Discovery of Drug-like Inhibitors of the Human Caf1/CNOT7 poly(A)-Selective Nuclease Using Compound Screening
Abstract
1. Introduction
2. Materials and Methods
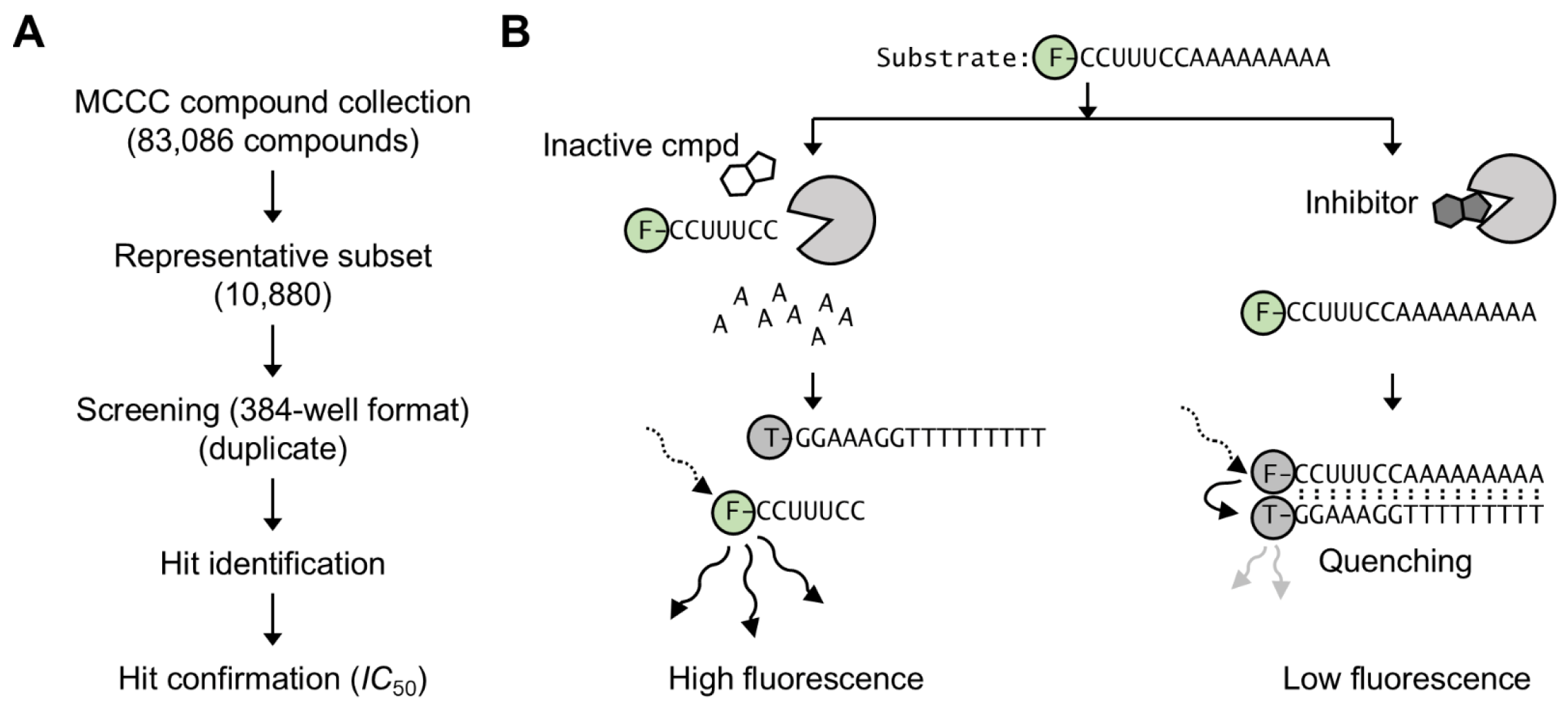
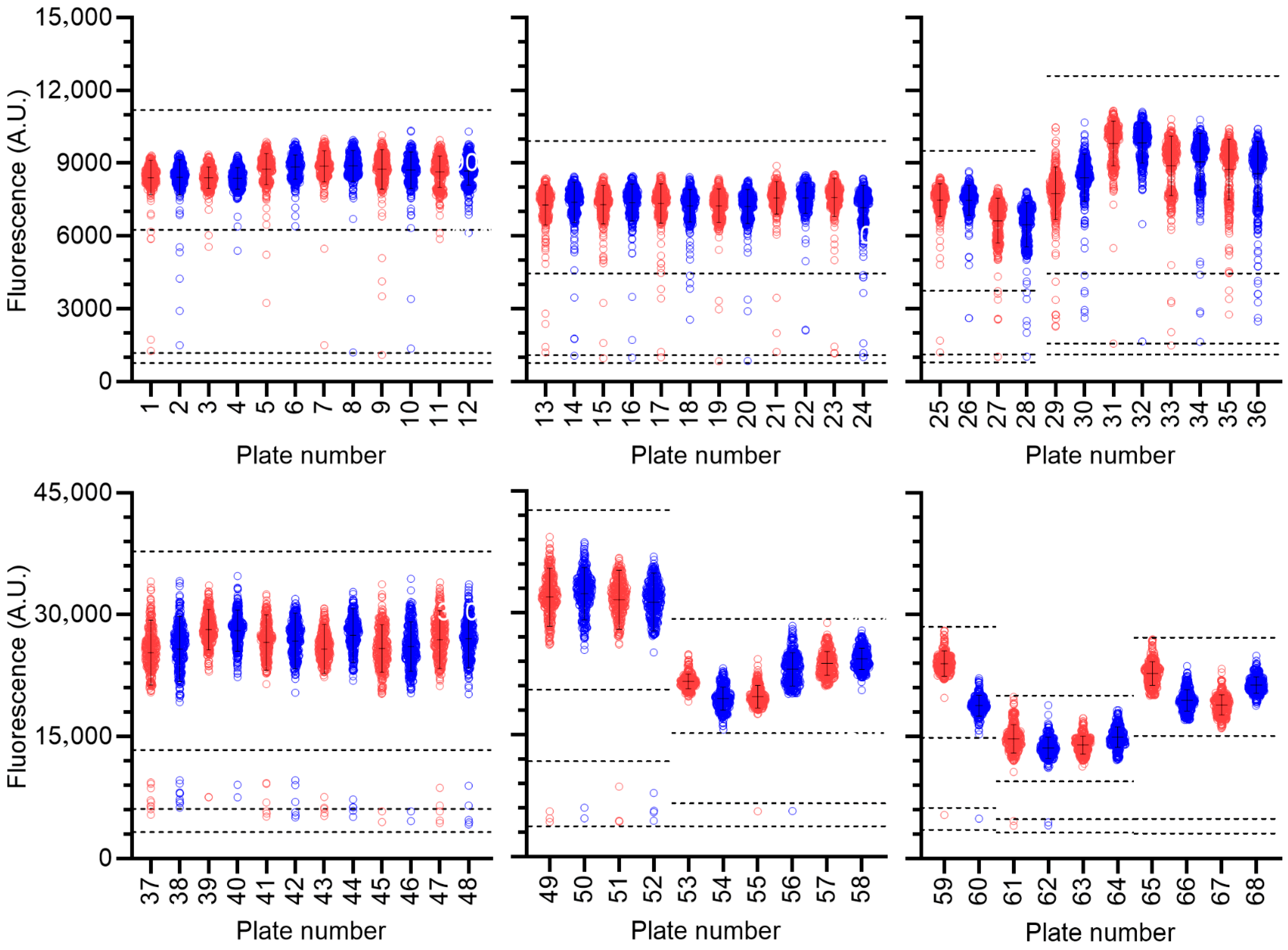
2.1. Compound Screening
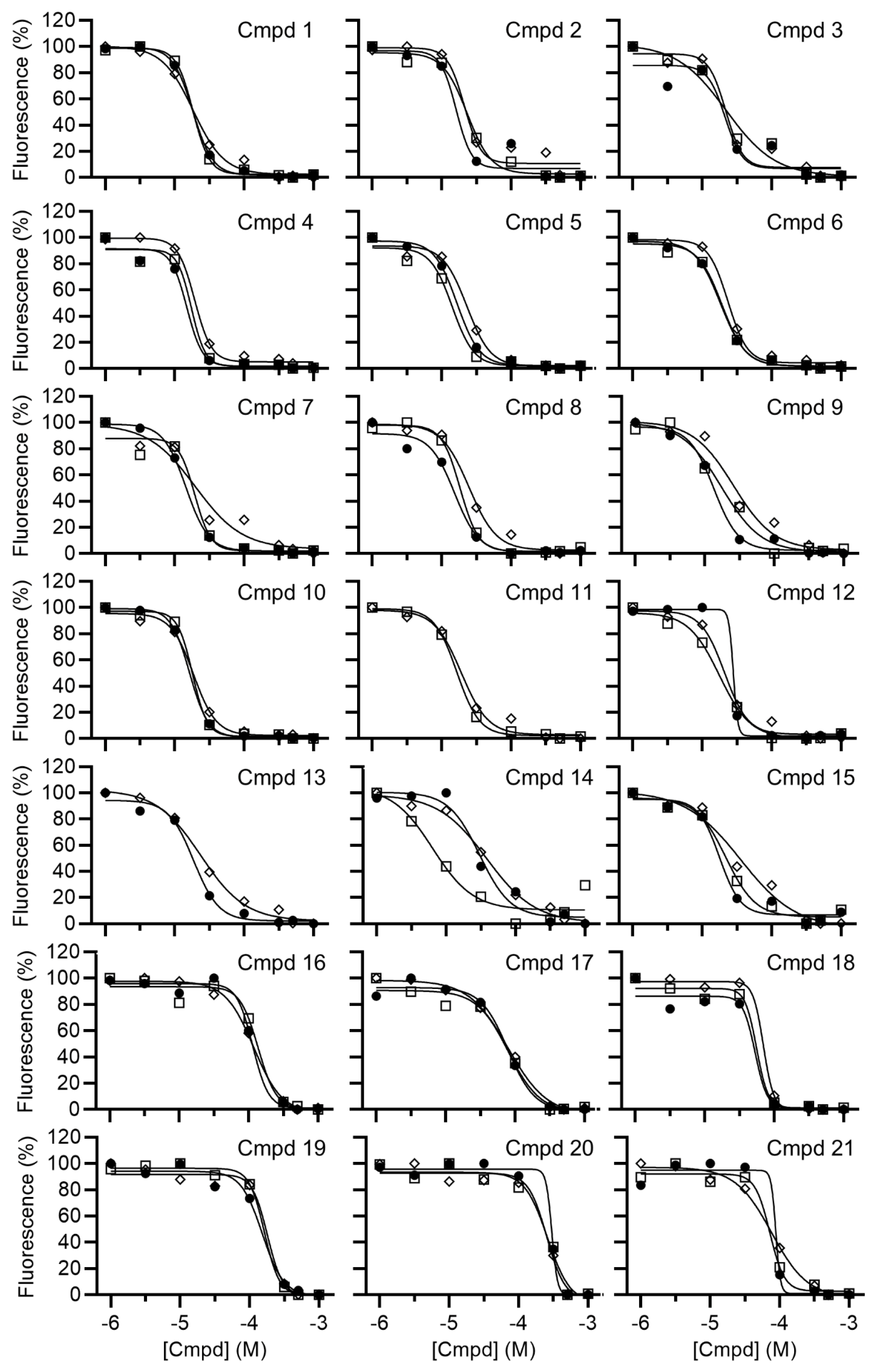
2.2. Determination of IC50 Values
2.3. Molecular Docking
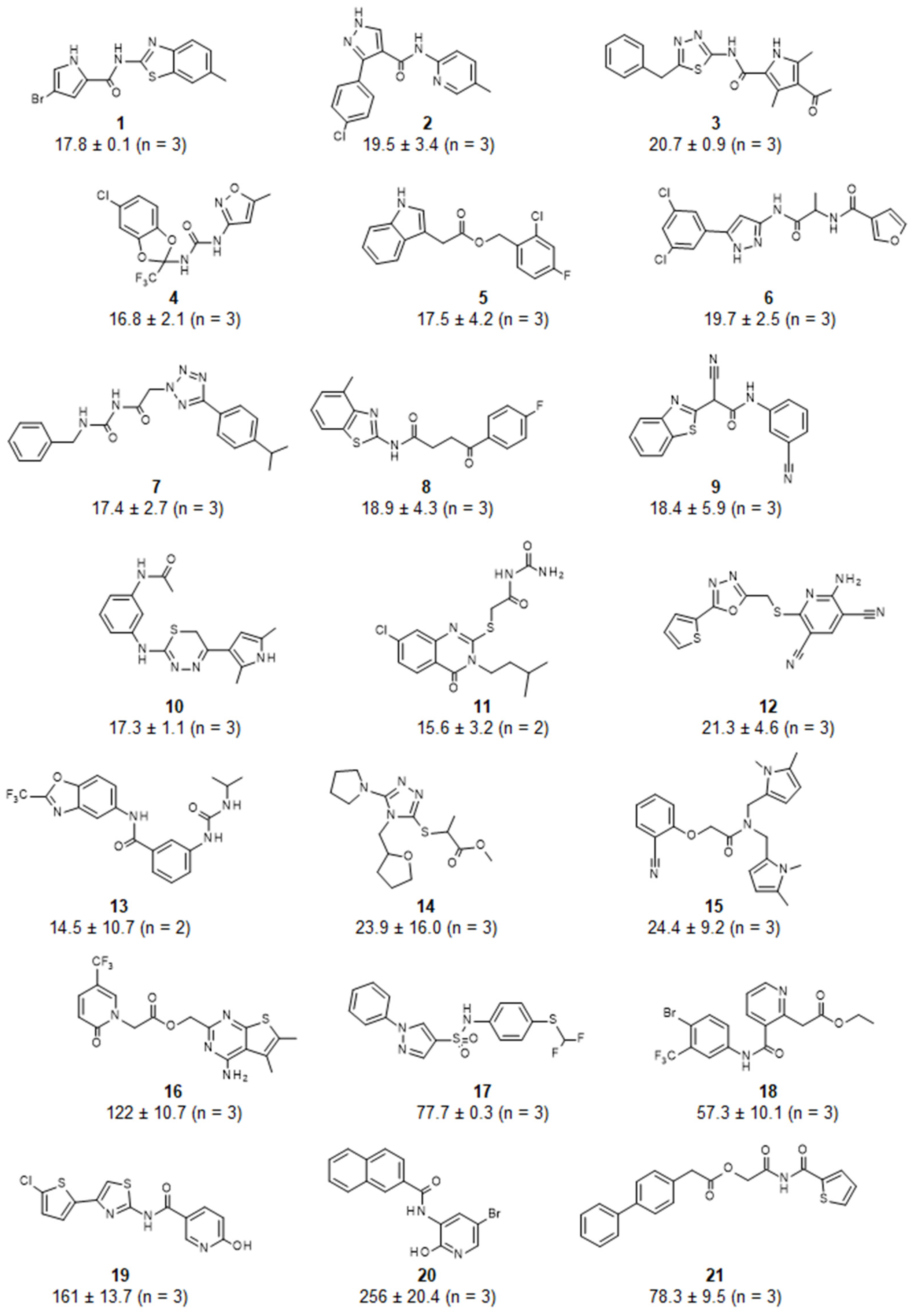
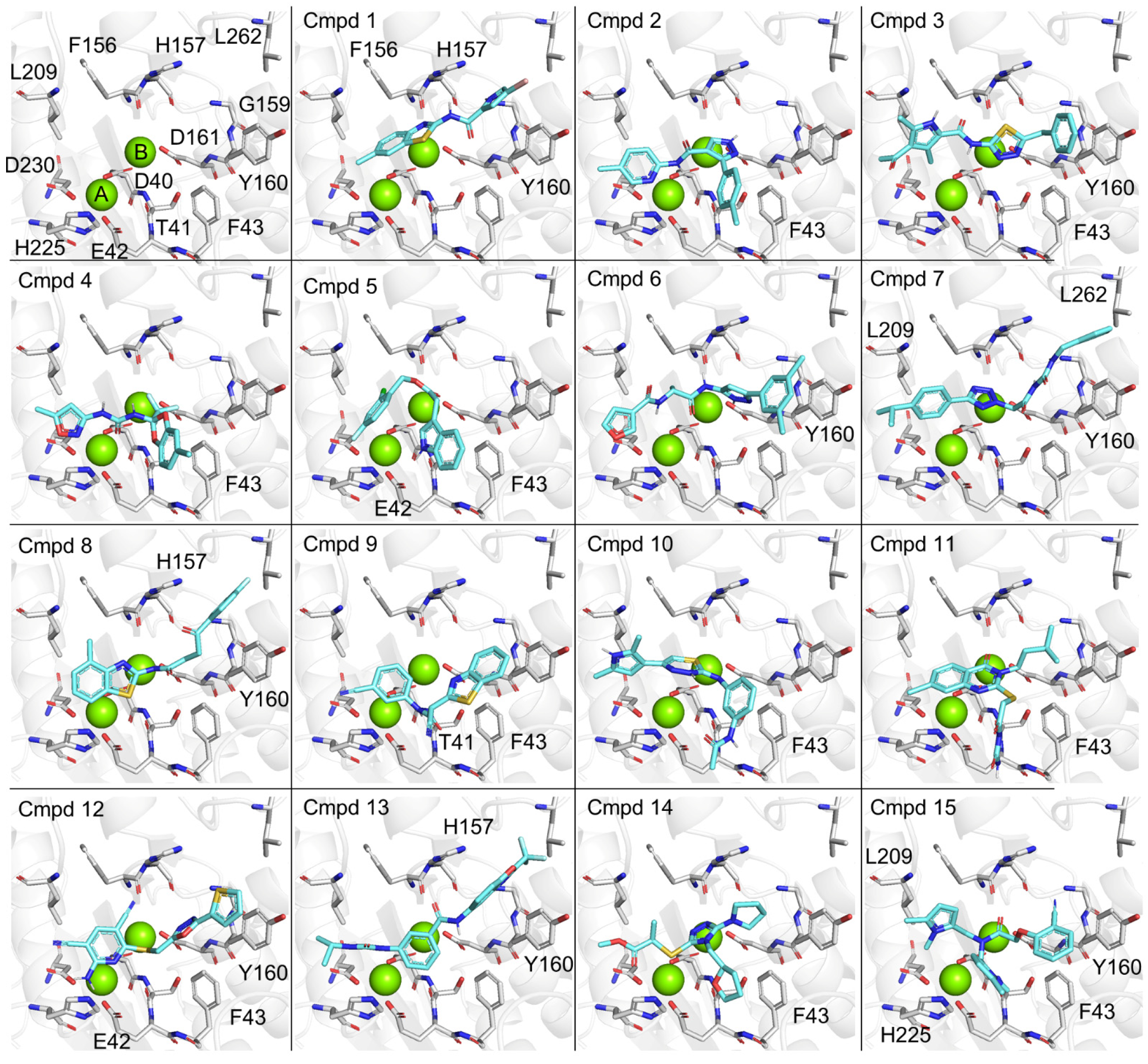
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Collart, M.A.; Panasenko, O.O. The Ccr4-Not Complex: Architecture and Structural Insights. Subcell Biochem. 2017, 83, 349–379. [Google Scholar]
- Pavanello, L.; Hall, M.; Winkler, G.S. Regulation of eukaryotic mRNA deadenylation and degradation by the Ccr4-Not complex. Front. Cell Dev. Biol. 2023, 11, 1153624. [Google Scholar] [CrossRef]
- Raisch, T.; Valkov, E. Regulation of the multisubunit CCR4-NOT deadenylase in the initiation of mRNA degradation. Curr. Opin. Struct. Biol. 2022, 77, 102460. [Google Scholar] [CrossRef]
- Krempl, C.; Lazzaretti, D.; Sprangers, R. A structural biology view on the enzymes involved in eukaryotic mRNA turnover. Biol. Chem. 2023, 404, 1101–1121. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.B.D.; Becker, T.; Denk, T.; Hashimoto, S.; Inada, T.; Beckmann, R. The ribosome as a platform to coordinate mRNA decay. Nucleic Acids Res. 2025, 53, gkaf049. [Google Scholar] [CrossRef]
- Absmeier, E.; Chandrasekaran, V.; O’Reilly, F.J.; Stowell, J.A.W.; Rappsilber, J.; Passmore, L.A. Specific recognition and ubiquitination of translating ribosomes by mammalian CCR4-NOT. Nat. Struct. Mol. Biol. 2023, 30, 1314–1322. [Google Scholar] [CrossRef]
- Buschauer, R.; Matsuo, Y.; Sugiyama, T.; Chen, Y.H.; Alhusaini, N.; Sweet, T.; Ikeuchi, K.; Cheng, J.; Matsuki, Y.; Nobuta, R.; et al. The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 2020, 368, eaay6912. [Google Scholar] [CrossRef]
- Zhu, X.; Cruz, V.E.; Zhang, H.; Erzberger, J.P.; Mendell, J.T. Specific tRNAs promote mRNA decay by recruiting the CCR4-NOT complex to translating ribosomes. Science 2024, 386, eadq8587. [Google Scholar] [CrossRef]
- Goldstrohm, A.C.; Wickens, M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008, 9, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.; Sheth, U. P bodies and the control of mRNA translation and degradation. Mol. Cell 2007, 25, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Wahle, E.; Winkler, G.S. RNA decay machines: Deadenylation by the Ccr4-Not and Pan2-Pan3 complexes. Biochim. Biophys. Acta 2013, 1829, 561–570. [Google Scholar] [CrossRef]
- Bianchin, C.; Mauxion, F.; Sentis, S.; Seraphin, B.; Corbo, L. Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA 2005, 11, 487–494. [Google Scholar] [CrossRef][Green Version]
- Horiuchi, M.; Takeuchi, K.; Noda, N.; Muroya, N.; Suzuki, T.; Nakamura, T.; Kawamura-Tsuzuku, J.; Takahasi, K.; Yamamoto, T.; Inagaki, F. Structural basis for the antiproliferative activity of the Tob-hCaf1 complex. J. Biol. Chem. 2009, 284, 13244–13255. [Google Scholar] [CrossRef] [PubMed]
- Basquin, J.; Roudko, V.V.; Rode, M.; Basquin, C.; Seraphin, B.; Conti, E. Architecture of the nuclease module of the yeast Ccr4-not complex: The Not1-Caf1-Ccr4 interaction. Mol. Cell 2012, 48, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Dupressoir, A.; Morel, A.P.; Barbot, W.; Loireau, M.P.; Corbo, L.; Heidmann, T. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genom. 2001, 2, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Pavanello, L.; Potapov, A.; Bartlam, M.; Winkler, G.S. Structure of the human Ccr4-Not nuclease module using X-ray crystallography and electron paramagnetic resonance spectroscopy distance measurements. Protein Sci. 2022, 31, 758–764. [Google Scholar] [CrossRef]
- Wang, H.; Morita, M.; Yang, X.; Suzuki, T.; Yang, W.; Wang, J.; Ito, K.; Wang, Q.; Zhao, C.; Bartlam, M.; et al. Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J. 2010, 29, 2566–2576. [Google Scholar] [CrossRef]
- Chen, Y.; Khazina, E.; Izaurralde, E.; Weichenrieder, O. Crystal structure and functional properties of the human CCR4-CAF1 deadenylase complex. Nucleic Acids Res. 2021, 49, 6489–6510. [Google Scholar] [CrossRef]
- Maryati, M.; Airhihen, B.; Winkler, G.S. The enzyme activities of Caf1 and Ccr4 are both required for deadenylation by the human Ccr4-Not nuclease module. Biochem. J. 2015, 469, 169–176. [Google Scholar] [CrossRef]
- Pekovic, F.; Rammelt, C.; Kubikova, J.; Metz, J.; Jeske, M.; Wahle, E. RNA binding proteins Smaug and Cup induce CCR4-NOT-dependent deadenylation of the nanos mRNA in a reconstituted system. Nucleic Acids Res. 2023, 51, 3950–3970. [Google Scholar] [CrossRef]
- Webster, M.W.; Chen, Y.H.; Stowell, J.A.W.; Alhusaini, N.; Sweet, T.; Graveley, B.R.; Coller, J.; Passmore, L.A. mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases. Mol. Cell 2018, 70, 1089–1100.e8. [Google Scholar] [CrossRef]
- Yi, H.; Park, J.; Ha, M.; Lim, J.; Chang, H.; Kim, V.N. PABP Cooperates with the CCR4-NOT Complex to Promote mRNA Deadenylation and Block Precocious Decay. Mol. Cell 2018, 70, 1081–1088.e5. [Google Scholar] [CrossRef]
- Winkler, G.S.; Balacco, D.L. Heterogeneity and complexity within the nuclease module of the Ccr4-Not complex. Front. Genet. 2013, 4, 296. [Google Scholar] [CrossRef] [PubMed]
- Aslam, A.; Mittal, S.; Koch, F.; Andrau, J.C.; Winkler, G.S. The Ccr4-Not Deadenylase Subunits CNOT7 and CNOT8 Have Overlapping Roles and Modulate Cell Proliferation. Mol. Biol. Cell 2009, 20, 3840–3850. [Google Scholar] [CrossRef]
- Mittal, S.; Aslam, A.; Doidge, R.; Medica, R.; Winkler, G.S. The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4-Not complex contribute to the prevention of cell death and senescence. Mol. Biol. Cell 2011, 22, 748–758. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, D.; Takahashi, A.; Yanagiya, A.; Yamaguchi, T.; Abe, T.; Kureha, T.; Kuba, K.; Kanegae, Y.; Furuta, Y.; Yamamoto, T.; et al. Essential functions of the CNOT7/8 catalytic subunits of the CCR4-NOT complex in mRNA regulation and cell viability. RNA Biol. 2020, 17, 403–416. [Google Scholar] [CrossRef]
- Berthet, C.; Morera, A.M.; Asensio, M.J.; Chauvin, M.A.; Morel, A.P.; Dijoud, F.; Magaud, J.P.; Durand, P.; Rouault, J.P. CCR4-associated factor CAF1 is an essential factor for spermatogenesis. Mol. Cell. Biol. 2004, 24, 5808–5820. [Google Scholar] [CrossRef]
- Nakamura, T.; Yao, R.; Ogawa, T.; Suzuki, T.; Ito, C.; Tsunekawa, N.; Inoue, K.; Ajima, R.; Miyasaka, T.; Yoshida, Y.; et al. Oligo-astheno-teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat. Genet. 2004, 36, 528–533. [Google Scholar] [CrossRef]
- Takahashi, A.; Adachi, S.; Morita, M.; Tokumasu, M.; Natsume, T.; Suzuki, T.; Yamamoto, T. Post-transcriptional Stabilization of Ucp1 mRNA Protects Mice from Diet-Induced Obesity. Cell Rep. 2015, 13, 2756–2767. [Google Scholar] [CrossRef] [PubMed]
- Washio-Oikawa, K.; Nakamura, T.; Usui, M.; Yoneda, M.; Ezura, Y.; Ishikawa, I.; Nakashima, K.; Noda, T.; Yamamoto, T.; Noda, M. Cnot7-null mice exhibit high bone mass phenotype and modulation of BMP actions. J. Bone Min. Res 2007, 22, 1217–1223. [Google Scholar] [CrossRef]
- Faraji, F.; Hu, Y.; Yang, H.H.; Lee, M.P.; Winkler, G.S.; Hafner, M.; Hunter, K.W. Post-transcriptional Control of Tumor Cell Autonomous Metastatic Potential by CCR4-NOT Deadenylase CNOT7. PLoS Genet. 2016, 12, e1005820. [Google Scholar] [CrossRef] [PubMed]
- Maryati, M.; Kaur, I.; Gopal, J.; Olotu-Umoren, L.; Oveh, B.; Hashmi, L.; Fischer, P.M.; Winkler, G.S. A fluorescence-based assay suitable for quantitative analysis of deadenylase enzyme activity. Nucleic Acids Res. 2014, 42, e30. [Google Scholar] [CrossRef] [PubMed]
- Kaur, I.; Jadhav, G.P.; Fischer, P.M.; Winkler, G.S. Discovery of Substituted 5-(2-Hydroxybenzoyl)-2-Pyridone Analogues as Inhibitors of the Human Caf1/CNOT7 Ribonuclease. Molecules 2024, 29, 4351. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, G.P.; Kaur, I.; Maryati, M.; Airhihen, B.; Fischer, P.M.; Winkler, G.S. Discovery, synthesis and biochemical profiling of purine-2,6-dione derivatives as inhibitors of the human poly(A)-selective ribonuclease Caf1. Bioorganic Med. Chem. Lett. 2015, 25, 4219–4224. [Google Scholar] [CrossRef]
- Volkamer, A.; Kuhn, D.; Rippmann, F.; Rarey, M. DoGSiteScorer: A web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics 2012, 28, 2074–2075. [Google Scholar] [CrossRef]
- Jonstrup, A.T.; Andersen, K.R.; Van, L.B.; Brodersen, D.E. The 1.4-A crystal structure of the S. pombe Pop2p deadenylase subunit unveils the configuration of an active enzyme. Nucleic Acids Res. 2007, 35, 3153–3164. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Bugnon, M.; Goullieux, M.; Rohrig, U.F.; Perez, M.A.S.; Daina, A.; Michielin, O.; Zoete, V. SwissParam 2023: A Modern Web-Based Tool for Efficient Small Molecule Parametrization. J. Chem. Inf. Model. 2023, 63, 6469–6475. [Google Scholar] [CrossRef]
- Zoete, V.; Cuendet, M.A.; Grosdidier, A.; Michielin, O. SwissParam: A fast force field generation tool for small organic molecules. J. Comput. Chem. 2011, 32, 2359–2368. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Bugnon, M.; Rohrig, U.F.; Goullieux, M.; Perez, M.A.S.; Daina, A.; Michielin, O.; Zoete, V. SwissDock 2024: Major enhancements for small-molecule docking with Attracting Cavities and AutoDock Vina. Nucleic Acids Res. 2024, 52, W324–W332. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39 (Suppl. S2), W270–W277. [Google Scholar] [CrossRef]
- Schoning-Stierand, K.; Diedrich, K.; Fahrrolfes, R.; Flachsenberg, F.; Meyder, A.; Nittinger, E.; Steinegger, R.; Rarey, M. ProteinsPlus: Interactive analysis of protein-ligand binding interfaces. Nucleic Acids Res. 2020, 48, W48–W53. [Google Scholar] [CrossRef]
- Stierand, K.; Rarey, M. Drawing the PDB: Protein-Ligand Complexes in Two Dimensions. ACS Med. Chem. Lett. 2010, 1, 540–545. [Google Scholar] [CrossRef]
- Stierand, K.; Maass, P.C.; Rarey, M. Molecular complexes at a glance: Automated generation of two-dimensional complex diagrams. Bioinformatics 2006, 22, 1710–1716. [Google Scholar] [CrossRef]
- Schrödinger, L.L.C. The PyMol Molecular Graphics System, Version 3.1.0 (Open Source). 2025.
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaur, I.; Hashmi, L.; Fischer, P.M.; Winkler, G.S. Discovery of Drug-like Inhibitors of the Human Caf1/CNOT7 poly(A)-Selective Nuclease Using Compound Screening. Biomolecules 2025, 15, 1563. https://doi.org/10.3390/biom15111563
Kaur I, Hashmi L, Fischer PM, Winkler GS. Discovery of Drug-like Inhibitors of the Human Caf1/CNOT7 poly(A)-Selective Nuclease Using Compound Screening. Biomolecules. 2025; 15(11):1563. https://doi.org/10.3390/biom15111563
Chicago/Turabian StyleKaur, Ishwinder, Lubna Hashmi, Peter M. Fischer, and Gerlof Sebastiaan Winkler. 2025. "Discovery of Drug-like Inhibitors of the Human Caf1/CNOT7 poly(A)-Selective Nuclease Using Compound Screening" Biomolecules 15, no. 11: 1563. https://doi.org/10.3390/biom15111563
APA StyleKaur, I., Hashmi, L., Fischer, P. M., & Winkler, G. S. (2025). Discovery of Drug-like Inhibitors of the Human Caf1/CNOT7 poly(A)-Selective Nuclease Using Compound Screening. Biomolecules, 15(11), 1563. https://doi.org/10.3390/biom15111563






