation | SMAD3; ZFP36L2 | 0.032 | 0045599 |
| Positive Regulation of G1/S Transition of Mitotic Cell Cycle | CCND2; CCND1 | 0.032 | 1900087 |
| Cellular Component | |||
| Cyclin-Dependent Protein Kinase Holoenzyme Complex | CCND2; CCND1; CDK5R1 | 0.001 | 0000307 |
| Serine/Threonine Protein Kinase Complex | CCND2; CCND1; CDK5R1 | 0.003 | 1902554 |
| Platelet Alpha Granule Lumen | SERPINA1; FN1 | 0.063 | 0031093 |
| Platelet Alpha Granule | SERPINA1; FN1 | 0.065 | 0031091 |
| Endoplasmic Reticulum Lumen | SERPINA1; FN1; CKAP4 | 0.065 | 0005788 |
| Intracellular Membrane-Bounded Organelle | SERPINA1; SMAD3; ZNF281; PRKAG1; ESRRG; ZC3H6; ZFP36L2; PURB; RAB11FIP1; CCND2; CCND1; BUB1; OGT; CDK5R1 | 0.065 | 0043231 |
| NSL Complex | OGT | 0.094 | 0044545 |
| Collagen-Containing Extracellular Matrix | SERPINA1; FN1; ANGPTL3 | 0.094 | 0062023 |
| Lamellar Body | CKAP4 | 0.094 | 0042599 |
| Nucleus | PURB; CCND2; SMAD3; CCND1; ZNF281; PRKAG1; ESRRG; ZC3H6; BUB1; OGT; ZFP36L2; CDK5R1 | 0.094 | 0005634 |
| Molecular Function | |||
| Cyclin-Dependent Protein Serine/Threonine Kinase Activator Activity | CCND2; CCND1; CDK5R1 | 2.22 | 0061575 |
| Protein Kinase Regulator Activity | CCND2; CCND1; PRKAG1; CDK5R1 | 6.52 | 0019887 |
| Cyclin-Dependent Protein Serine/Threonine Kinase Regulator Activity | CCND2; CCND1; CDK5R1 | 7.93 | 0016538 |
| Protein Serine/Threonine Kinase Activator Activity | CCND2; CCND1; CDK5R1 | 0.002 | 0043539 |
| Kinase Binding | CCND2; SMAD3; CCND1; PRKAG1; CDK5R1 | 0.005 | 0019900 |
| Protein Kinase Activator Activity | CCND2; CCND1; CDK5R1 | 0.005 | 0030295 |
| Protein Kinase Binding | CCND2; SMAD3; CCND1; PRKAG1; CDK5R1 | 0.007 | 0019901 |
| Kinase Activity | GK5; CDK5R1 | 0.057 | 0016301 |
| cAMP-dependent Protein Kinase Activity | PRKAG1 | 0.057 | 0004691 |
| DEAD/H-box RNA Helicase Binding | SMAD3 | 0.057 | 0017151 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tahmasebi, S.; Amani, D.; Salimi, B.; Adcock, I.M.; Mortaz, E. LncRNA-Mediated miR-145 Sponging Drives FN1 and CCND1 Expression: Prognostic and Therapeutic Targets in NSCLC. Biomolecules 2025, 15, 1564. https://doi.org/10.3390/biom15111564
Tahmasebi S, Amani D, Salimi B, Adcock IM, Mortaz E. LncRNA-Mediated miR-145 Sponging Drives FN1 and CCND1 Expression: Prognostic and Therapeutic Targets in NSCLC. Biomolecules. 2025; 15(11):1564. https://doi.org/10.3390/biom15111564
Chicago/Turabian StyleTahmasebi, Safa, Davar Amani, Babak Salimi, Ian M. Adcock, and Esmaeil Mortaz. 2025. "LncRNA-Mediated miR-145 Sponging Drives FN1 and CCND1 Expression: Prognostic and Therapeutic Targets in NSCLC" Biomolecules 15, no. 11: 1564. https://doi.org/10.3390/biom15111564
APA StyleTahmasebi, S., Amani, D., Salimi, B., Adcock, I. M., & Mortaz, E. (2025). LncRNA-Mediated miR-145 Sponging Drives FN1 and CCND1 Expression: Prognostic and Therapeutic Targets in NSCLC. Biomolecules, 15(11), 1564. https://doi.org/10.3390/biom15111564








