Integrative Physiological, Metabolomic and Transcriptomic Analyses Uncover the Mechanisms Underlying Differential Responses of Two Anubias Genotypes to Low-Temperature Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatment
2.2. Measurement of Physiological Indicators
2.3. Non-Targeted Metabolomics Analysis Based on UPLC-MS/MS
2.3.1. Metabolite Extraction
2.3.2. LC-MS/MS Analysis
2.3.3. Data Preprocessing, Annotation and Differential Metabolite Screening
2.4. RNA-Sequence Analysis
2.4.1. Library Construction and Sequencing
2.4.2. Quality Control and De Novo Assembly
2.4.3. Gene Annotation and Differential Expression Analysis
2.5. Correlation Network Analysis
2.6. Statistical Analysis
3. Results
3.1. Effects of Low Temperature on Leaf Phenotype and Physiological Indicators
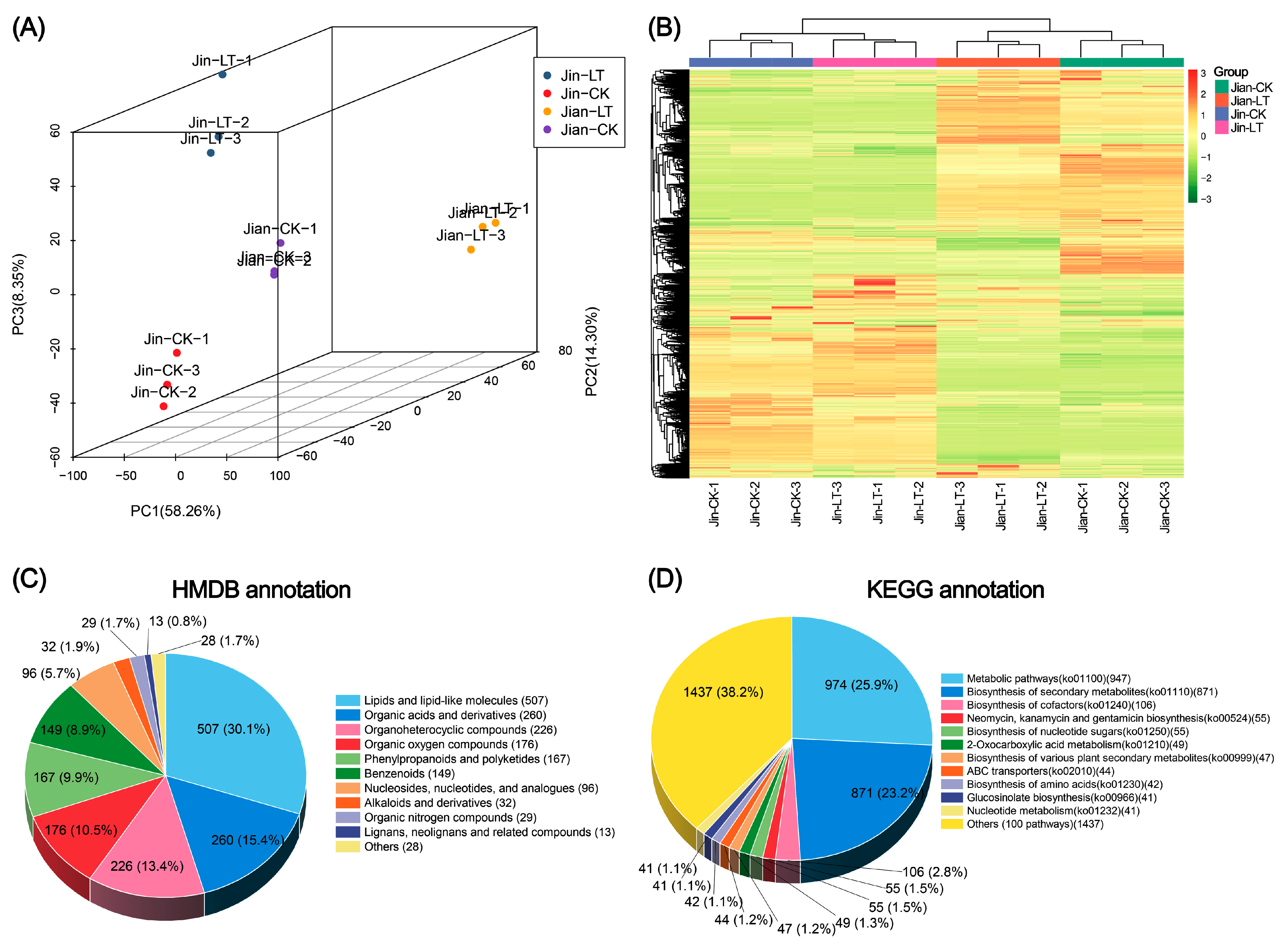
3.2. Non-Targeted Metabolite Detection, Identification, and Annotation
3.3. Metabolic Changes Under Low-Temperature Stress
3.4. Transcriptomic Analysis Under Low-Temperature Stress
3.4.1. RNA-Seq Quality Assessment, Assembly, and Annotation
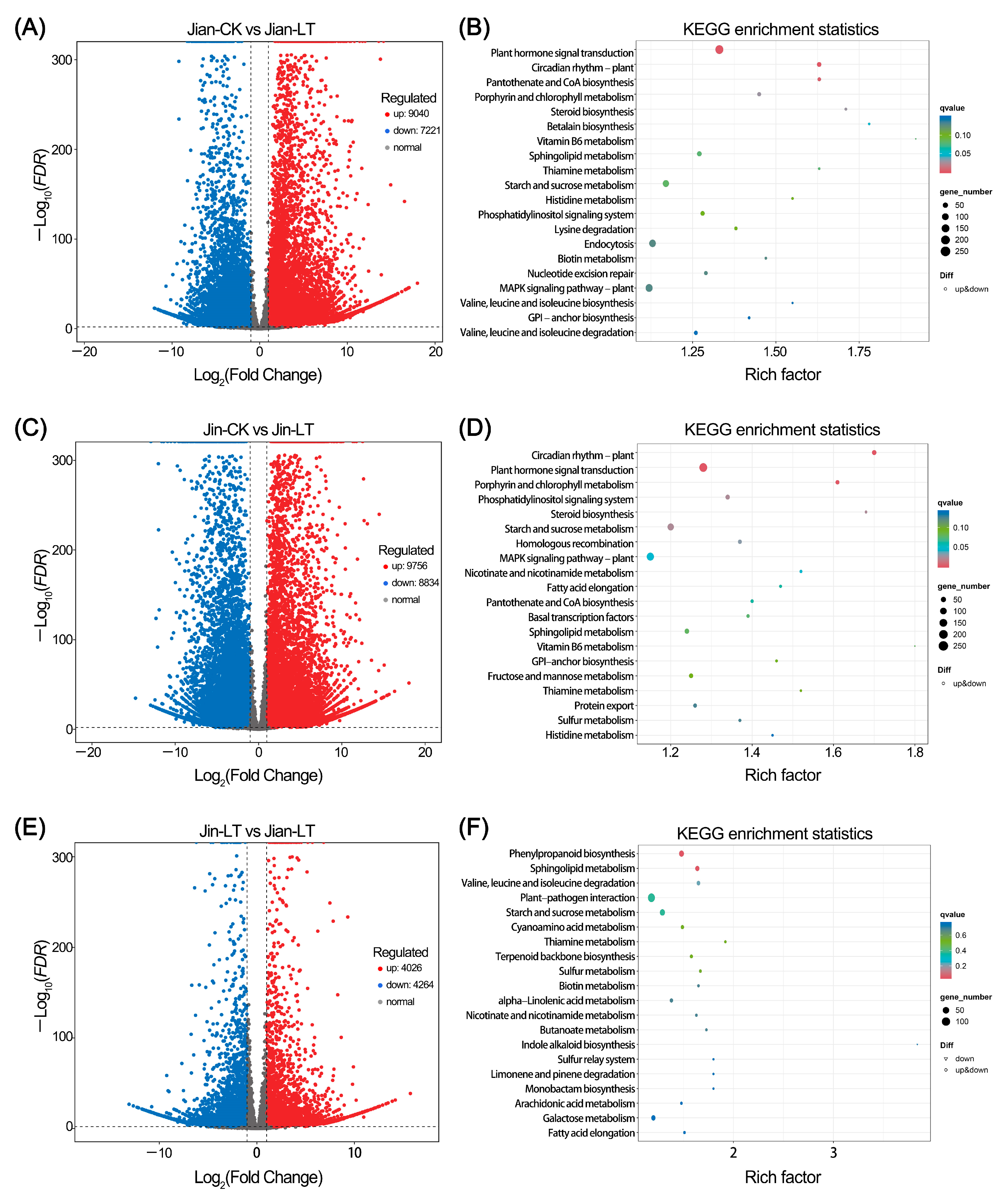
3.4.2. Identification of Differentially Expressed Genes (DEGs)
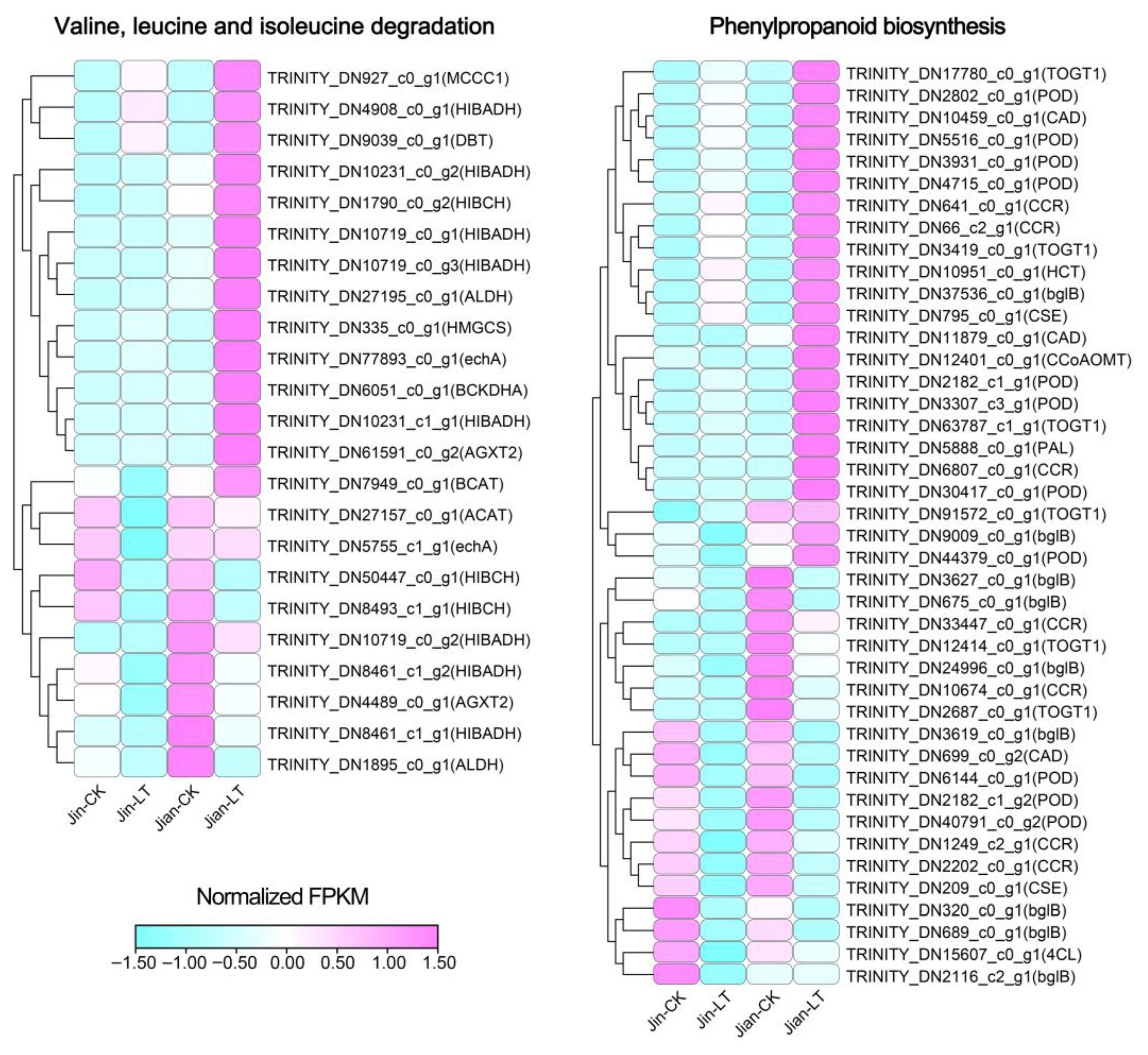
3.4.3. Differences in Gene Numbers in Commonly Enriched Pathways of DEGs Under Control and Low-Temperature Stress
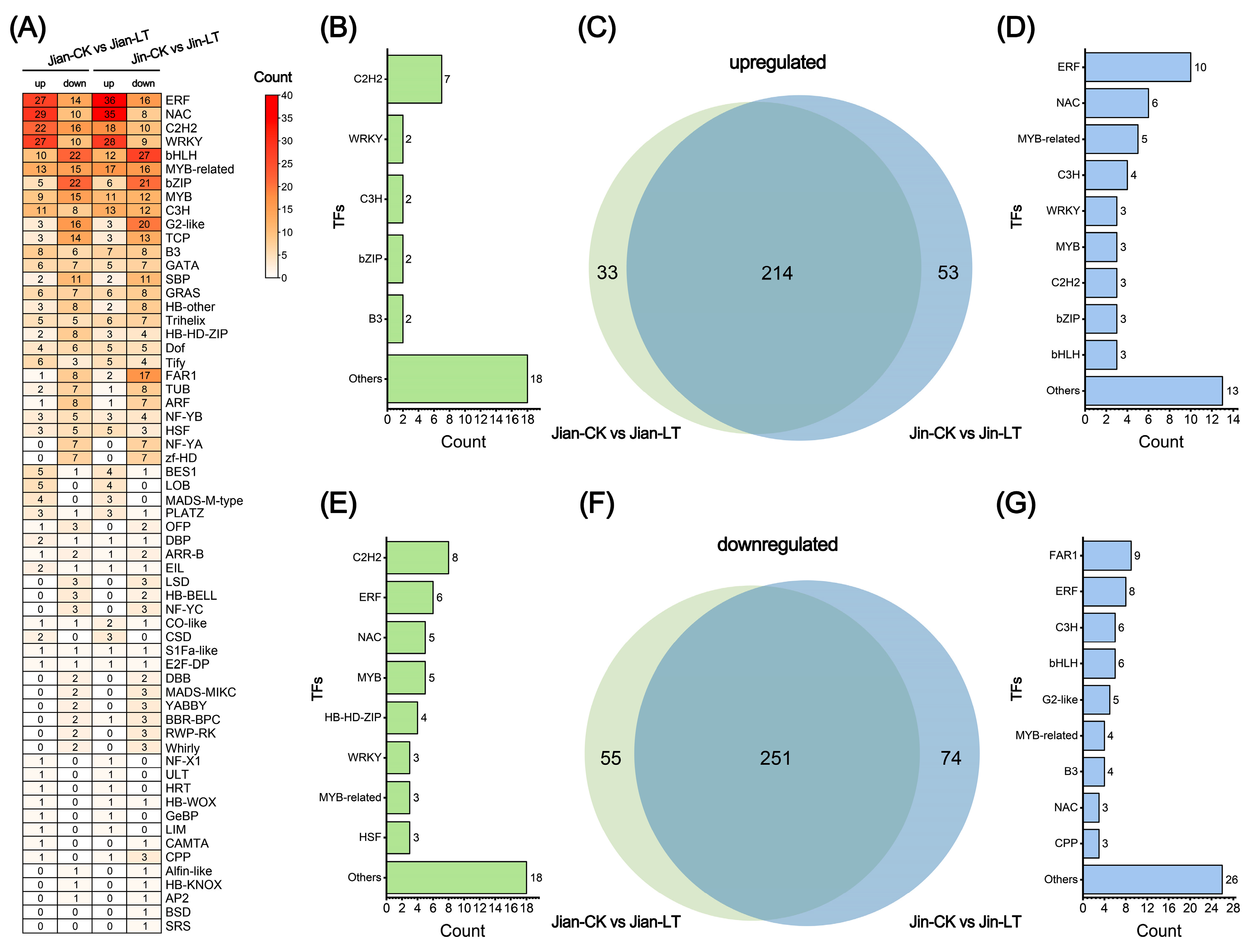
3.4.4. Analysis of Differentially Expressed Transcription Factors
3.5. Correlation Network Analysis Between Transcription Factors and Key Differential Metabolic Pathways
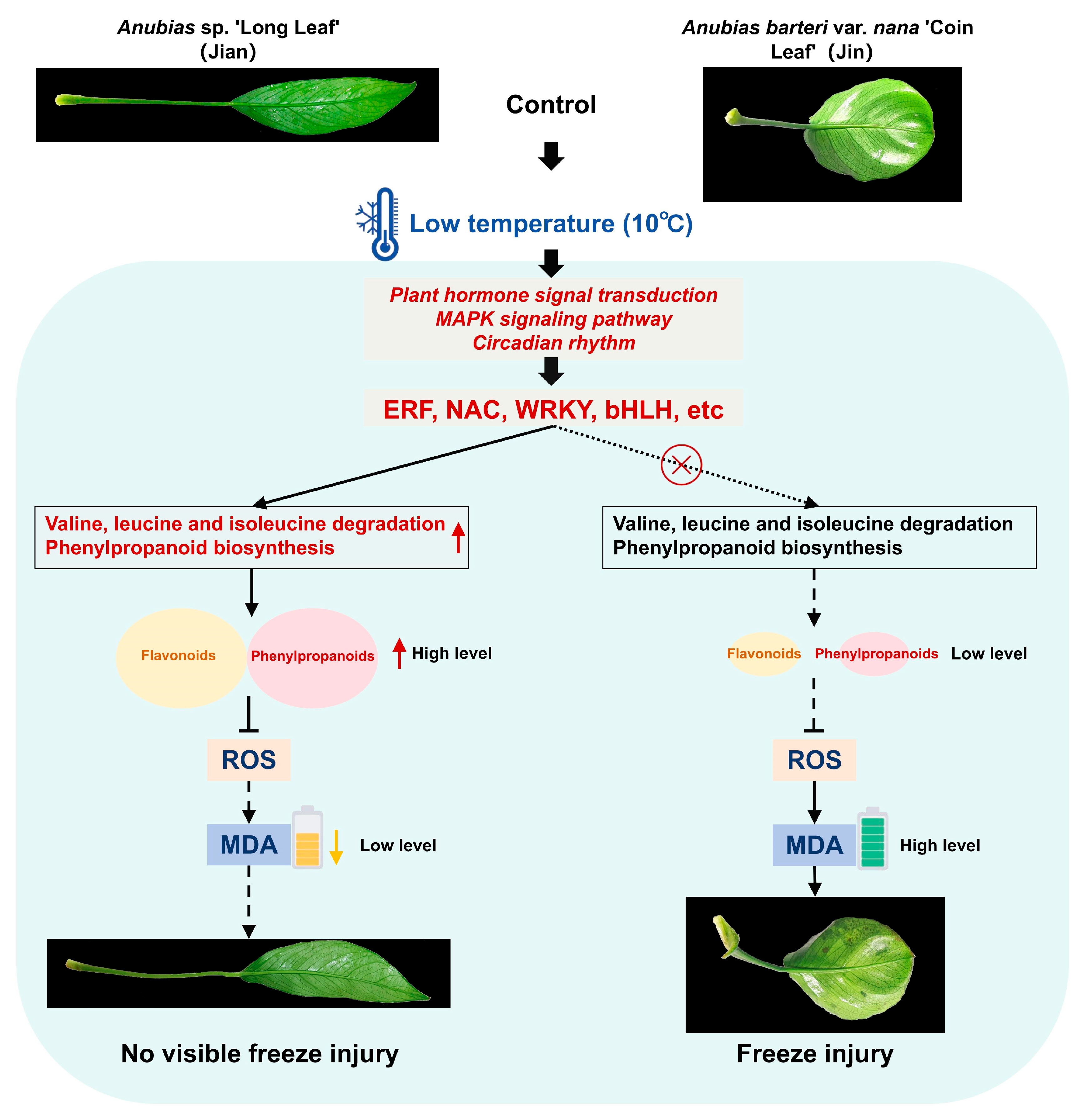
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goswami, A.K.; Maurya, N.K.; Goswami, S.; Bardhan, K.; Singh, S.K.; Prakash, J.; Pradhan, S.; Kumar, A.; Chinnusamy, V.; Kumar, P.; et al. Physio-biochemical and molecular stress regulators and their crosstalk for low-temperature stress responses in fruit crops: A review. Front. Plant Sci. 2022, 13, 1022167. [Google Scholar] [CrossRef]
- Dou, N.; Li, L.; Fang, Y.; Fan, S.; Wu, C. Comparative physiological and transcriptome analyses of tolerant and susceptible cultivars reveal the molecular mechanism of cold tolerance in Anthurium andraeanum. Int. J. Mol. Sci. 2023, 25, 250. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, H.; Zhao, L.; Gu, C.; Na, Y.; Xie, B.; Cheng, S.; Pan, G. Application of brassinolide alleviates cold stress at the booting stage of rice. J. Integr. Agric. 2020, 19, 975–987. [Google Scholar] [CrossRef]
- Zhou, M.; Li, W.; Zheng, Y.; Lin, P.; Yao, X.; Lin, J. Cbrci35, a cold responsive peroxidase from capsella bursa-pastoris regulates reactive oxygen species homeostasis and enhances cold tolerance in tobacco. Front. Plant Sci. 2016, 7, 1599. [Google Scholar] [CrossRef]
- Geng, J.; Wei, T.; Wang, Y.; Huang, X.; Liu, J. Overexpression of ptrbhlh, a basic helix-loop-helix transcription factor from poncirus trifoliata, confers enhanced cold tolerance in pummelo (Citrus grandis) by modulation of H2O2 level via regulating a CAT gene. Tree Physiol. 2019, 39, 2045–2054. [Google Scholar] [CrossRef]
- Xu, J.; Duan, X.; Yang, J.; Beeching, J.R.; Zhang, P. Coupled expression of cu/zn-superoxide dismutase and catalase in cassava improves tolerance against cold and drought stresses. Plant Signal. Behav. 2013, 8, e24525. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Q.; Wu, J.; Zheng, X.; Zheng, S.; Sun, X.; Qiu, Q.; Lu, T. Gene knockout study reveals that cytosolic ascorbate peroxidase 2 (OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS ONE 2013, 8, e57472. [Google Scholar] [CrossRef]
- Tian, J.; Ma, Y.; Chen, Y.; Chen, X.; Wei, A. Plant hormone response to low-temperature stress in cold-tolerant and cold-sensitive varieties of Zanthoxylum bungeanum maxim. Front. Plant Sci. 2022, 13, 847202. [Google Scholar] [CrossRef]
- Xu, Y.; Fu, X. Reprogramming of plant central metabolism in response to abiotic stresses: A metabolomics view. Int. J. Mol. Sci. 2022, 23, 5716. [Google Scholar] [CrossRef]
- Chen, D.; Mubeen, B.; Hasnain, A.; Rizwan, M.; Adrees, M.; Naqvi, S.; Iqbal, S.; Kamran, M.; El-Sabrout, A.M.; Elansary, H.O.; et al. Role of promising secondary metabolites to confer resistance against environmental stresses in crop plants: Current scenario and future perspectives. Front. Plant Sci. 2022, 13, 881032. [Google Scholar]
- Huang, X.; Shi, H.; Hu, Z.; Liu, A.; Amombo, E.; Chen, L.; Fu, J. Aba is involved in regulation of cold stress response in bermudagrass. Front. Plant Sci. 2017, 8, 1613. [Google Scholar] [CrossRef]
- Lata, C.; Prasad, M. Role of drebs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 62, 4731–4748. [Google Scholar] [CrossRef]
- Mega, R.; Meguro-Maoka, A.; Endo, A.; Shimosaka, E.; Murayama, S.; Nambara, E.; Seo, M.; Kanno, Y.; Abrams, S.R.; Sato, Y. Sustained low abscisic acid levels increase seedling vigor under cold stress in rice (Oryza sativa L.). Sci. Rep. 2015, 5, 13819. [Google Scholar] [CrossRef]
- Rahman, A. Auxin: A regulator of cold stress response. Physiol. Plant. 2013, 147, 28–35. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate regulates the inducer of cbf expression-c-repeat binding factor/dre binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef]
- Chaudhuri, A.; Halder, K.; Abdin, M.Z.; Majee, M.; Datta, A. Abiotic stress tolerance in plants: Brassinosteroids navigate competently. Int. J. Mol. Sci. 2022, 23, 14577. [Google Scholar] [CrossRef]
- Fu, X.; Feng, Y.Q.; Zhang, X.W.; Zhang, Y.Y.; Bi, H.G.; Ai, X.Z. Salicylic acid is involved in rootstock-scion communication in improving the chilling tolerance of grafted cucumber. Front. Plant Sci. 2021, 12, 693344. [Google Scholar] [CrossRef]
- Kaplan, F.; Kopka, J.; Haskell, D.W.; Zhao, W.; Schiller, K.C.; Gatzke, N.; Sung, D.Y.; Guy, C.L. Exploring the temperature-stress metabolome of arabidopsis. Plant Physiol. 2004, 136, 4159–4168. [Google Scholar] [CrossRef]
- Morsy, M.R.; Jouve, L.; Hausman, J.F.; Hoffmann, L.; Stewart, J.M. Alteration of oxidative and carbohydrate metabolism under abiotic stress in two rice (Oryza sativa L.) Genotypes contrasting in chilling tolerance. J. Plant Physiol. 2007, 164, 157–167. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Fialko, A.V.; Yugay, Y.A. Involvement of epigenetic factors in flavonoid accumulation during plant cold adaptation. Plant Physiol. Biochem. 2024, 216, 109096. [Google Scholar] [CrossRef]
- Yu, Y.; Lv, Y.; Shi, Y.; Li, T.; Chen, Y.; Zhao, D.; Zhao, Z. The role of phyto-melatonin and related metabolites in response to stress. Molecules 2018, 23, 1887. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. Cold1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Y.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.; Zhu, S.; Wan, H.; Yang, Y.; et al. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 2021, 14, 315–329. [Google Scholar] [CrossRef]
- Peng, Y.; Ming, Y.; Jiang, B.; Zhang, X.; Fu, D.; Lin, Q.; Zhang, X.; Wang, Y.; Shi, Y.; Gong, Z.; et al. Differential phosphorylation of Ca2+-permeable channel CYCLIC NUCLEOTIDE-GATED CHANNEL20 modulates calcium-mediated freezing tolerance in arabidopsis. Plant Cell 2024, 36, 4356–4371. [Google Scholar] [CrossRef]
- de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, P.; Si, T.; Hsu, C.C.; Wang, L.; Zayed, O.; Yu, Z.; Zhu, Y.; Dong, J.; Tao, W.A.; et al. MAP kinase cascades regulate the cold response by modulating ice1 protein stability. Dev. Cell 2017, 43, 618–629. [Google Scholar] [CrossRef]
- Lu, X.; Yang, L.; Yu, M.; Lai, J.; Wang, C.; Mcneil, D.; Zhou, M.; Yang, C. A novel Zea mays ssp. Mexicana L. MYC-type ICE-like transcription factor gene ZmmICE1, enhances freezing tolerance in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2017, 113, 78–88. [Google Scholar] [CrossRef]
- Ritonga, F.N.; Ngatia, J.N.; Wang, Y.; Khoso, M.A.; Farooq, U.; Chen, S. AP2/ERF, an important cold stress-related transcription factor family in plants: A review. Physiol. Mol. Biol. Plants 2021, 27, 1953–1968. [Google Scholar] [CrossRef]
- Wu, C.L.; Lin, L.F.; Hsu, H.C.; Huang, L.F.; Hsiao, C.D.; Chou, M.L. Saussurea involucrata (snow lotus) ICE1 and ICE2 orthologues involved in regulating cold stress tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 10850. [Google Scholar] [CrossRef]
- Zuo, Z.F.; Kang, H.G.; Hong, Q.C.; Park, M.Y.; Sun, H.J.; Kim, J.; Song, P.S.; Lee, H.Y. A novel basic helix-loop-helix transcription factor, ZjICE2 from Zoysia japonica confers abiotic stress tolerance to transgenic plants via activating the DREB/CBF regulon and enhancing ros scavenging. Plant Mol. Biol. 2020, 102, 447–462. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, H.S.; Bahk, S.; An, J.; Yoo, Y.; Kim, J.Y.; Chung, W.S. Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res. 2017, 45, 6613–6627. [Google Scholar] [CrossRef]
- Kong, X.; Zhou, Q.; Zhou, X.; Wei, B.; Ji, S. Transcription factor CaNAC1 regulates low-temperature-induced phospholipid degradation in green bell pepper. J. Exp. Bot. 2020, 71, 1078–1091. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, H.; Yang, X.; Li, Q.; Ling, J.; Wang, H.; Gu, X.; Huang, S.; Jiang, W. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an aba-dependent manner. Plant Physiol. Biochem. 2016, 108, 478–487. [Google Scholar] [CrossRef]
- Brunel, S. Pathway analysis: Aquatic plants imported in 10 eppo countries. Bulletin OEPP 2009, 39, 201–213. [Google Scholar] [CrossRef]
- Li, L.; Liu, C.; Hou, K.; Liu, W. Comparative analyses of plastomes of four Anubias (Araceae) taxa, tropical aquatic plants endemic to africa. Genes 2022, 13, 2043. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Wei, J.; Chen, Y.; Zeng, C.; Deng, C.; Zeng, P.; Tang, Y.; Zhou, Q.; Huang, Y.; Zhu, Q. Codon usage patterns and genomic variation analysis of chloroplast genomes provides new insights into the evolution of Aroideae. Sci. Rep. 2025, 15, 4314–4333. [Google Scholar] [CrossRef]
- Kanchanapoom, K.; Chunui, P.; Kanchanapoom, K. Micropropagation of Anubias barteri var. nana from shoot tip culture and the analysis of ploidy stability. Not. Bot. Horti Agrobo. 2012, 40, 148–151. [Google Scholar]
- Xu, C.L.; Zhao, C.B.; Ding, S.; Zhang, J.F.; Xie, H.; Huang, C.X. First report of root-knot nematode Meloidogyne arenaria infesting roots of Anubias barteri in guangdong, china. Plant Dis. 2012, 96, 773. [Google Scholar] [CrossRef]
- Garibaldi, A.; Minuto, G.; Nicoletti, R.; Gullino, M.L. First report of a blight caused by Rhizoctonia solani on Anubias heterophylla in Italy. Plant Dis. 2003, 87, 1005. [Google Scholar] [CrossRef]
- Chen, A.; Han, R.; Li, D.; Ling, L.; Luo, H.; Tang, S. A Comparison of two methods for electrical conductivity about plant leaves. J. Guangdong Educ. Insistite 2010, 30, 88–91. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the kegg orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chung, W. Unraveling new functions of superoxide dismutase using yeast model system: Beyond its conventional role in superoxide radical scavenging. J. Microbiol. 2017, 55, 409–416. [Google Scholar] [CrossRef]
- Liu, X.; Sui, L.; Huang, Y.; Geng, C.; Yin, B. Physiological and visible injury responses in different growth stages of winter wheat to ozone stress and the protection of spermidine. Atmos. Pollut. Res. 2015, 6, 596–604. [Google Scholar] [CrossRef]
- García-Caparrós, P.; De Filippis, L.; Gul, A.; Hasanuzzaman, M.; Ozturk, M.; Altay, V.; Lao, M.T. Oxidative stress and antioxidant metabolism under adverse environmental conditions: A review. Bot. Rev. 2021, 87, 421–466. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, F.; Sayed, M.A.; Shen, X.; Zhou, L.; Liu, X.; Sun, X.; Chen, S.; Wu, Y.; Lu, L.; et al. Integrated transcriptomic and metabolomic data reveal the cold stress responses molecular mechanisms of two coconut varieties. Front. Plant Sci. 2024, 15, 1353352. [Google Scholar] [CrossRef]
- Liu, J.; Wang, T.; Weng, Y.; Liu, B.; Gao, Q.; Ji, W.; Wang, Z.; Wang, Y.; Ma, X. Identification and characterization of regulatory pathways controlling dormancy under lower temperature in alfalfa (Medicago sativa L.). Front. Plant Sci. 2022, 13, 872839. [Google Scholar] [CrossRef]
- Li, Q.; Song, J. Analysis of widely targeted metabolites of the euhalophyte Suaeda salsa under saline conditions provides new insights into salt tolerance and nutritional value in halophytic species. BMC Plant Biol. 2019, 19, 388. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Luobu, Z.; Zhuoga, D.; Wei, X.; Tang, Y. Physiological and broadly targeted metabolomic analyses of barley (hordeum vulgare l.) In response to low-temperature stress. BMC Genom. 2025, 26, 617–618. [Google Scholar] [CrossRef] [PubMed]
- Subki, A. The Role of Thiamine in Plants and Current Perspectives in Crop Improvement; IntechOpen: London, UK, 2018. [Google Scholar]
- Jaiwal, P.K.; Chhillar, A.K.; Chaudhary, D.; Jaiwal, R. Nutritional Quality Improvement in Plants; Springer International Publishing AG: Cham, Switzerland, 2019. [Google Scholar]
- Tunc-Ozdemir, M.; Miller, G.; Song, L.; Kim, J.; Sodek, A.; Koussevitzky, S.; Misra, A.N.; Mittler, R.; Shintani, D. Thiamin confers enhanced tolerance to oxidative stress in Arabidopsis. Plant Physiol. 2009, 151, 421–432. [Google Scholar] [CrossRef]
- Konecny, T.; Nikoghosyan, M.; Binder, H. Machine learning extracts marks of thiamine’s role in cold acclimation in the transcriptome of Vitis vinifera. Front. Plant Sci. 2023, 14, 1303542. [Google Scholar] [CrossRef]
- Meng, D.; Yu, X.; Ma, L.; Hu, J.; Liang, Y.; Liu, X.; Yin, H.; Liu, H.; He, X.; Li, D. Transcriptomic response of chinese yew (Taxus chinensis) to cold stress. Front. Plant Sci. 2017, 8, 468. [Google Scholar] [CrossRef]
- Back, K.; Tan, D.X.; Reiter, R.J. Melatonin biosynthesis in plants: Multiple pathways catalyze tryptophan to melatonin in the cytoplasm or chloroplasts. J. Pineal Res. 2016, 61, 426–437. [Google Scholar] [CrossRef]
- Ren, C.; Cheng, T.; Jia, J.; Cao, L.; Zhang, W.; Zhang, S.; Li, W.; Zhang, Y.; Yu, G. Exogenous tryptophan enhances cold resistance of soybean seedlings by promoting melatonin biosynthesis. Physiol. Plantarum 2025, 177, e70189. [Google Scholar] [CrossRef]
- Zhao, Y.; Qi, T.; Cui, C.; Liu, X.; Liu, R.; Liu, S.; Song, Y.; Li, Y.; Lv, H. Effects of different storage temperatures on the quality and metabolome of maize with high moisture content. Food Sci. Technol. 2024, 214, 117117. [Google Scholar] [CrossRef]
- Araújo, W.L.; Tohge, T.; Ishizaki, K.; Leaver, C.J.; Fernie, A.R. Protein degradation—An alternative respiratory substrate for stressed plants. Trends Plant Sci. 2011, 16, 489–498. [Google Scholar] [CrossRef]
- Peng, C.; Uygun, S.; Shiu, S.; Last, R.L. The impact of the branched-chain ketoacid dehydrogenase complex on amino acid homeostasis in Arabidopsis. Plant Physiol. 2015, 169, 1807–1820. [Google Scholar] [CrossRef] [PubMed]
- Latimer, S.; Li, Y.; Nguyen, T.T.H.; Soubeyrand, E.; Fatihi, A.; Elowsky, C.G.; Block, A.; Pichersky, E.; Basset, G.J. Metabolic reconstructions identify plant 3-methylglutaconyl-coa hydratase that is crucial for branched-chain amino acid catabolism in mitochondria. Plant J. Cell Mol. Biol. 2018, 95, 358–370. [Google Scholar] [CrossRef]
- Wang, C.; Quangkiet, T.; Li, W.; Jian, B.; Yang, X. Integration of transcriptome and metabolome reveals candidate metabolites responding to drought stress in sugarcane. Genomics 2025, 117, 111092. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lian, Y.; Guo, H.; Li, C.; Ren, Y.; Xin, Z.; Lin, T.; Wang, Z. Network analysis of metabolomics, transcriptome and hormones reveals propionic acid-mediated novel survival strategy against drought in wheat. Physiol. Plantarum 2024, 176, e14551. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, Q.; Yong, X.; Wu, M.; Jiang, B.; Jia, Y.; Ma, J.; Mou, L.; Tang, S.; Pan, Y. Effects of water deficit on two cultivars of Hibiscus mutabilis: A comprehensive study on morphological, physiological, and metabolic responses. Plant Physiol. Bioch. 2024, 217, 109269. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Li, H.; Zhang, Z.; Yang, Y.; Jin, Z.; Chen, W.; Tang, D.; Wei, C.; Tang, Q. Characterization of the difference between day and night temperatures on the growth, photosynthesis, and metabolite accumulation of tea seedlings. Int. J. Mol. Sci. 2023, 24, 6718. [Google Scholar] [CrossRef]
- Mehrotra, S.; Verma, S.; Kumar, S.; Kumari, S.; Mishra, B.N. Transcriptional regulation and signalling of cold stress response in plants: An overview of current understanding. Environ. Exp. Bot. 2020, 180, 104243. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Y.; Wei, L.; Liu, W.; Chen, J.; Zhang, J.; Yang, Z.; Huang, S.; Zhou, Y. Integrative Physiological, Metabolomic and Transcriptomic Analyses Uncover the Mechanisms Underlying Differential Responses of Two Anubias Genotypes to Low-Temperature Stress. Biomolecules 2025, 15, 1520. https://doi.org/10.3390/biom15111520
Luo Y, Wei L, Liu W, Chen J, Zhang J, Yang Z, Huang S, Zhou Y. Integrative Physiological, Metabolomic and Transcriptomic Analyses Uncover the Mechanisms Underlying Differential Responses of Two Anubias Genotypes to Low-Temperature Stress. Biomolecules. 2025; 15(11):1520. https://doi.org/10.3390/biom15111520
Chicago/Turabian StyleLuo, Yanyu, Liguo Wei, Weiguang Liu, Jiwei Chen, Jinzhong Zhang, Zhijian Yang, Shaoli Huang, and Yiwei Zhou. 2025. "Integrative Physiological, Metabolomic and Transcriptomic Analyses Uncover the Mechanisms Underlying Differential Responses of Two Anubias Genotypes to Low-Temperature Stress" Biomolecules 15, no. 11: 1520. https://doi.org/10.3390/biom15111520
APA StyleLuo, Y., Wei, L., Liu, W., Chen, J., Zhang, J., Yang, Z., Huang, S., & Zhou, Y. (2025). Integrative Physiological, Metabolomic and Transcriptomic Analyses Uncover the Mechanisms Underlying Differential Responses of Two Anubias Genotypes to Low-Temperature Stress. Biomolecules, 15(11), 1520. https://doi.org/10.3390/biom15111520






