Effects of Bile on Pathogenic Vibrio, Aeromonas, and Clostridioides spp. Toxin Effector Domains
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Expression and Purification
2.2. Bile Salt Preparation
2.3. Cell Culture
2.4. Cytotoxicity Assays
2.5. Liquid Chromatography Coupled Tandem Mass Spectrometry (LC-MS/MS)
2.6. Collisional Quenching
2.7. Guanidinium Chloride Induced Denaturation
2.8. Circular Dichroism (CD) Spectroscopy
2.9. Reconstruction of the Secondary Structure Composition
2.10. High-Speed Ultracentrifugation
2.11. Native Polyacrylamide Gel Electrophoresis (Native PAGE)
2.12. Cysteine Protease Domain (CPD) Cleavage Activity Assays
2.13. Limited Proteolysis
2.14. TMR-5-Maleimide and DCP-Rho1 Labeling Assays
2.15. ACDMARTX Actin Crosslinking Activity Assays
2.16. VopF-Mediated Actin Polymerization
2.17. Statistical Analysis
3. Results
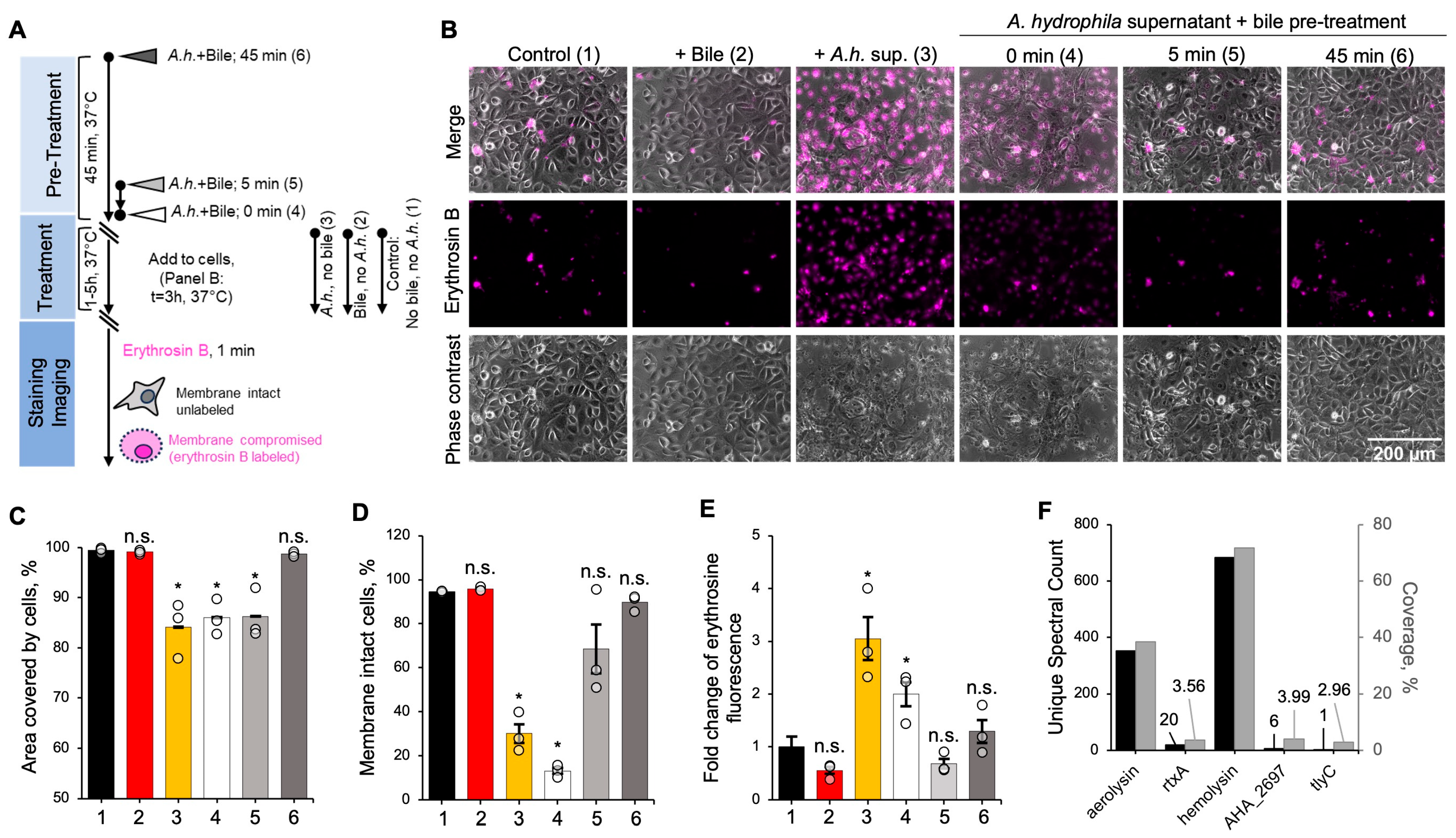
3.1. Bile Inactivates A. hydrophila Exotoxins and Protects Cultured Enterocytes
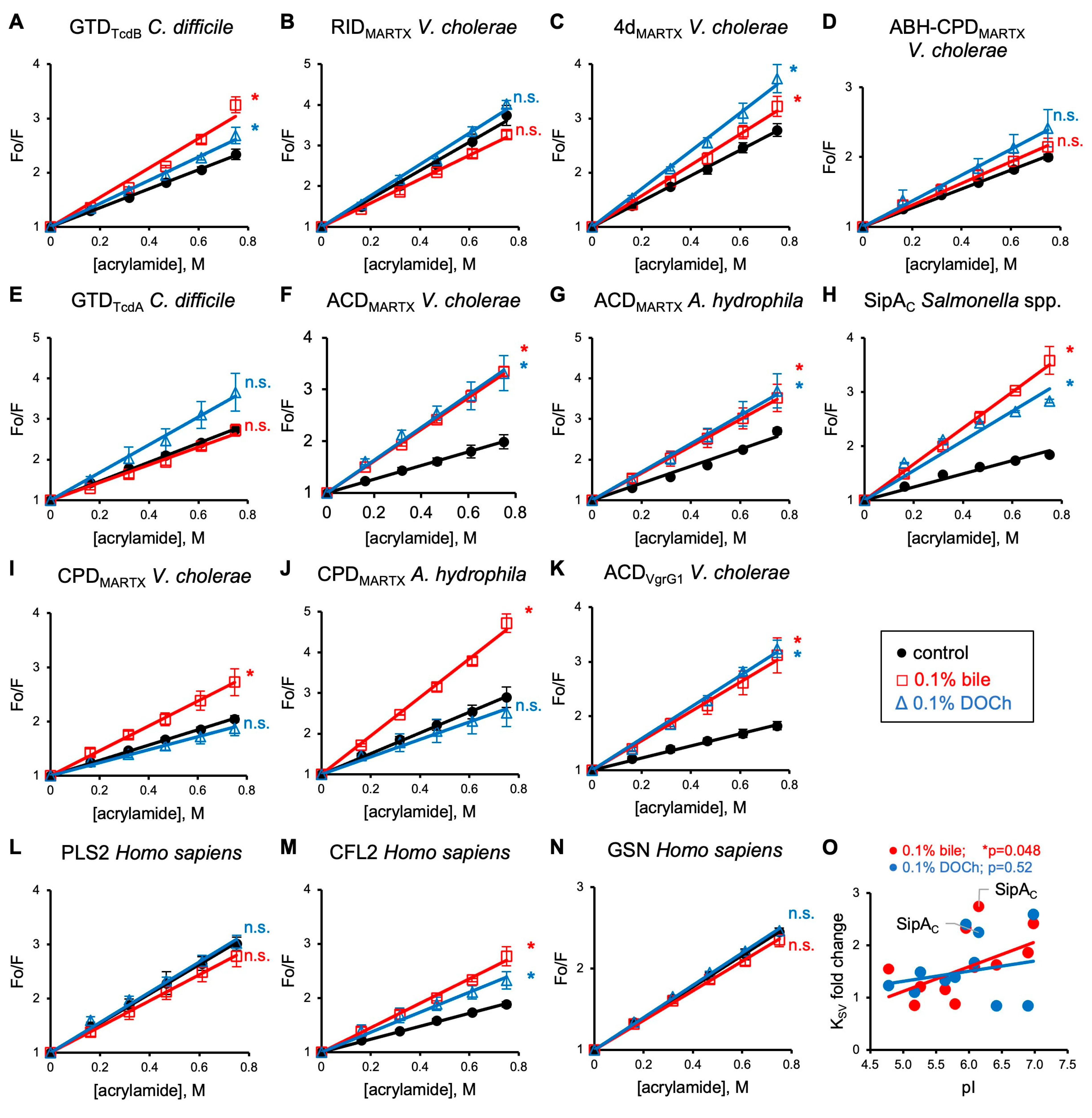
3.2. Bile and Deoxycholate Promote Exposure of Effectors’ Tryptophan Residues
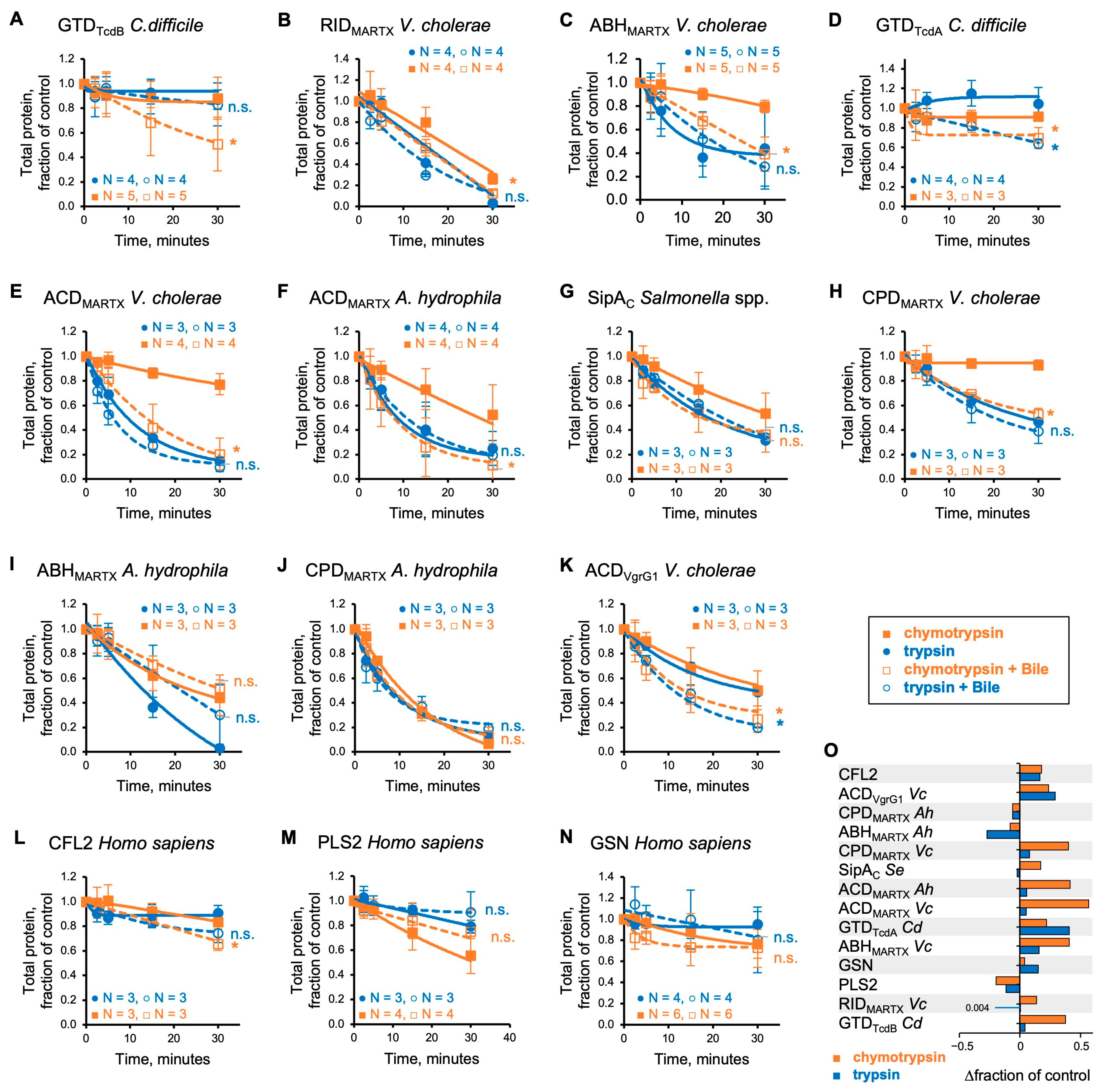
3.3. Bile Facilitates Proteolytic Cleavage of Bacterial Effectors at Hydrophobic Residues
3.4. Chemical Unfolding of Bacterial Effectors by Guanidinium Chloride Is Only Mildly Affected by Bile
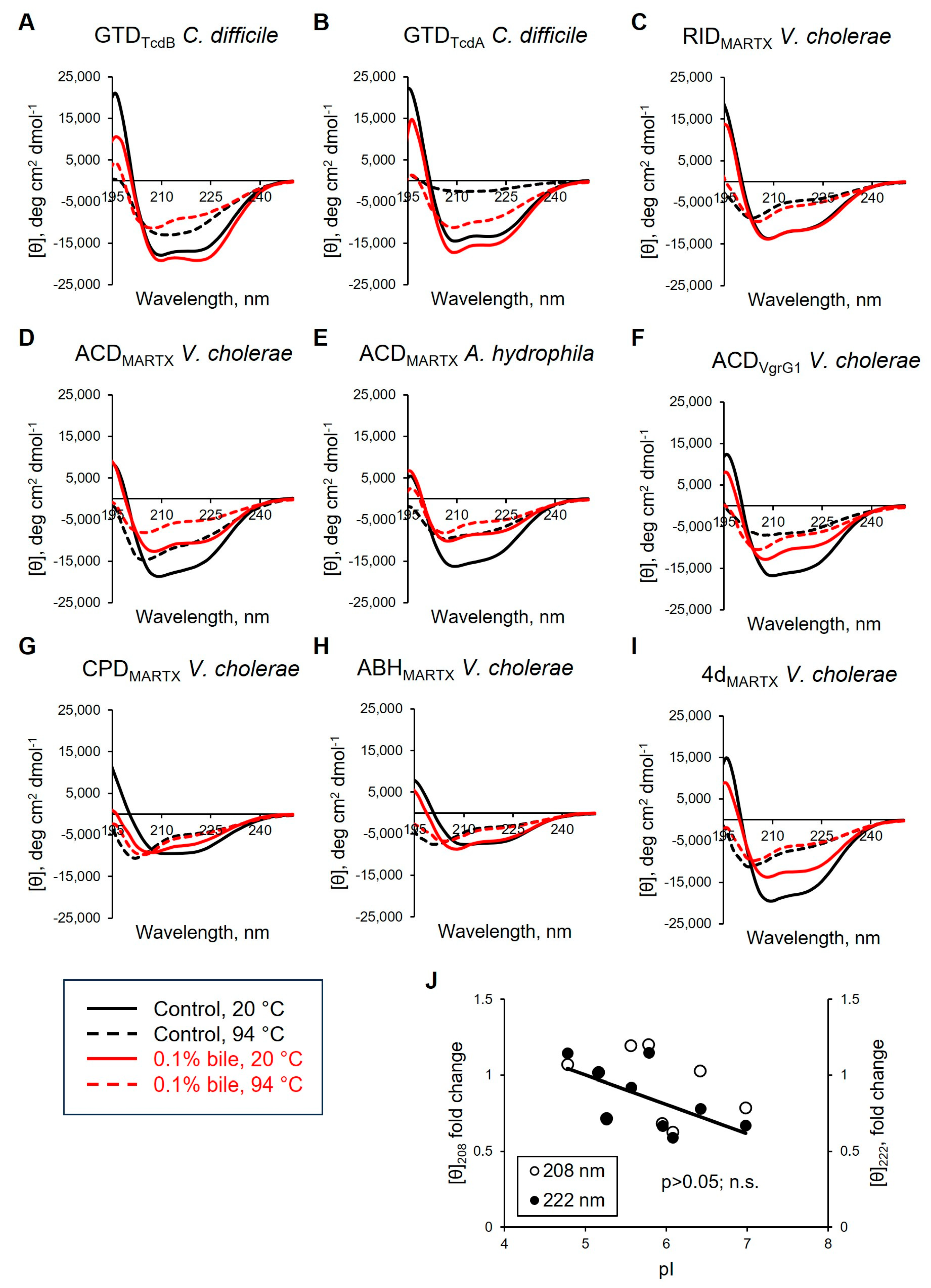
3.5. Effects of Bile and Deoxycholate on the Secondary Structure Elements of Bacterial Effectors
3.6. Bile Potentiates Precipitation of Effector Proteins
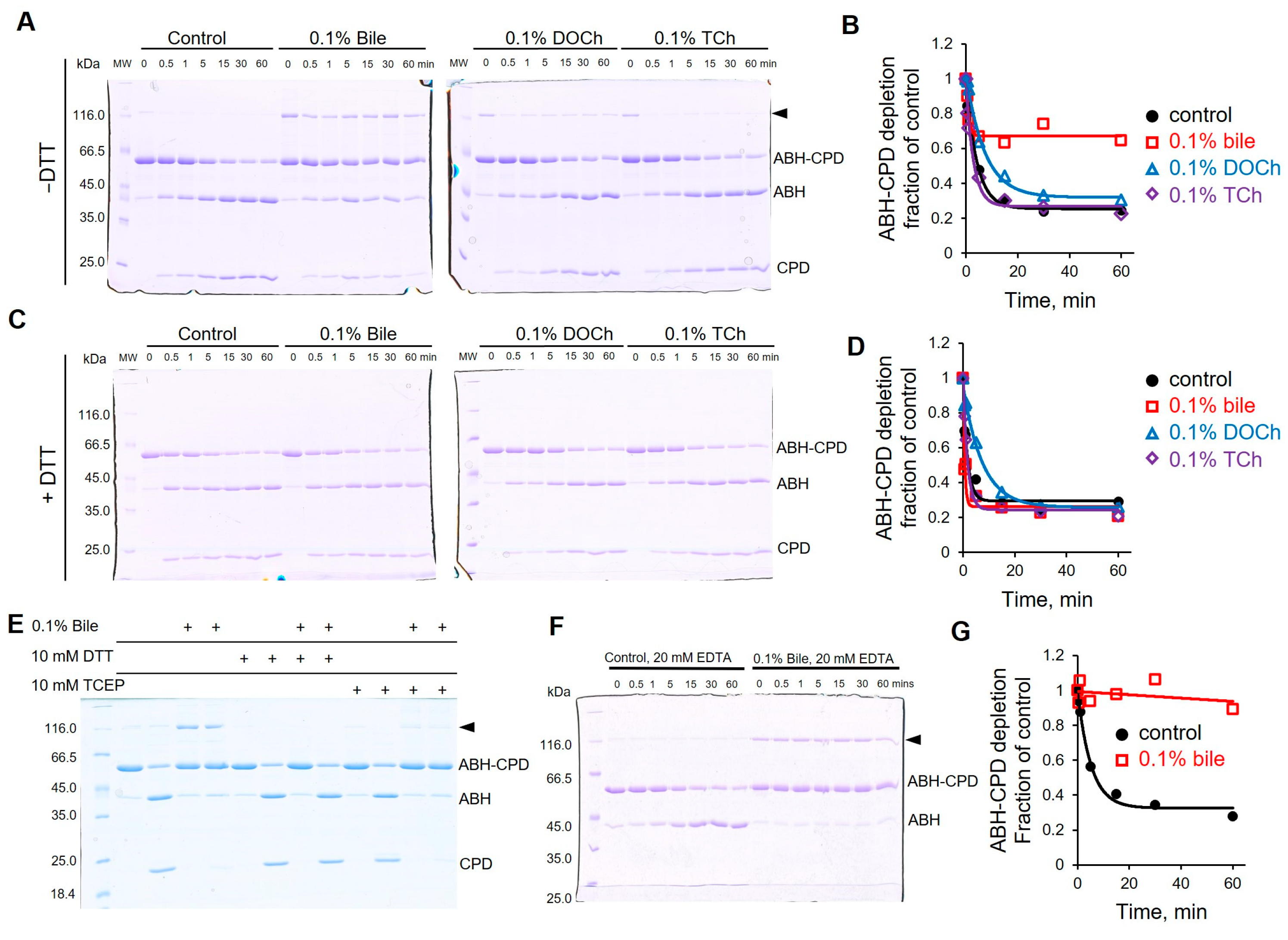
3.7. Bile Inhibits the Specific Activity of CPD by Promoting Oxidation of the Catalytic Cysteine
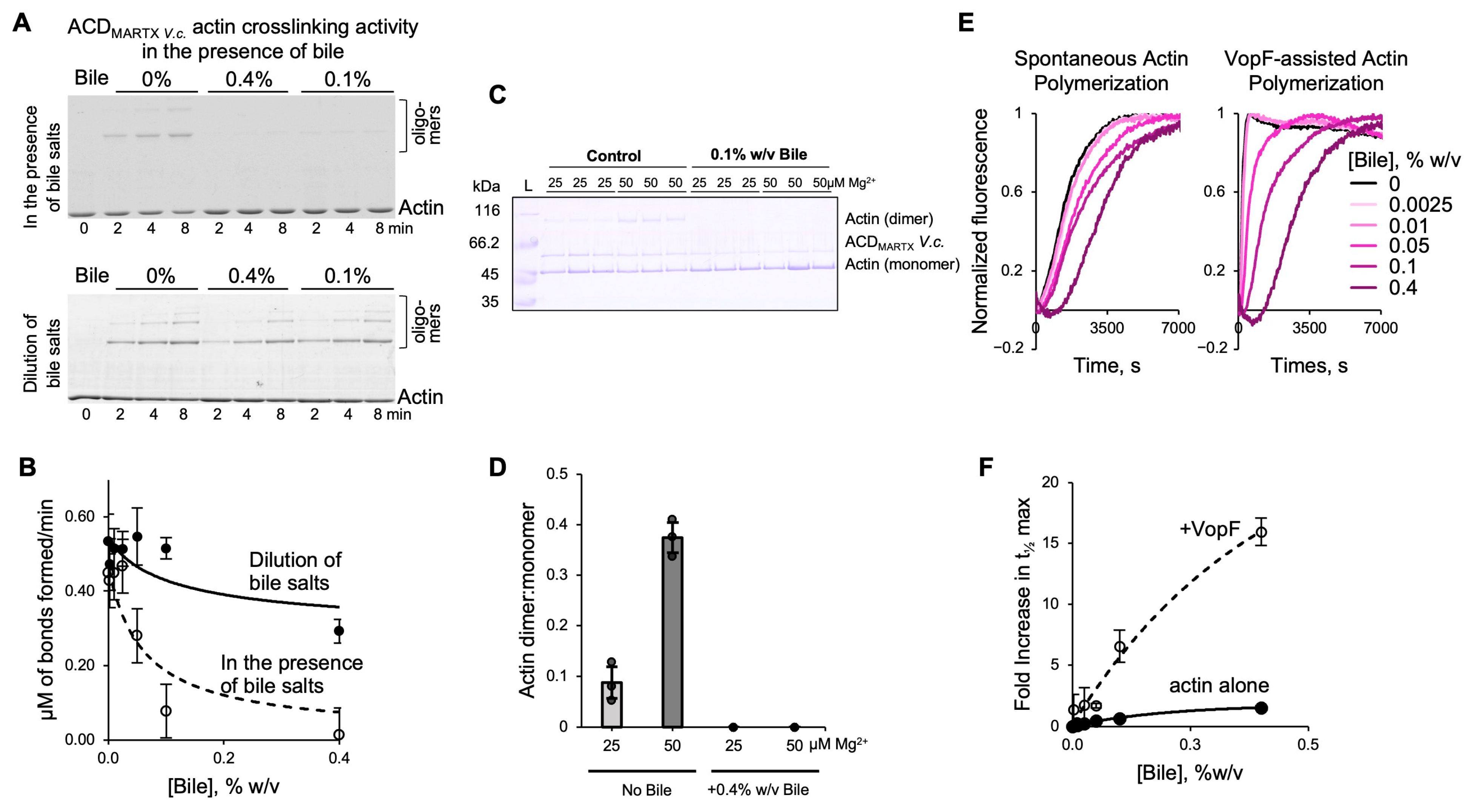
3.8. Bile Inhibits the Activities of V. cholerae Effectors ACD and VopF
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABH | α/β-hydrolase domain |
| ACD | Actin crosslinking domain |
| β-ME | β-mercaptoethanol |
| CD | Circular dichroism |
| CFL2 | Cofillin 2 |
| CID | Collision-induced dissociation |
| CPD | Cysteine protease domain |
| DCP | Diethyl chlorophosphite |
| DMEM | Dulbecco’s modified Eagle medium |
| DOCh | Deoxycholate |
| DTT | Dithiothreitol |
| EF | Edema factor |
| EDTA | Ethylenediaminetetraacetic acid |
| FBS | Fetal bovine serum |
| GdmCl | Guanidinium hydrochloride |
| GSH | Reduced glutathione |
| GSN | Gelsolin |
| GSSG | Oxidized glutathione |
| GTD | Glycosyltransferase domain |
| HD5 | Human defensin 5 |
| HD6 | Human defensin 6 |
| HNP1 | Human neutrophil peptide 1 |
| HNP2 | Human neutrophil peptide 2 |
| HNP3 | Human neutrophil peptide 3 |
| IAA | Iodoacetamide |
| IP6 | Inositol-hexakisphosphate |
| KSV | Stern–Volmer coefficient |
| LatB | Latrunculin B |
| LB | Luria–Bertani medium |
| LF | Lethal factor |
| LPS | Lipopolysaccharide |
| MARTX | Multifunctional autoprocessing repeats-in-toxin |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NEM | N-ethylmaleimide |
| PBS | Phosphate-buffered saline |
| pI | Isoelectric point |
| PA | Protective antigen |
| PLS2 | Plastin 2 |
| PMSF | Phenylmethylsulfonyl fluoride |
| RID | Rho-inactivation domain |
| SDS | Sodium dodecyl sulfate |
| SipAC | SipA C-terminal domain |
| T1SS | Type I secretion system |
| T2SS | Type II secretion system |
| T3SS | Type III secretion system |
| T4SS | Type IV secretion system |
| T6SS | Type VI secretion system |
| TCEP | Tris(2-carboxyethyl)phosphine |
| TCh | Taurocholate |
| TEAB | Triethylammonium bicarbonate |
| TMR | Tetramethylrhodamine |
References
- Barthold, L.; Heber, S.; Schmidt, C.Q.; Gradl, M.; Weidinger, G.; Barth, H.; Fischer, S. Human A-Defensin-6 Neutralizes Clostridioides Difficile Toxins TcdA and TcdB by Direct Binding. Int. J. Mol. Sci. 2022, 23, 4509. [Google Scholar] [CrossRef] [PubMed]
- Kling, C.; Sommer, A.; Almeida-hernandez, Y.; Rodr, A.; Perez-erviti, J.A.; Bhadane, R.; Ständker, L.; Wiese, S.; Barth, H.; Pupo-meriño, M.; et al. Inhibition of Pertussis Toxin by Human α -Defensins-1 and -5: Differential Mechanisms of Action. Int. J. Mol. Sci. 2023, 24, 10557. [Google Scholar] [CrossRef] [PubMed]
- Kling, C.; Pulliainen, A.T.; Barth, H.; Ernst, K. Human Peptides α -Defensin-1 and -5 Inhibit Pertussis Toxin. Toxins 2021, 13, 480. [Google Scholar] [CrossRef]
- Kudryashova, E.; Quintyn, R.; Seveau, S.; Lu, W.; Wysocki, V.H.; Kudryashov, D.S. Human Defensins Facilitate Local Unfolding of Thermodynamically Unstable Regions of Bacterial Protein Toxins. Immunity 2014, 41, 709–721. [Google Scholar] [CrossRef]
- Kim, C.; Gajendran, N.; Mittrucker, H.-W.; Weiward, M.; Song, Y.-H.; Hurwitz, R.; Wilmanns, M.; Fischer, G.; Kaufmann, S.H.E. Human Alpha-Defensins Neutralize Anthrax Lethal Toxin and Protect against Its Fatal Consequences. Proc. Natl. Acad. Sci. USA 2005, 102, 4830–4835. [Google Scholar] [CrossRef]
- Kim, C.; Slavinskaya, Z.; Merrill, A.R.; Kaufmann, S.H.E. Human α-Defensins Neutralize Toxins of the Mono-ADP- Ribosyltransferase Family. Biochem. J. 2006, 399, 225–229. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Jung, G.; Ruchala, P.; Wang, W.; Micewicz, E.D.; Waring, A.J.; Gillespie, E.J.; Bradley, K.A.; Ratner, A.J.; Rest, R.F.; et al. Human Alpha-Defensins Inhibit Hemolysis Mediated by Cholesterol-Dependent Cytolysins. Infect. Immun. 2009, 77, 4028–4040. [Google Scholar] [CrossRef]
- Kudryashova, E.; Seveau, S.M.; Kudryashov, D.S. Targeting and Inactivation of Bacterial Toxins by Human Defensins. Biol. Chem. 2017, 398, 1069–1085. [Google Scholar] [CrossRef]
- Kintzer, A.F.; Thoren, K.L.; Sterling, H.J.; Dong, K.C.; Feld, G.K.; Tang, I.I.; Zhang, T.T.; Williams, E.R.; Berger, J.M.; Krantz, B.A. The Protective Antigen Component of Anthrax Toxin Forms Functional Octameric Complexes. J. Mol. Biol. 2009, 392, 614–629. [Google Scholar] [CrossRef]
- Qa’dan, M.; Christensen, K.A.; Zhang, L.; Roberts, T.M.; Collier, R.J. Membrane Insertion by Anthrax Protective Antigen in Cultured Cells. Mol. Cell. Biol. 2005, 25, 5492–5498. [Google Scholar] [CrossRef]
- Kudryashova, E.; Heisler, D.; Zywiec, A.; Kudryashov, D.S. Thermodynamic Properties of the Effector Domains of MARTX Toxins Suggest Their Unfolding for Translocation across the Host Membrane. Mol. Microbiol. 2014, 92, 1056–1071. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Lu, W. α-Defensins in Human Innate Immunity. Immunol. Rev. 2012, 245, 84–112. [Google Scholar] [CrossRef]
- Ganz, T.; Selsted, M.E.; Szklarek, D.; Harwig, S.S.; Daher, K.; Bainton, D.F.; Lehrer, R.I. Defensins. Natural Peptide Antibiotics of Human Neutrophils. J. Clin. Investig. 1985, 76, 1427–1435. [Google Scholar] [CrossRef] [PubMed]
- Rajabi, M.; Ericksen, B.; Wu, X.; De Leeuw, E.; Zhao, L.; Pazgier, M.; Lu, W. Functional Determinants of Human Enteric α-Defensin HD5: Crucial Role for Hydrophobicity at Dimer Interface. J. Biol. Chem. 2012, 287, 21615–21627. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Pazgier, M.; De Leeuw, E.; Rajabi, M.; Li, J.; Zou, G.; Jung, G.; Yuan, W.; Lu, W.Y.; Lehrer, R.I.; et al. Trp-26 Imparts Functional Versatility to Human α-Defensin HNP. J. Biol. Chem. 2010, 285, 16275–16285. [Google Scholar] [CrossRef] [PubMed]
- de Leeuw, E.; Rajabi, M.; Zou, G.; Pazgier, M.; Lu, W. Selective Arginines Are Important for the Antibacterial Activity and Host Cell Interaction of Human α-Defensin 5. FEBS Lett. 2009, 583, 2507–2512. [Google Scholar] [CrossRef]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting Dysbiosis, Bile-Acid Dysmetabolism and Gut Inflammation in Inflammatory Bowel Diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef]
- Begley, M.; Gahan, C.G.M.; Hill, C. The Interaction between Bacteria and Bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef]
- Cremers, C.M.; Knoefler, D.; Vitvitsky, V.; Banerjee, R.; Jakob, U. Bile Salts Act as Effective Protein-Unfolding Agents and Instigators of Disulfide Stress in Vivo. Proc. Natl. Acad. Sci. USA 2014, 111, E1610–E1619. [Google Scholar] [CrossRef]
- Wibbenmeyer, J.A.; Provenzano, D.; Landry, C.F.; Klose, K.E.; Delcour, A.H. Vibrio cholerae OmpU and OmpT Porins Are Differentially Affected by Bile. Infect. Immun. 2002, 70, 121–126. [Google Scholar] [CrossRef]
- Hung, D.T.; Mekalanos, J.J. Bile Acids Induce Cholera Toxin Expression in Vibrio cholerae in a ToxT-Independent Manner. Proc. Natl. Acad. Sci. USA 2005, 102, 3028–3033. [Google Scholar] [CrossRef]
- Alam, A.; Tam, V.; Hamilton, E.; Dziejman, M. VttRa and VttRB Encode ToxR Family Proteins That Mediate Bile-Induced Expression of Type Three Secretion System Genes in a Non-O1/Non-O139 Vibrio cholerae Strain. Infect. Immun. 2010, 78, 2554–2570. [Google Scholar] [CrossRef]
- Gupta, S.; Chowdhury, R. Bile Affects Production of Virulence Factors and Motility of Vibrio cholerae. Infect. Immun. 1997, 65, 1131–1134. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.A.; Hamilton, E.; Dziejman, M. The Vibrio cholerae Trh Gene Is Coordinately Regulated in Vitro with Type Iii Secretion System Genes by VttrA/VttrB but Does Not Contribute to Caco2-Bbe Cell Cytotoxicity. Infect. Immun. 2012, 80, 4444–4455. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Liu, Z.; Hughes, C.; Stern, A.M.; Wang, H.; Zhong, Z.; Kan, B.; Fenical, W.; Zhu, J. Bile Salt-Induced Intermolecular Disulfide Bond Formation Activates Vibrio cholerae Virulence. Proc. Natl. Acad. Sci. USA 2013, 110, 2348–2353. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Dutta, P.K.; Chowdhury, R. Effect of Fatty Acids and Cholesterol Present in Bile on Expression of Virulence Factors and Motility of Vibrio cholerae. Infect. Immun. 2007, 75, 1946–1953. [Google Scholar] [CrossRef]
- Li, P.; Rivera-Cancel, G.; Kinch, L.N.; Salomon, D.; Tomchick, D.R.; Grishin, N.V.; Orth, K. Bile Salt Receptor Complex Activates a Pathogenic Type III Secretion System. eLife 2016, 5, e15718. [Google Scholar] [CrossRef]
- Gotoh, K.; Kodama, T.; Hiyoshi, H.; Izutsu, K.; Park, K.S.; Dryselius, R.; Akeda, Y.; Honda, T.; Iida, T. Bile Acid-Induced Virulence Gene Expression of Vibrio Parahaemolyticus Reveals a Novel Therapeutic Potential for Bile Acid Sequestrants. PLoS ONE 2010, 5, e13365. [Google Scholar] [CrossRef]
- Livny, J.; Zhou, X.; Mandlik, A.; Hubbard, T.; Davis, B.M.; Waldor, M.K. Comparative RNA-Seq Based Dissection of the Regulatory Networks and Environmental Stimuli Underlying Vibrio Parahaemolyticus Gene Expression during Infection. Nucleic Acids Res. 2014, 42, 12212–12223. [Google Scholar] [CrossRef]
- Johnson, R.; Ravenhall, M.; Pickard, D.; Dougan, G.; Byrne, A.; Frankel, G. Comparison of Salmonella enterica Serovars Typhi and Typhimurium Reveals Typhoidal Serovar-Specific Responses to Bile. Infect. Immun. 2018, 86, e00490-17. [Google Scholar] [CrossRef]
- Wang, Y.; Nordhues, B.A.; Zhong, D.; De Guzman, R.N. NMR Characterization of the Interaction of the Salmonella Type III Secretion System Protein SipD and Bile Salts. Biochemistry 2010, 49, 4220–4226. [Google Scholar] [CrossRef] [PubMed]
- Annaba, F.; Sarwar, Z.; Gill, R.K.; Ghosh, A.; Saksena, S.; Borthakur, A.; Hecht, G.A.; Dudeja, P.K.; Alrefai, W.A. Enteropathogenic Escherichia Coli Inhibits Ileal Sodium-Dependent Bile Acid Transporter ASBT. Am. J. Physiol.-Lung Cell. Mol. Physiol. Gastrointest. Liver Physiol. 2012, 302, 1216–1222. [Google Scholar] [CrossRef] [PubMed]
- De Jesus, M.C.; Urban, A.A.; Marasigan, M.E.; Foster, D.E.B. Acid and Bile-Salt Stress of Enteropathogenic Escherichia Coli Enhances Adhesion to Epithelial Cells and Alters Glycolipid Receptor Binding Specificity. J. Infect. Dis. 2005, 192, 1430–1440. [Google Scholar] [CrossRef] [PubMed]
- Nicklasson, M.; Sjöling, Å.; von Mentzer, A.; Qadri, F.; Svennerholm, A.M. Expression of Colonization Factor CS5 of Enterotoxigenic Escherichia Coli (ETEC) Is Enhanced in Vivo and by the Bile Component Na Glycocholate Hydrate. PLoS ONE 2012, 7, e35827. [Google Scholar] [CrossRef]
- Grewal, H.M.S.; Valvatne, H.; Bhan, M.K.; Van Dijk, L.; Gaastra, W.; Sommerfelt, H. A New Putative Fimbrial Colonization Factor, CS19, of Human Enterotoxigenic Escherichia Coli. Infect. Immun. 1997, 65, 507–513. [Google Scholar] [CrossRef]
- Tam, J.; Icho, S.; Utama, E.; Orrell, K.E.; Gómez-Biagi, R.F.; Theriot, C.M.; Kroh, H.K.; Rutherford, S.A.; Borden Lacy, D.; Melnyk, R.A. Intestinal Bile Acids Directly Modulate the Structure and Function of C. Difficile TcdB Toxin. Proc. Natl. Acad. Sci. USA 2020, 117, 6792–6800. [Google Scholar] [CrossRef]
- Spudich, J.A.; Watt, S. The Regulation of Rabbit Skeletal Muscle Contraction. J. Biol. Chem. 1971, 246, 4866–4871. [Google Scholar] [CrossRef]
- Heisler, D.B.; Kudryashova, E.; Grinevich, D.O.; Suarez, C.; Winkleman, J.D.; Birukov, K.G.; Kotha, S.R.; Parinandi, N.L.; Vavylonis, D.; Kovar, D.R.; et al. ACD Toxin-Produced Actin Oligomers Poison Formin-Controlled Actin Polymerization. Science 2015, 349, 535–540. [Google Scholar] [CrossRef]
- Niedzialkowska, E.; Runyan, L.A.; Kudryashova, E.; Egelman, E.H.; Kudryashov, D.S. Stabilization of F-Actin by Salmonella Effector SipA Resembles the Structural Effects of Inorganic Phosphate and Phalloidin. Struct. Des. 2024, 32, 725–738. [Google Scholar] [CrossRef]
- Kudryashova, E.; Ankita; Ulrichs, H.; Shekhar, S.; Kudryashov, D.S. Pointed-End Processive Elongation of Actin Filaments by Vibrio Effectors VopF and VopL. Sci. Adv. 2022, 8, eadc9239. [Google Scholar] [CrossRef]
- Pruitt, R.N.; Chumbler, N.M.; Rutherford, S.A.; Farrow, M.A.; Friedman, D.B.; Spiller, B.; Lacy, D.B. Structural Determinants of Clostridium Difficile Toxin A Glucosyltransferase Activity. J. Biol. Chem. 2012, 287, 8013–8020. [Google Scholar] [CrossRef]
- Yang, G.; Zhou, B.; Wang, J.; He, X.; Sun, X.; Nie, W.; Tzipori, S.; Feng, H. Expression of Recombinant Clostridium Difficile Toxin A and B in Bacillus Megaterium. BMC Microbiol. 2008, 8, 192. [Google Scholar] [CrossRef]
- Schwebach, C.L.; Kudryashova, E.; Agrawal, R.; Zheng, W.; Egelman, E.H.; Kudryashov, D.S. Allosteric Regulation Controls Actin-Bundling Properties of Human Plastins. Nat. Struct. Mol. Biol. 2022, 29, 519–528. [Google Scholar] [CrossRef]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ Ecosystem: An Open Platform for Biomedical Image Analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Kong, A.T.; Leprevost, F.V.; Avtonomov, D.M.; Mellacheruvu, D.; Nesvizhskii, A.I. MSFragger: Ultrafast and Comprehensive Peptide Identification in Mass Spectrometry-Based Proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.L.; Yu, F.; Teo, G.C.; Li, K.; Demichev, V.; Ralser, M.; Nesvizhskii, A.I. MSBooster: Improving Peptide Identification Rates Using Deep Learning-Based Features. Nat. Commun. 2023, 14, 4539. [Google Scholar] [CrossRef] [PubMed]
- Gessulat, S.; Schmidt, T.; Zolg, D.P.; Samaras, P.; Schnatbaum, K.; Zerweck, J.; Knaute, T.; Rechenberger, J.; Delanghe, B.; Huhmer, A.; et al. Prosit: Proteome-Wide Prediction of Peptide Tandem Mass Spectra by Deep Learning. Nat. Methods 2019, 16, 509–518. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Varadi, M.; Bertoni, D.; Magana, P.; Paramval, U.; Pidruchna, I.; Radhakrishnan, M.; Tsenkov, M.; Nair, S.; Mirdita, M.; Yeo, J.; et al. AlphaFold Protein Structure Database in 2024: Providing Structure Coverage for over 214 Million Protein Sequences. Nucleic Acids Res. 2024, 52, D368–D375. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Micsonsai, A.; Moussong, E.; Wien, F.; Boros, E.; Vadaszi, H.; Murvai, N.; Lee, Y.-H.; Molnar, T.; Matthieu, R.; Goto, Y.; et al. BeStSel: Webserver for Secondary Structure and Fold Prediction for Protein CD Spectroscopy. Nucleic Acids Res. 2022, 50, W90–W98. [Google Scholar] [CrossRef]
- Kudryashova, E.; Kalda, C.; Kudryashov, D.S. Glutamyl Phosphate Is an Activated Intermediate in Actin Crosslinking by Actin Crosslinking Domain (ACD) Toxin. PLoS ONE 2012, 7, e45721. [Google Scholar] [CrossRef] [PubMed]
- Cordero, C.L.; Kudryashov, D.S.; Reisler, E.; Fullner Satchell, K.J. The Actin Cross-Linking Domain of the Vibrio cholerae RTX Toxin Directly Catalyzes the Covalent Cross-Linking of Actin. J. Biol. Chem. 2006, 281, 32366–32374. [Google Scholar] [CrossRef] [PubMed]
- Grim, C.J.; Kozlov, E.V.; Sha, J.; Fitts, E.C.; van Lier, C.J.; Kirtley, M.L.; Joseph, S.J.; Read, T.D.; Burd, E.M.; Tall, B.D.; et al. Characterization of Aeromonas hydrophila Wound Pathotypes by Comparative Genomic and Functional Analyses of Virulence Genes. mBio 2013, 4, e00064-13. [Google Scholar] [CrossRef]
- Tomás, J.M. The Main Aeromonas Pathogenic Factors. ISRN Microbiol. 2012, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen-Ivey, C.R.; Figueras, M.J.; McGarey, D.; Liles, M.R. Virulence Factors of Aeromonas hydrophila: In the Wake of Reclassification. Front. Microbiol. 2016, 7, 1337. [Google Scholar] [CrossRef]
- Eftink, M.R.; Ghiron, C.A. Exposure of Tryptophanyl Residues in Proteins Quantitative Determination by Fluorescence Quenching Studies. Biochemistry 1976, 15, 672–680. [Google Scholar] [CrossRef]
- Robic, S.; Linscott, K.B.; Aseem, M.; Humphreys, E.A.; McCartha, S.R. Bile Acids as Modulators of Enzyme Activity and Stability. Protein J. 2011, 30, 539–545. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using Circular Dichroism Collected as a Function of Temperature to Determine the Thermodynamics of Protein Unfolding and Binding Interactions. Nat. Protoc. 2007, 1, 2527–2535. [Google Scholar] [CrossRef]
- Lupardus, P.J.; Shen, A.; Bogyo, M.; Garcia, K.C. Small Molecule—Induced Allosteric Cysteine Protease Domain. Science 2008, 322, 265–268. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, B.S.; Choi, S.; Lee, E.Y.; Park, S.; Hwang, J.; Kwon, Y.; Hyun, J.; Lee, C.; Kim, J.F.; et al. Makes Caterpillars Floppy-like Effector-Containing MARTX Toxins Require Host ADP-Ribosylation Factor (ARF) Proteins for Systemic Pathogenicity. Proc. Natl. Acad. Sci. USA 2019, 116, 18031–18040. [Google Scholar] [CrossRef] [PubMed]
- Satchell, K.J.F. Actin Crosslinking Toxins of Gram-Negative Bacteria. Toxins 2009, 1, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Malarkani, K.; Sarkar, I.; Selvam, S. Denaturation Studies on Bovine Serum Albumin–Bile Salt System: Bile Salt Stabilizes Bovine Serum Albumin through Hydrophobicity. J. Pharm. Anal. 2018, 8, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Egerer, M.; Giesemann, T.; Jank, T.; Fullner Satchell, K.J.; Aktories, K. Auto-Catalytic Cleavage of Clostridium Difficile Toxins A and B Depends on Cysteine Protease Activity. J. Biol. Chem. 2007, 282, 25314–25321. [Google Scholar] [CrossRef]
- Sheahan, K.L.; Cordero, C.L.; Satchell, K.J.F. Autoprocessing of the Vibrio cholerae RTX Toxin by the Cysteine Protease Domain. EMBO J. 2007, 26, 2552–2561. [Google Scholar] [CrossRef]
- Kudryashova, E.; Heisler, D.B.; Williams, B.; Harker, A.J.; Shafer, K.; Quinlan, M.E.; Kovar, D.R.; Vavylonis, D.; Kudryashov, D.S. Actin Cross-Linking Toxin Is a Universal Inhibitor of Tandem-Organized and Oligomeric G-Actin Binding Proteins. Curr. Biol. 2018, 28, 1536–1547.e9. [Google Scholar] [CrossRef]
- Kudryashov, D.S.; Cordero, C.L.; Reisler, E.; Fullner Satchell, K.J. Characterization of the Enzymatic Activity of the Actin Cross-Linking Domain from the Vibrio cholerae MARTXVc Toxin. J. Biol. Chem. 2008, 283, 445–452. [Google Scholar] [CrossRef]
- Kudryashov, D.S.; Oztug Durer, Z.A.; Ytterberg, A.J.; Sawaya, M.R.; Pashkov, I.; Prochazkova, K.; Yeates, T.O.; Ogorzalek Loo, R.R.; Loo, J.A.; Fullner Satchell, K.J.; et al. Connecting Actin Monomers by Iso-Peptide Bond Is a Toxicity Mechanism of the Vibrio cholerae MARTX Toxin. Proc. Natl. Acad. Sci. USA 2008, 105, 18537–18542. [Google Scholar] [CrossRef]
- Burke, T.A.; Harker, A.J.; Dominguez, R.; Kovar, D.R. The Bacterial Virulence Factors VopL and VopF Nucleate Actin from the Pointed End. J. Cell Biol. 2017, 216, 1267–1276. [Google Scholar] [CrossRef]
- Dubois, T.; Tremblay, Y.D.N.; Hamiot, A.; Martin-Verstraete, I.; Deschamps, J.; Monot, M.; Briandet, R.; Dupuy, B. A Microbiota-Generated Bile Salt Induces Biofilm Formation in Clostridium Difficile. npj Biofilms Microbiomes 2019, 5, 14. [Google Scholar] [CrossRef]
- Darkoh, C.; Brown, E.L.; Kaplan, H.B.; DuPont, H.L. Bile Salt Inhibition of Host Cell Damage by Clostridium Difficile Toxins. PLoS ONE 2013, 8, e79631. [Google Scholar] [CrossRef]
- Icho, S.; Ward, J.S.; Tam, J.; Kociolek, L.; Theriot, C.M.; Melynk, R.A. Intestinal Bile Acids Provide a Surmountable Barrier against C. Difficile TcdB-Induced Disease Pathogenesis. Proc. Natl. Acad. Sci. USA 2023, 120, e2301252120. [Google Scholar] [CrossRef]
- Pike, C.M.; Tam, J.; Melnyk, R.A.; Theriot, C.M. Tauroursodeoxycholic Acid Inhibits Clostridioides Difficile Toxin-Induced Apoptosis. Infect. Immun. 2022, 90, e0015322. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.J.; Elliott, J.L.; Collier, R.J. Anthrax Protective Antigen: Prepore-to-Pore Conversion. Biochemistry 1999, 38, 10432–10441. [Google Scholar] [CrossRef] [PubMed]
- Krantz, B.A.; Trivedi, A.D.; Cunningham, K.; Christensen, K.A.; Collier, R.J. Acid-Induced Unfolding of the Amino-Terminal Domains of the Lethal and Edema Factors of Anthrax Toxin. J. Mol. Biol. 2004, 344, 739–756. [Google Scholar] [CrossRef] [PubMed]
- Gubensäk, N.; Sagmeister, T.; Buhlheller, C.; Geronimo, B.D.; Wagner, G.E.; Petrowitsch, L.; Gräwert, M.; Rotzinger, M.; Berger, T.M.I.; Schäfer, J.; et al. Vibrio cholerae’s ToxRS Bile Sensing System. eLife 2023, 12, e88721. [Google Scholar] [CrossRef]
- Kinch, L.N.; Cong, Q.; Jaishankar, J.; Orth, K. Co-Component Signal Transduction Systems: Fast-Evolving Virulence Regulation Cassettes Discovered in Enteric Bacteria. Proc. Natl. Acad. Sci. USA 2022, 119, e2203176119. [Google Scholar] [CrossRef]
- Xue, Y.; Tu, F.; Shi, M.; Wu, C.Q.; Ren, G.; Wang, X.; Fang, W.; Song, H.; Yang, M. Redox Pathway Sensing Bile Salts Activates Virulence Gene Expression in Vibrio cholerae. Mol. Microbiol. 2016, 102, 909–924. [Google Scholar] [CrossRef]
- Fischer, S.; Marc, A.Ü.; Papatheodorou, P.; Mittler, C.H.A.; Ziener, U.; Hägele, M.; Schwan, C.; Müller, M.; Kleger, A.; Benz, R.; et al. Human Peptide α-Defensin-1 Interferes with Clostridioides Difficile Toxins TcdA, TcdB, and CDT. FASEB 2020, 34, 6244–6261. [Google Scholar] [CrossRef]
- Giesemann, T.; Guttenberg, G.; Aktories, K. Human α-Defensins Inhibit Clostridium Difficile Toxin B. Gastroenterology 2008, 134, 2049–2058. [Google Scholar] [CrossRef]
- Kudryashova, E.; Koneru, P.C.; Kvaratskhelia, M.; Strömstedt, A.A.; Lu, W.; Kudryashov, D.S. Thermodynamic Instability of Viral Proteins Is a Pathogen-Associated Molecular Pattern Targeted by Human Defensins. Sci. Rep. 2016, 6, 32499. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taylor, J.E.; Heisler, D.B.; Choudhary, E.; Kudryashova, E.; Kudryashov, D.S. Effects of Bile on Pathogenic Vibrio, Aeromonas, and Clostridioides spp. Toxin Effector Domains. Biomolecules 2025, 15, 1539. https://doi.org/10.3390/biom15111539
Taylor JE, Heisler DB, Choudhary E, Kudryashova E, Kudryashov DS. Effects of Bile on Pathogenic Vibrio, Aeromonas, and Clostridioides spp. Toxin Effector Domains. Biomolecules. 2025; 15(11):1539. https://doi.org/10.3390/biom15111539
Chicago/Turabian StyleTaylor, Jaylen E., David B. Heisler, Eshan Choudhary, Elena Kudryashova, and Dmitri S. Kudryashov. 2025. "Effects of Bile on Pathogenic Vibrio, Aeromonas, and Clostridioides spp. Toxin Effector Domains" Biomolecules 15, no. 11: 1539. https://doi.org/10.3390/biom15111539
APA StyleTaylor, J. E., Heisler, D. B., Choudhary, E., Kudryashova, E., & Kudryashov, D. S. (2025). Effects of Bile on Pathogenic Vibrio, Aeromonas, and Clostridioides spp. Toxin Effector Domains. Biomolecules, 15(11), 1539. https://doi.org/10.3390/biom15111539







