Dynamic Executors of Bacterial Signals: Functional Versatility and Regulatory Networks of c-di-GMP Effectors
Abstract
1. Introduction
2. RNA-Based c-di-GMP Effectors: Riboswitches and Their Mechanisms
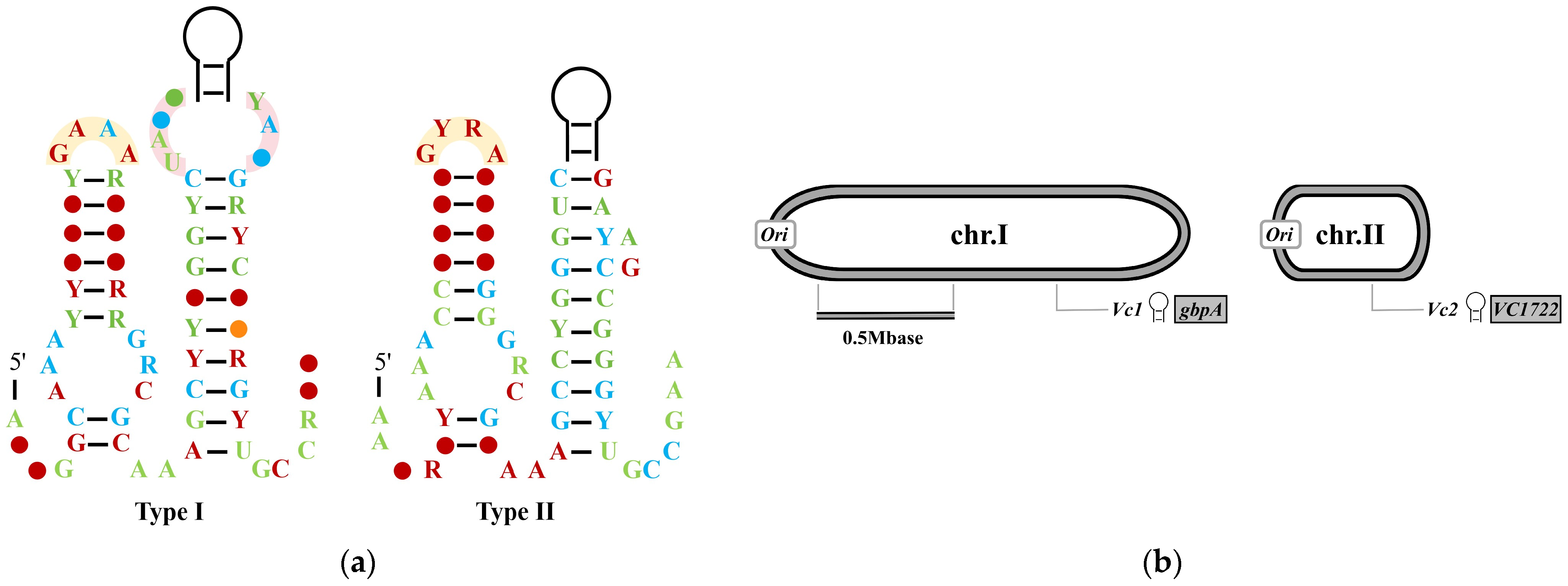
2.1. c-di-GMP-I Riboswitches: Structural Basis for Regulating Pathogenic Gene Expression in V. cholerae
2.2. c-di-GMP-II Riboswitches: Controlling GTP-Mediated RNA Self-Splicing in Clostridioides difficile
3. c-di-GMP-Responsive Transcription Factors: Structural Determinants and Regulatory Roles in Bacterial Physiology
4. The PilZ Domain Fold: c-di-GMP Binding and Allosteric Regulation
5. GGDEF/EAL Domain Proteins: Degenerate Domains and Triggered Phosphodiesterases
5.1. Structural Basis of Degenerate Domain Proteins: Conserved c-di-GMP Binding Motifs Distinct from Catalytic Sites
5.2. Triggered Phosphodiesterases as Transcriptional Switches: Targeting Factor Binding for Gene Regulation
6. The Role of c-di-GMP Effectors with High Functional Specificity in Bacterial Life Activities
7. Conclusions and Outlook
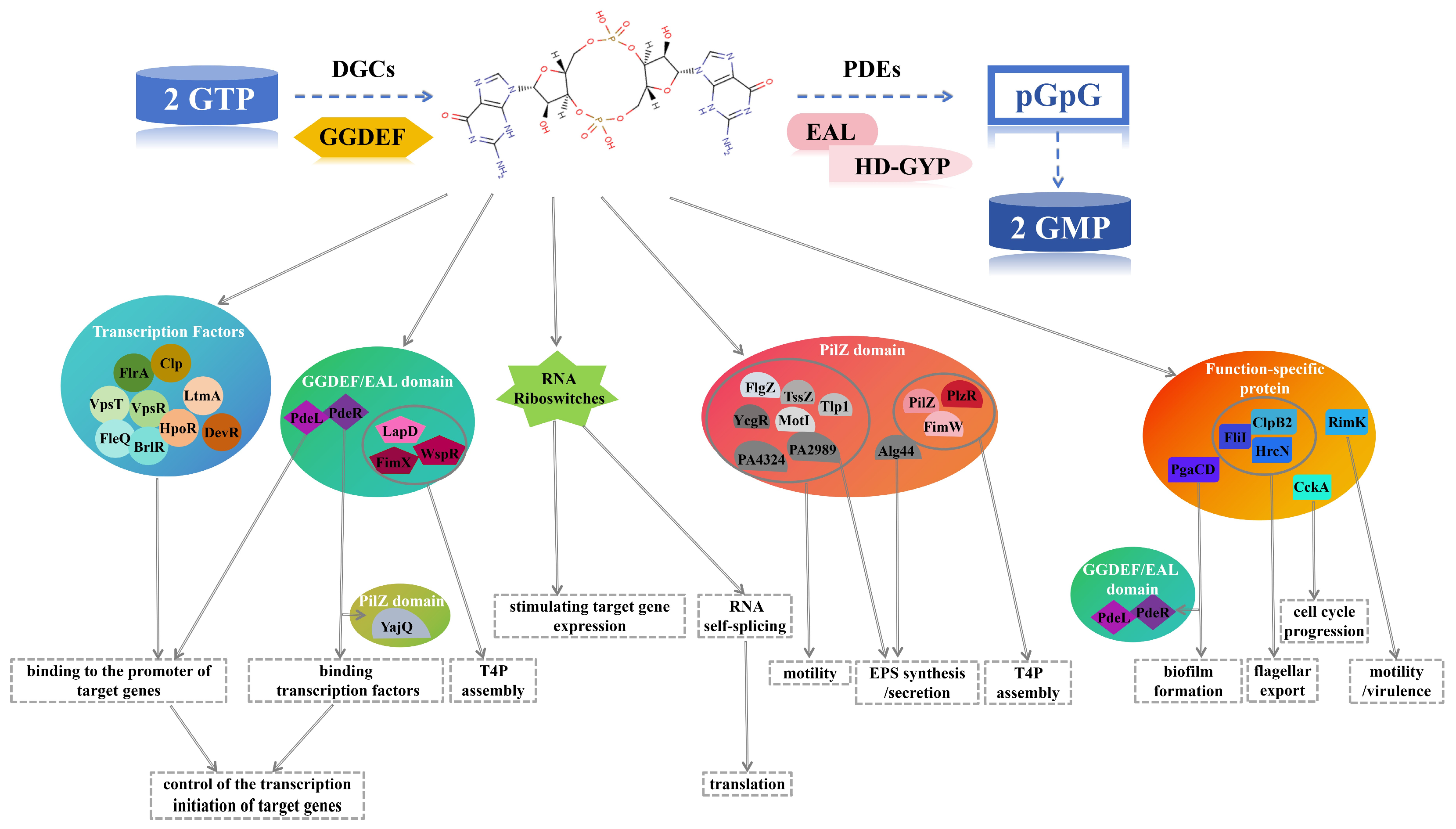
- The evolution and divergence of effector domains lay the molecular foundation for their functional diversity. For example, c-di-GMP acts as an allosteric activator of cellulose synthase by binding to the conserved active site in the PilZ domain, subsequently stimulating cellulose synthesis. Effectors containing degenerate GGDEF or EAL domains, although losing their intrinsic enzymatic activity as DGCs or PDEs, can still bind to c-di-GMP due to their specific c-di-GMP-binding conserved motifs. Trigger PDEs with normally catalytically active EAL domains regulate the expression of biofilm-related genes through allosteric regulation and product inhibition.
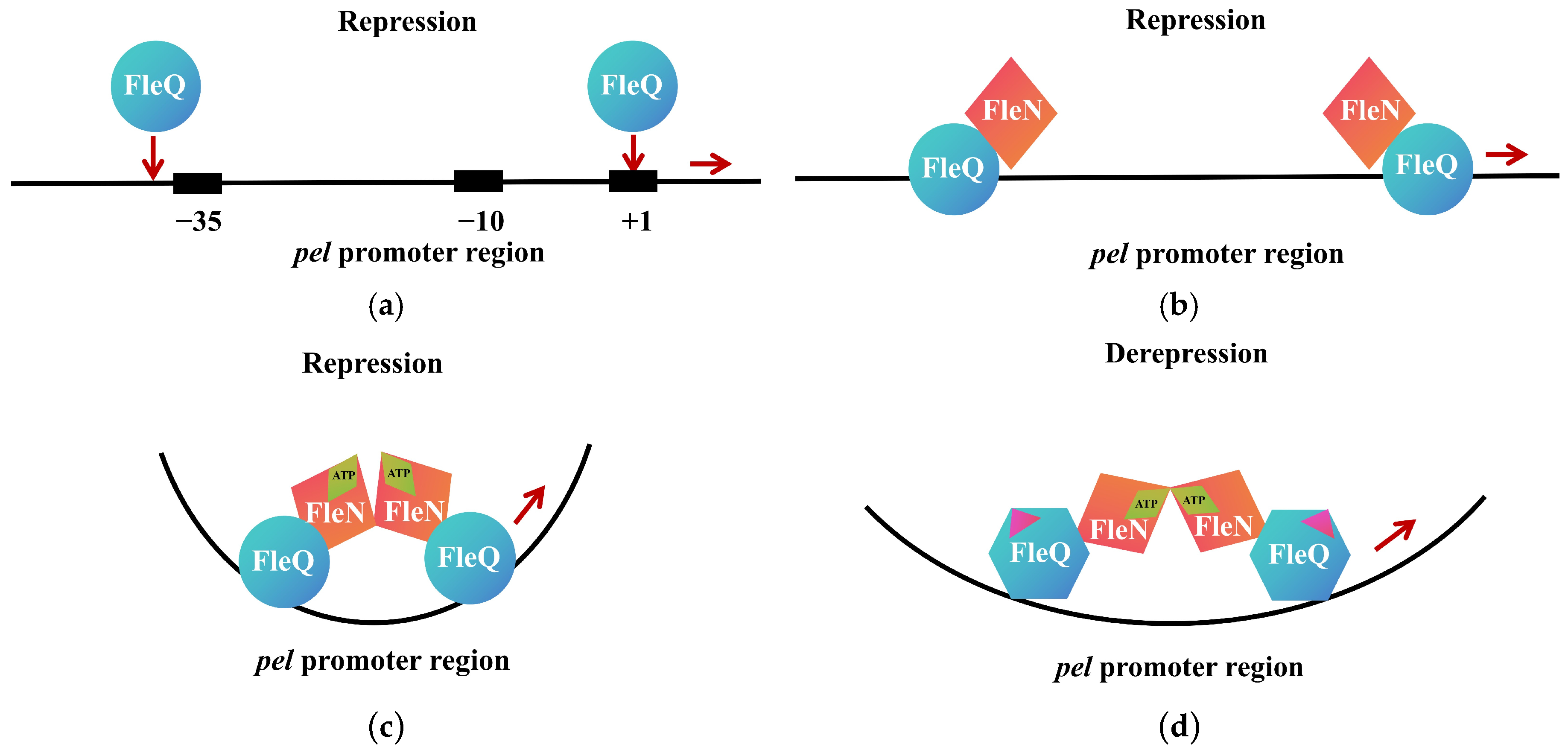
- A single effector can integrate multiple signaling pathways (Figure 3). For instance, PilZ is involved in two distinct signaling pathways: one regulating EPS synthesis and another integrating T3SS and flagellar functions. The transcription factor FleQ, on the one hand, undergoes changes in its quaternary structure and transcriptional activity upon binding to c-di-GMP, thereby repressing the expression of flagellar-related genes; on the other hand, FleQ cooperates with FleN to regulate EPS production. Additionally, another transcription factor, VpsT, inhibits flagellar assembly under high c-di-GMP levels and suppresses biofilm formation under high cell density conditions.
- Functional redundancy among effectors may constitute a bacterial barrier against antibiotic resistance (Table 1). For example, the integration and activity regulation of bacterial motility-related flagella involve multiple effectors with distinct structures, such as FlgZ, YcgR, and FliI. Transcription factors such as FleQ, GGDEF domain-containing proteins such as PelD, PilZ domain-containing proteins such as BcsA, Alg44, and PA2989, as well as functionally specific proteins such as PgaCD, synergistically regulate the synthesis and secretion of EPS through distinct c-di-GMP regulatory networks.
| Effectors | Types | Sources | c-di-GMP Binding Domain | Binding Affinity (KD) | Functional Effects of c-di-GMP Binding | References |
|---|---|---|---|---|---|---|
| c-di-GMP-I | RNA Riboswitches | V. cholerae | Type I GEMM RNA (Vc2) | 1 nM | Activate the expression of genes associated with intestinal colonization | [33,34,35,36] |
| c-di-GMP-II | RNA Riboswitches | C. difficile | mRNA 5′-region | 200 pM | Determine the site of RNA self-splicing | [31,32,39,40,41] |
| FleQ | Transcription Factors | P. aeruginosa | Receptor domain(REC) at N-terminus | 7 μM | Repress flagellar -related genes expression, activate EPS production | [5,42,43,44,45] |
| BrlR | Transcription Factors | P. aeruginosa | Gyrl-like Domain at C-terminus | 2.2 μM | Enhance the high-level drug resistance of biofilms | [132] |
| FlrA | Transcription Factors | V. cholerae | REC at N-terminus | 0.38 μM | Interfere with flagellar biosynthesis | [49] |
| VpsT | Transcription Factors | V. cholerae | REC, HTH | 3.2 μM | Promote biofilm synthesis and repress flagellar assembly genes expression | [50,51,52] |
| VpsR | Transcription Factors | V. cholerae | predicted ATP binding domain | 1.6 μM | Activate biofilm synthesis | [51] |
| LtmA | Transcription Factors | M. smegmatis | TetR-type HTH domain | 0.83 μM | Positively regulate the expression of redox gene clusters, enhance bacterial resistance to hydrogen peroxide | [54,55,56,57] |
| HpoR | Transcription Factors | M. smegmatis | --- | 1.78 μM | Negatively regulate the expression of redox gene clusters, increase bacterial sensitivity to hydrogen peroxide | [56,57] |
| DevR | Transcription Factors | M. smegmatis | C-terminus | 1.96 μM | Increase survival rate under oxidative stress | [58] |
| XcCLP | Transcription Factors | X. campestris | β-barrel domain at N-terminus | 3.5 μM | Interfere with downstream virulence genes expression | [59,60] |
| BldD | Transcription Factors | Streptomyces coelicolor | --- | 2.5 μM | Inhibit sporulation genes expression | [129] |
| MrkH | Transcription Factors | Klebsiella pneumoniae | PilZ domain at N-terminus | 0.107 μM | Promote biofilm formation | [131] |
| FsnR | Transcription Factors | Stenotrophomonas maltophilia | REC, HTH | 3.43 μM | Induce flagellar genes expression | [133] |
| CdbA | Transcription Factors | Myxococcus xanthus | RHH DNA binding domain | ~0.083 μM | Promote chromosome formation | [134] |
| BcsA | PilZ domain | K. xylinus | PilZ | 0.98 μM | Regulate cellulose synthesis | [61,62,63,64] |
| Alg44 | PilZ domain | P. aeruginosa | PilZ | --- | Regulate alginate synthesis | [69,70] |
| FimW | PilZ domain | P. aeruginosa | PilZ | --- | Regulate T4P assembly | [72] |
| PilZ | PilZ domain | P. aeruginosa | PilZ | --- | Regulate T4P assembly, mediate bacterial twitching motility, regulate EPS synthesis | [76,77] |
| PlzR | PilZ domain | P. aeruginosa | PilZ | --- | Regulate T4P assembly | [78] |
| FlgZ (PA2560) | PilZ domain | P. aeruginosa | PilZ | --- | Inhibits bacterial swarming | [81] |
| MapZ | PilZ domain | P. aeruginosa | PilZ | --- | Regulate the switching of flagellar motors | [82] |
| HapZ | PilZ domain | P. aeruginosa | PilZ | 2.0 μM | Regulate two-component signal transduction | [83] |
| TssZ (PA0012) | PilZ domain | P. aeruginosa | PilZ | --- | Affect bacterial swarming motility and T6SS-mediated bacterial killing activity | [68] |
| PA4324 | PilZ domain | P. aeruginosa | PilZ | Affect bacterial swarming motility and virulence | [84] | |
| PA2989 | PilZ domain | P. aeruginosa | PilZ | Affect bacterial swarming motility and EPS | [85] | |
| YcgR | PilZ domain | E.coli | PilZ | --- | Impair flagellar function and inhibit bacterial motility | [86,87] |
| MotI | PilZ domain | B. subtilis | PilZ | --- | Impair flagellar function and inhibit bacterial motility | [89,90] |
| DgrA | PilZ domain | C.crescentus | PilZ | --- | Impair flagellar function and inhibit bacterial motility | [91] |
| YajQ (XC_3703) | PilZ domain | X. campestris | PilZ | 2.0 μM | Activate virulence genes expression, enhance bacterial pathogenicity | [92,93] |
| CdgL | PilZ domain | L. enzymogenes | PilZ | --- | Activate HSAF expression | [94] |
| Tlp1 | PilZ domain | Azospirillum brasilense | PilZ | --- | promote the bacterial continuous motility | [135] |
| Cbp1 | PilZ domain | Azorhizobium caulinodans | PilZ | 14.94 μM | Regulate bacterial motility, biofilm, virulence | [136] |
| CdbS | PilZ domain | M. xanthus | PilZ | ~1.4 μM | Interfere with bacterial chromosome organization and accelerate cell death under heat stress | [137] |
| PlzC/PlzD | PilZ domain | V. cholerae | PilZ | 0.1–0.3 μM | Regulate bacterial motility, biofilm, virulence | [138,139] |
| PopA | GGDEF/EAL domain | C.crescentus | GGDEF | 2 μM | Regulate cell cycle | [16,95,96] |
| PleD | GGDEF/EAL domain | C.crescentus | GGDEF | --- | --- | [97,98] |
| WspR | GGDEF/EAL domain | C. crescentus, P. aeruginosa | GGDEF | --- | Regulate biofilm formation | [97,98] |
| PelD | GGDEF/EAL domain | P. aeruginosa | GGDEF | 0.5–1.9 μM | Produced Pel polysaccharide | [140] |
| FimX | GGDEF/EAL domain | P. aeruginosa, Xanthomonas citri | EAL | 0.1–0.2 μM | Regulate bacterial twitching motility, T4P synthesis, biofilm formation, virulence genes expression | [99,100,101] |
| LapD | GGDEF/EAL domain | Pseudomonas fluorescens | EAL | 1.9 μM | Control cell adhesion and biofilm formation | [102,103,104,105] |
| PdeR | GGDEF/EAL domain | E.coli | EAL | --- | Regulate biofilm formation | [106,107] |
| PdeL | GGDEF/EAL domain | E.coli | EAL | --- | Regulate biofilm formation | [110,111] |
| PgaC/PgaD | Function- specific effector | E.coli | --- | --- | Induce biofilm formation | [113,114,115] |
| CobBL | Function- specific effector | E.coli | N-terminus | 4.7 μM | Control energy metabolism, chemotaxis, and DNA supercoiling | [141,142] |
| FliI | Function- specific effector | P. fluorescens | --- | --- | Control flagellar export | [121,122,123] |
| HrcN | Function- specific effector | Pseudomonas syringae | --- | --- | Control flagellar export | [11,19] |
| ClpB2 | Function- specific effector | P. syringae | --- | --- | Control flagellar export | [11,19] |
| RimK | Function- specific effector | P. fluorescens, P. syringae | --- | --- | Affect bacterial motility, virulence, colonization, infection | [21] |
| CckA | Function- specific effector | C.crescentus | --- | 4.7 μM | Affect cell cycle progression and cell replication | [125,126,127,128] |
| GlgX | Function- specific effector | Streptomyces venezuelae | C-terminus | ~8 μM | Activate the catalytic activity of the enzyme to hydrolyze glycogen | [143] |
| MshE | Function- specific effector | V. cholerae | N-terminus | 0.014–2 μM | Participate in MSHA pili assembly | [22,23,24] |
| HxrA | Function- specific effector | P. aeruginosa | N-terminus | --- | Participate in MSHA pili assembly | [22,23,24] |
| LonA | Function- specific effector | V. cholerae | --- | --- | Regulate cell division, biofilm formation, flagellar motility | [144] |
| TfoY | Function- specific effector | V. cholerae | N-terminus | --- | Regulate bacterial motility, participate in T6SS | [144] |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| c-di-GMP | Cyclic diguanosine monophosphate |
| DGC | Diguanylate cyclase |
| PDE | Phosphodiesterase |
| EPS | Exopolysaccharides |
| GTP | Guanosine triphosphate |
| GMP | Guanosine monophosphate |
| MSHA | Mannose-sensitive haemagglutinin |
| ORFs | Open reading frames |
| GEMM | Genes for the Environment, Membranes and Motility |
| KD | Dissociation constant |
| EBP | Enhancer-binding protein |
| AAA+ | ATPase Associated with diverse cellular Activities |
| HTH | Helix-turn-helix |
| CAP | Catabolite activation protein |
| CRP | cAMP receptor protein |
| FNR | Fumarate nitrate reductase regulator |
| TM | Transmembrane |
| GT | Glycosyltransferase |
| T4P | Type IV pili |
| QS | Quorum-sensing |
| HSAF | Heat-stable antifungal factor |
| poly-GlcNAc | Poly-β-1,6-N-acetylglucosamine |
| T3SS | Type III secretion systems |
| T6SS | Type VI secretion systems |
References
- Ross, P.; Weinhouse, H.; Aloni, Y.; Michaeli, D.; Weinberger-Ohana, P.; Mayer, R.; Braun, S.; de Vroom, E.; van der Marel, G.A.; van Boom, J.H.; et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 1987, 325, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Jeong, G.J.; Tabassum, N.; Kim, Y.M. Functional diversity of c-di-GMP receptors in prokaryotic and eukaryotic systems. Cell Commun. Signal 2023, 21, 259. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, D.A.; Miller, S.I. Cyclic di-GMP as a bacterial second messenger. Microbiology 2004, 150, 2497–2502. [Google Scholar] [CrossRef]
- Jenal, U. Cyclic di-guanosine-monophosphate comes of age: A novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 2004, 7, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Jenal, U.; Reinders, A.; Lori, C. Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 2017, 15, 271–284. [Google Scholar] [CrossRef]
- Ryu, M.H.; Fomicheva, A.; Moskvin, O.V.; Gomelsky, M. Optogenetic Module for Dichromatic Control of c-di-GMP Signaling. J. Bacteriol. 2017, 199, e00014-17. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, Y.; Gao, T.; Zhang, Y.; Wang, Q. C-di-GMP turnover influences motility and biofilm formation in Bacillus amyloliquefaciens PG12. Res. Microbiol. 2018, 169, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.; Paul, R.; Samoray, D.; Amiot, N.C.; Giese, B.; Jenal, U.; Schirmer, T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. USA 2004, 101, 17084–17089. [Google Scholar] [CrossRef]
- Tchigvintsev, A.; Xu, X.; Singer, A.; Chang, C.; Brown, G.; Proudfoot, M.; Cui, H.; Flick, R.; Anderson, W.F.; Joachimiak, A.; et al. Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J. Mol. Biol. 2010, 402, 524–538. [Google Scholar] [CrossRef]
- Chou, S.H.; Galperin, M.Y. Diversity of Cyclic Di-GMP-Binding Proteins and Mechanisms. J. Bacteriol. 2016, 198, 32–46. [Google Scholar] [CrossRef]
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef]
- Hengge, R.; Galperin, M.Y.; Ghigo, J.M.; Gomelsky, M.; Green, J.; Hughes, K.T.; Jenal, U.; Landini, P. Systematic Nomenclature for GGDEF and EAL Domain-Containing Cyclic Di-GMP Turnover Proteins of Escherichia coli. J. Bacteriol. 2016, 198, 7–11. [Google Scholar] [CrossRef]
- Tschowri, N.; Schumacher, M.A.; Schlimpert, S.; Chinnam, N.B.; Findlay, K.C.; Brennan, R.G.; Buttner, M.J. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 2014, 158, 1136–1147. [Google Scholar] [CrossRef]
- Habazettl, J.; Allan, M.G.; Jenal, U.; Grzesiek, S. Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout. J. Biol. Chem. 2011, 286, 14304–14314. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Zeng, W. Structures of the activator of K. pneumonia biofilm formation, MrkH, indicates PilZ domains involved in c-di-GMP and DNA binding. Proc. Natl. Acad. Sci. USA 2016, 113, 10067–10072. [Google Scholar] [CrossRef]
- Duerig, A.; Abel, S.; Folcher, M.; Nicollier, M.; Schwede, T.; Amiot, N.; Giese, B.; Jenal, U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes. Dev. 2009, 23, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Hengge, R. Trigger phosphodiesterases as a novel class of c-di-GMP effector proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20150498. [Google Scholar] [CrossRef] [PubMed]
- Steiner, S.; Lori, C.; Boehm, A.; Jenal, U. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J. 2013, 32, 354–368. [Google Scholar] [CrossRef] [PubMed]
- Trampari, E.; Stevenson, C.E.; Little, R.H.; Wilhelm, T.; Lawson, D.M.; Malone, J.G. Bacterial rotary export ATPases are allosterically regulated by the nucleotide second messenger cyclic-di-GMP. J. Biol. Chem. 2015, 290, 24470–24483. [Google Scholar] [CrossRef]
- Little, R.H.; Grenga, L.; Saalbach, G.; Howat, A.M.; Pfeilmeier, S.; Trampari, E.; Malone, J.G. Adaptive Remodeling of the Bacterial Proteome by Specific Ribosomal Modification Regulates Pseudomonas Infection and Niche Colonisation. PLoS Genet. 2016, 12, e1005837. [Google Scholar] [CrossRef]
- Lori, C.; Ozaki, S.; Steiner, S.; Böhm, R.; Abel, S.; Dubey, B.N.; Schirmer, T.; Hiller, S.; Jenal, U. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 2015, 523, 236–239. [Google Scholar] [CrossRef]
- Roelofs, K.G.; Jones, C.J.; Helman, S.R.; Shang, X.; Orr, M.W.; Goodson, J.R.; Galperin, M.Y.; Yildiz, F.H.; Lee, V.T. Systematic Identification of Cyclic-di-GMP Binding Proteins in Vibrio cholerae Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems. PLoS Pathog. 2015, 11, e1005232. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.J.; Utada, A.; Davis, K.R.; Thongsomboon, W.; Zamorano Sanchez, D.; Banakar, V.; Cegelski, L.; Wong, G.C.; Yildiz, F.H. C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in Vibrio cholerae. PLoS Pathog. 2015, 11, e1005068. [Google Scholar] [CrossRef]
- Wang, Y.C.; Chin, K.H.; Tu, Z.L.; He, J.; Jones, C.J.; Sanchez, D.Z.; Yildiz, F.H.; Galperin, M.Y.; Chou, S.H. Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain. Nat. Commun. 2016, 7, 12481. [Google Scholar] [CrossRef]
- Wiesmann, C.L.; Wang, N.R.; Zhang, Y.; Liu, Z.; Haney, C.H. Origins of symbiosis: Shared mechanisms underlying microbial pathogenesis, commensalism and mutualism of plants and animals. FEMS Microbiol. Rev. 2023, 47, fuac048. [Google Scholar] [CrossRef] [PubMed]
- Krasteva, P.V.; Giglio, K.M.; Sondermann, H. Sensing the messenger: The diverse ways that bacteria signal through c-di-GMP. Protein Sci. 2012, 21, 929–948. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.; Belasco, J.G. Riboswitch control of bacterial RNA stability. Mol. Microbiol. 2021, 116, 361–365. [Google Scholar] [CrossRef]
- Pavlova, N.; Kaloudas, D.; Penchovsky, R. Riboswitch distribution, structure, and function in bacteria. Gene 2019, 708, 38–48. [Google Scholar] [CrossRef]
- Kulshina, N.; Baird, N.J.; Ferré-D’Amaré, A.R. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat. Struct. Mol. Biol. 2009, 16, 1212–1217. [Google Scholar] [CrossRef]
- Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009, 7, 263–273. [Google Scholar] [CrossRef]
- Hengge, R. Cyclic-di-GMP reaches out into the bacterial RNA world. Sci. Signal 2010, 3, pe44. [Google Scholar] [CrossRef]
- Lee, E.R.; Baker, J.L.; Weinberg, Z.; Sudarsan, N.; Breaker, R.R. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 2010, 329, 845–848. [Google Scholar] [CrossRef]
- Weinberg, Z.; Barrick, J.E.; Yao, Z.; Roth, A.; Kim, J.N.; Gore, J.; Wang, J.X.; Lee, E.R.; Block, K.F.; Sudarsan, N.; et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007, 35, 4809–4819. [Google Scholar] [CrossRef]
- Sudarsan, N.; Lee, E.R.; Weinberg, Z.; Moy, R.H.; Kim, J.N.; Link, K.H.; Breaker, R.R. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 2008, 321, 411–413. [Google Scholar] [CrossRef] [PubMed]
- Meibom, K.L.; Blokesch, M.; Dolganov, N.A.; Wu, C.Y.; Schoolnik, G.K. Chitin induces natural competence in Vibrio cholerae. Science 2005, 310, 1824–1827. [Google Scholar] [CrossRef]
- Kirn, T.J.; Jude, B.A.; Taylor, R.K. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 2005, 438, 863–866. [Google Scholar] [CrossRef] [PubMed]
- Breaker, R.R.; Atilho, R.M.; Malkowski, S.N.; Nelson, J.W.; Sherlock, M.E. The Biology of Free Guanidine As Revealed by Riboswitches. Biochemistry 2017, 56, 345–347. [Google Scholar] [CrossRef]
- Kavita, K.; Narunsky, A.; Mohsen, J.J.; Mahadeshwar, I.; Mohsen, M.G.; Chang, Y.S.; Breaker, R.R. Guanidine aptamers are present in vertebrate RNAs associated with calcium signaling and neuromuscular function. Nat. Commun. 2025, 16, 7362. [Google Scholar] [CrossRef] [PubMed]
- Soukup, G.A.; Breaker, R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 1999, 5, 1308–1325. [Google Scholar] [CrossRef]
- Regulski, E.E.; Breaker, R.R. In-line probing analysis of riboswitches. Methods Mol. Biol. 2008, 419, 53–67. [Google Scholar] [CrossRef]
- Vicens, Q.; Cech, T.R. Atomic level architecture of group I introns revealed. Trends Biochem. Sci. 2006, 31, 41–51. [Google Scholar] [CrossRef]
- Baraquet, C.; Harwood, C.S. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc. Natl. Acad. Sci. USA 2013, 110, 18478–18483. [Google Scholar] [CrossRef] [PubMed]
- Hickman, J.W.; Harwood, C.S. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2008, 69, 376–389. [Google Scholar] [CrossRef] [PubMed]
- Francke, C.; Groot Kormelink, T.; Hagemeijer, Y.; Overmars, L.; Sluijter, V.; Moezelaar, R.; Siezen, R.J. Comparative analyses imply that the enigmatic Sigma factor 54 is a central controller of the bacterial exterior. BMC Genom. 2011, 12, 385. [Google Scholar] [CrossRef]
- Matsuyama, B.Y.; Krasteva, P.V.; Baraquet, C.; Harwood, C.S.; Sondermann, H.; Navarro, M.V. Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2016, 113, E209–E218. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, N.; Arora, S.K.; Ramphal, R. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 357–364. [Google Scholar] [CrossRef]
- Baraquet, C.; Murakami, K.; Parsek, M.R.; Harwood, C.S. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 2012, 40, 7207–7218. [Google Scholar] [CrossRef]
- Torres-Sánchez, L.; Sana, T.G.; Decossas, M.; Hashem, Y.; Krasteva, P.V. Structures of the P. aeruginosa FleQ-FleN master regulators reveal large-scale conformational switching in motility and biofilm control. Proc. Natl. Acad. Sci. USA 2023, 120, e2312276120. [Google Scholar] [CrossRef]
- Srivastava, D.; Hsieh, M.L.; Khataokar, A.; Neiditch, M.B.; Waters, C.M. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol. Microbiol. 2013, 90, 1262–1276. [Google Scholar] [CrossRef]
- Krasteva, P.V.; Fong, J.C.; Shikuma, N.J.; Beyhan, S.; Navarro, M.V.; Yildiz, F.H.; Sondermann, H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 2010, 327, 866–868. [Google Scholar] [CrossRef]
- Srivastava, D.; Harris, R.C.; Waters, C.M. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J. Bacteriol. 2011, 193, 6331–6341. [Google Scholar] [CrossRef]
- Waters, C.M.; Lu, W.; Rabinowitz, J.D.; Bassler, B.L. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 2008, 190, 2527–2536. [Google Scholar] [CrossRef]
- Rutherford, S.T.; van Kessel, J.C.; Shao, Y.; Bassler, B.L. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes. Dev. 2011, 25, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; He, Z.G. LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Res. 2012, 40, 11292–11307. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, L.; Zhang, H.; He, Z.G. Cyclic di-GMP triggers the hypoxic adaptation of Mycobacterium bovis through a metabolic switching regulator ArgR. Environ. Microbiol. 2022, 24, 4382–4400. [Google Scholar] [CrossRef]
- Li, W.; Hu, L.; Xie, Z.; Xu, H.; Li, M.; Cui, T.; He, Z.G. Cyclic di-GMP integrates functionally divergent transcription factors into a regulation pathway for antioxidant defense. Nucleic Acids Res. 2018, 46, 7270–7283. [Google Scholar] [CrossRef]
- Li, W.; Li, M.; Hu, L.; Zhu, J.; Xie, Z.; Chen, J.; He, Z.G. HpoR, a novel c-di-GMP effective transcription factor, links the second messenger’s regulatory function to the mycobacterial antioxidant defense. Nucleic Acids Res. 2018, 46, 3595–3611. [Google Scholar] [CrossRef]
- Hu, Q.; Zhang, J.; Chen, Y.; Hu, L.; Li, W.; He, Z.G. Cyclic di-GMP co-activates the two-component transcriptional regulator DevR in Mycobacterium smegmatis in response to oxidative stress. J. Biol. Chem. 2019, 294, 12729–12742. [Google Scholar] [CrossRef] [PubMed]
- Chin, K.H.; Lee, Y.C.; Tu, Z.L.; Chen, C.H.; Tseng, Y.H.; Yang, J.M.; Ryan, R.P.; McCarthy, Y.; Dow, J.M.; Wang, A.H.; et al. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J. Mol. Biol. 2010, 396, 646–662. [Google Scholar] [CrossRef] [PubMed]
- He, Y.W.; Ng, A.Y.; Xu, M.; Lin, K.; Wang, L.H.; Dong, Y.H.; Zhang, L.H. Xanthomonas campestris cell-cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol. Microbiol. 2007, 64, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Amikam, D.; Galperin, M.Y. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 2006, 22, 3–6. [Google Scholar] [CrossRef]
- Morgan, J.L.; Strumillo, J.; Zimmer, J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 2013, 493, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Slabaugh, E.; Davis, J.K.; Haigler, C.H.; Yingling, Y.G.; Zimmer, J. Cellulose synthases: New insights from crystallography and modeling. Trends Plant Sci. 2014, 19, 99–106. [Google Scholar] [CrossRef]
- Morgan, J.L.; McNamara, J.T.; Zimmer, J. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat. Struct. Mol. Biol. 2014, 21, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Merighi, M.; Lee, V.T.; Hyodo, M.; Hayakawa, Y.; Lory, S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 2007, 65, 876–895. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.E.; Diepold, A.; Kuchma, S.L.; Scott, J.E.; Ha, D.G.; Orazi, G.; Armitage, J.P.; O’Toole, G.A. PilZ Domain Protein FlgZ Mediates Cyclic Di-GMP-Dependent Swarming Motility Control in Pseudomonas aeruginosa. J. Bacteriol. 2016, 198, 1837–1846. [Google Scholar] [CrossRef]
- Li, T.N.; Chin, K.H.; Liu, J.H.; Wang, A.H.; Chou, S.H. XC1028 from Xanthomonas campestris adopts a PilZ domain-like structure without a c-di-GMP switch. Proteins 2009, 75, 282–288. [Google Scholar] [CrossRef]
- Cheng, T.; Cheang, Q.W.; Xu, L.; Sheng, S.; Li, Z.; Shi, Y.; Zhang, H.; Pang, L.M.; Liu, D.X.; Yang, L.; et al. A PilZ domain protein interacts with the transcriptional regulator HinK to regulate type VI secretion system in Pseudomonas aeruginosa. J. Biol. Chem. 2024, 300, 105741. [Google Scholar] [CrossRef]
- Xu, L.; Xin, L.; Zeng, Y.; Yam, J.K.; Ding, Y.; Venkataramani, P.; Cheang, Q.W.; Yang, X.; Tang, X.; Zhang, L.H.; et al. A cyclic di-GMP-binding adaptor protein interacts with a chemotaxis methyltransferase to control flagellar motor switching. Sci. Signal 2016, 9, ra102. [Google Scholar] [CrossRef]
- Petrova, O.E.; Gupta, K.; Liao, J.; Goodwine, J.S.; Sauer, K. Divide and conquer: The Pseudomonas aeruginosa two-component hybrid SagS enables biofilm formation and recalcitrance of biofilm cells to antimicrobial agents via distinct regulatory circuits. Environ. Microbiol. 2017, 19, 2005–2024. [Google Scholar] [CrossRef]
- Valentini, M.; Filloux, A. Biofilms and Cyclic di-GMP (c-di-GMP) Signaling: Lessons from Pseudomonas aeruginosa and Other Bacteria. J. Biol. Chem. 2016, 291, 12547–12555. [Google Scholar] [CrossRef] [PubMed]
- Laventie, B.J.; Sangermani, M.; Estermann, F.; Manfredi, P.; Planes, R.; Hug, I.; Jaeger, T.; Meunier, E.; Broz, P.; Jenal, U. A Surface-Induced Asymmetric Program Promotes Tissue Colonization by Pseudomonas aeruginosa. Cell Host Microbe 2019, 25, 140–152.e146. [Google Scholar] [CrossRef] [PubMed]
- Gacesa, P. Bacterial alginate biosynthesis--recent progress and future prospects. Microbiology 1998, 144 Pt 5, 1133–1143. [Google Scholar] [CrossRef]
- Yu, H.; Head, N.E. Persistent infections and immunity in cystic fibrosis. Front. Biosci. 2002, 7, 442–457. [Google Scholar] [CrossRef] [PubMed]
- Dinh, T.; Paulsen, I.T.; Saier, M.H., Jr. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J. Bacteriol. 1994, 176, 3825–3831. [Google Scholar] [CrossRef]
- Mattick, J.S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002, 56, 289–314. [Google Scholar] [CrossRef]
- Burrows, L.L. Pseudomonas aeruginosa twitching motility: Type IV pili in action. Annu. Rev. Microbiol. 2012, 66, 493–520. [Google Scholar] [CrossRef]
- Hendrix, H.; Itterbeek, A.; Longin, H.; Delanghe, L.; Vriens, E.; Vallino, M.; Lammens, E.M.; Haque, F.; Yusuf, A.; Noben, J.P.; et al. PlzR regulates type IV pili assembly in Pseudomonas aeruginosa via PilZ binding. Nat. Commun. 2024, 15, 8717. [Google Scholar] [CrossRef]
- Toutain, C.M.; Zegans, M.E.; O’Toole, G.A. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 771–777. [Google Scholar] [CrossRef]
- Kuchma, S.L.; Delalez, N.J.; Filkins, L.M.; Snavely, E.A.; Armitage, J.P.; O’Toole, G.A. Cyclic di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa PA14 requires the MotAB stator. J. Bacteriol. 2015, 197, 420–430. [Google Scholar] [CrossRef]
- Bense, S.; Bruchmann, S.; Steffen, A.; Stradal, T.E.B.; Häussler, S.; Düvel, J. Spatiotemporal control of FlgZ activity impacts Pseudomonas aeruginosa flagellar motility. Mol. Microbiol. 2019, 111, 1544–1557. [Google Scholar] [CrossRef]
- Sheng, S.; Xin, L.; Yam, J.K.H.; Salido, M.M.; Khong, N.Z.J.; Liu, Q.; Chea, R.A.; Li, H.Y.; Yang, L.; Liang, Z.X.; et al. The MapZ-Mediated Methylation of Chemoreceptors Contributes to Pathogenicity of Pseudomonas aeruginosa. Front. Microbiol. 2019, 10, 67. [Google Scholar] [CrossRef]
- Park, S.; Dingemans, J.; Gowett, M.; Sauer, K. Glucose-6-Phosphate Acts as an Extracellular Signal of SagS To Modulate Pseudomonas aeruginosa c-di-GMP Levels, Attachment, and Biofilm Formation. mSphere 2021, 6, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Sheng, S. Preliminary screening of biological phenotypes of single PilZ domain protein PA0012 and PA4324 mutants in Pseudomonas aeruginosa. J. Zunyi Med. Univ. 2024, 47, 38–46. [Google Scholar] [CrossRef]
- Liu, Q. Study on Pathogenicity-Related Biological Functions of PilZ Domain-Containing Protein PA2989 in Pseudomonas Aeruginosa. Master’s Thesis, South China Agricultural University, Guangzhou, China, 2019. [Google Scholar] [CrossRef]
- Boehm, A.; Kaiser, M.; Li, H.; Spangler, C.; Kasper, C.A.; Ackermann, M.; Kaever, V.; Sourjik, V.; Roth, V.; Jenal, U. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 2010, 141, 107–116. [Google Scholar] [CrossRef]
- Fang, X.; Gomelsky, M. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 2010, 76, 1295–1305. [Google Scholar] [CrossRef]
- Zorraquino, V.; García, B.; Latasa, C.; Echeverz, M.; Toledo-Arana, A.; Valle, J.; Lasa, I.; Solano, C. Coordinated cyclic-di-GMP repression of Salmonella motility through YcgR and cellulose. J. Bacteriol. 2013, 195, 417–428. [Google Scholar] [CrossRef]
- Gao, X.; Mukherjee, S.; Matthews, P.M.; Hammad, L.A.; Kearns, D.B.; Dann, C.E., 3rd. Functional characterization of core components of the Bacillus subtilis cyclic-di-GMP signaling pathway. J. Bacteriol. 2013, 195, 4782–4792. [Google Scholar] [CrossRef]
- Subramanian, S.; Gao, X.; Dann, C.E., 3rd; Kearns, D.B. MotI (DgrA) acts as a molecular clutch on the flagellar stator protein MotA in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2017, 114, 13537–13542. [Google Scholar] [CrossRef] [PubMed]
- Christen, M.; Christen, B.; Allan, M.G.; Folcher, M.; Jenö, P.; Grzesiek, S.; Jenal, U. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 2007, 104, 4112–4117. [Google Scholar] [CrossRef]
- An, S.Q.; Caly, D.L.; McCarthy, Y.; Murdoch, S.L.; Ward, J.; Febrer, M.; Dow, J.M.; Ryan, R.P. Novel cyclic di-GMP effectors of the YajQ protein family control bacterial virulence. PLoS Pathog. 2014, 10, e1004429. [Google Scholar] [CrossRef]
- Han, S.; Shen, D.; Wang, Y.C.; Chou, S.H.; Gomelsky, M.; Gao, Y.G.; Qian, G. A YajQ-LysR-like, cyclic di-GMP-dependent system regulating biosynthesis of an antifungal antibiotic in a crop-protecting bacterium, Lysobacter enzymogenes. Mol. Plant Pathol. 2020, 21, 218–229. [Google Scholar] [CrossRef]
- Xu, L.; Wu, P.; Wright, S.J.; Du, L.; Wei, X. Bioactive Polycyclic Tetramate Macrolactams from Lysobacter enzymogenes and Their Absolute Configurations by Theoretical ECD Calculations. J. Nat. Prod. 2015, 78, 1841–1847. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, S.; Schalch-Moser, A.; Zumthor, L.; Manfredi, P.; Ebbensgaard, A.; Schirmer, T.; Jenal, U. Activation and polar sequestration of PopA, a c-di-GMP effector protein involved in Caulobacter crescentus cell cycle control. Mol. Microbiol. 2014, 94, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.C.; Joshi, K.K.; Zik, J.J.; Trinh, K.; Kamajaya, A.; Chien, P.; Ryan, K.R. Cell cycle-dependent adaptor complex for ClpXP-mediated proteolysis directly integrates phosphorylation and second messenger signals. Proc. Natl. Acad. Sci. USA 2014, 111, 14229–14234. [Google Scholar] [CrossRef]
- De, N.; Navarro, M.V.; Raghavan, R.V.; Sondermann, H. Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J. Mol. Biol. 2009, 393, 619–633. [Google Scholar] [CrossRef]
- De, N.; Pirruccello, M.; Krasteva, P.V.; Bae, N.; Raghavan, R.V.; Sondermann, H. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 2008, 6, e67. [Google Scholar] [CrossRef]
- Kazmierczak, B.I.; Lebron, M.B.; Murray, T.S. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 2006, 60, 1026–1043. [Google Scholar] [CrossRef]
- Llontop, E.E.; Cenens, W.; Favaro, D.C.; Sgro, G.G.; Salinas, R.K.; Guzzo, C.R.; Farah, C.S. The PilB-PilZ-FimX regulatory complex of the Type IV pilus from Xanthomonas citri. PLoS Pathog. 2021, 17, e1009808. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Sliusarenko, O.; Kazmierczak, B.I. Interaction of the cyclic-di-GMP binding protein FimX and the Type 4 pilus assembly ATPase promotes pilus assembly. PLoS Pathog. 2017, 13, e1006594. [Google Scholar] [CrossRef]
- Newell, P.D.; Monds, R.D.; O’Toole, G.A. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. USA 2009, 106, 3461–3466. [Google Scholar] [CrossRef]
- Cooley, R.B.; Smith, T.J.; Leung, W.; Tierney, V.; Borlee, B.R.; O’Toole, G.A.; Sondermann, H. Cyclic Di-GMP-Regulated Periplasmic Proteolysis of a Pseudomonas aeruginosa Type Vb Secretion System Substrate. J. Bacteriol. 2016, 198, 66–76. [Google Scholar] [CrossRef]
- Rybtke, M.; Berthelsen, J.; Yang, L.; Høiby, N.; Givskov, M.; Tolker-Nielsen, T. The LapG protein plays a role in Pseudomonas aeruginosa biofilm formation by controlling the presence of the CdrA adhesin on the cell surface. Microbiologyopen 2015, 4, 917–930. [Google Scholar] [CrossRef]
- Collins, A.J.; Smith, T.J.; Sondermann, H.; O’Toole, G.A. From Input to Output: The Lap/c-di-GMP Biofilm Regulatory Circuit. Annu. Rev. Microbiol. 2020, 74, 607–631. [Google Scholar] [CrossRef] [PubMed]
- Pesavento, C.; Becker, G.; Sommerfeldt, N.; Possling, A.; Tschowri, N.; Mehlis, A.; Hengge, R. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes. Dev. 2008, 22, 2434–2446. [Google Scholar] [CrossRef] [PubMed]
- Lindenberg, S.; Klauck, G.; Pesavento, C.; Klauck, E.; Hengge, R. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J. 2013, 32, 2001–2014. [Google Scholar] [CrossRef]
- Sommerfeldt, N.; Possling, A.; Becker, G.; Pesavento, C.; Tschowri, N.; Hengge, R. Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology (Reading) 2009, 155, 1318–1331. [Google Scholar] [CrossRef]
- Sundriyal, A.; Massa, C.; Samoray, D.; Zehender, F.; Sharpe, T.; Jenal, U.; Schirmer, T. Inherent regulation of EAL domain-catalyzed hydrolysis of second messenger cyclic di-GMP. J. Biol. Chem. 2014, 289, 6978–6990. [Google Scholar] [CrossRef]
- Reinders, A.; Hee, C.S.; Ozaki, S.; Mazur, A.; Boehm, A.; Schirmer, T.; Jenal, U. Expression and Genetic Activation of Cyclic Di-GMP-Specific Phosphodiesterases in Escherichia coli. J. Bacteriol. 2016, 198, 448–462. [Google Scholar] [CrossRef]
- Spangler, C.; Böhm, A.; Jenal, U.; Seifert, R.; Kaever, V. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J. Microbiol. Methods 2010, 81, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Borlee, B.R.; Goldman, A.D.; Murakami, K.; Samudrala, R.; Wozniak, D.J.; Parsek, M.R. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 2010, 75, 827–842. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Rice, J.D.; Goller, C.; Pannuri, A.; Taylor, J.; Meisner, J.; Beveridge, T.J.; Preston, J.F., 3rd; Romeo, T. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J. Bacteriol. 2008, 190, 3670–3680. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Preston, J.F., 3rd; Romeo, T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 2004, 186, 2724–2734. [Google Scholar] [CrossRef]
- Tagliabue, L.; Antoniani, D.; Maciąg, A.; Bocci, P.; Raffaelli, N.; Landini, P. The diguanylate cyclase YddV controls production of the exopolysaccharide poly-N-acetylglucosamine (PNAG) through regulation of the PNAG biosynthetic pgaABCD operon. Microbiology (Reading) 2010, 156, 2901–2911. [Google Scholar] [CrossRef] [PubMed]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef]
- Afsharmanesh, H.; Ahmadzadeh, M.; Sharifi-Tehrani, A.; Javan-Nikkhah, M.; Ghazanfari, K. Detection of phlD gene in some fluorescent pseudomonads isolated from Iran and its relative with antifungal activities. Commun. Agric. Appl. Biol. Sci. 2007, 72, 941–950. [Google Scholar]
- Lindeberg, M.; Cunnac, S.; Collmer, A. The evolution of Pseudomonas syringae host specificity and type III effector repertoires. Mol. Plant Pathol. 2009, 10, 767–775. [Google Scholar] [CrossRef]
- Xin, X.F.; He, S.Y. Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 2013, 51, 473–498. [Google Scholar] [CrossRef]
- Lugtenberg, B.J.; Dekkers, L.; Bloemberg, G.V. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 2001, 39, 461–490. [Google Scholar] [CrossRef]
- Macnab, R.M. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 2004, 1694, 207–217. [Google Scholar] [CrossRef]
- Minamino, T.; Imada, K.; Namba, K. Mechanisms of type III protein export for bacterial flagellar assembly. Mol. Biosyst. 2008, 4, 1105–1115. [Google Scholar] [CrossRef]
- Minamino, T.; Namba, K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 2008, 451, 485–488. [Google Scholar] [CrossRef]
- Abel, S.; Chien, P.; Wassmann, P.; Schirmer, T.; Kaever, V.; Laub, M.T.; Baker, T.A.; Jenal, U. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol. Cell 2011, 43, 550–560. [Google Scholar] [CrossRef]
- Kirkpatrick, C.L.; Viollier, P.H. Decoding Caulobacter development. FEMS Microbiol. Rev. 2012, 36, 193–205. [Google Scholar] [CrossRef]
- Dubey, B.N.; Lori, C.; Ozaki, S.; Fucile, G.; Plaza-Menacho, I.; Jenal, U.; Schirmer, T. Cyclic di-GMP mediates a histidine kinase/phosphatase switch by noncovalent domain cross-linking. Sci. Adv. 2016, 2, e1600823. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.E.; Tropini, C.; Jonas, K.; Tsokos, C.G.; Huang, K.C.; Laub, M.T. Spatial gradient of protein phosphorylation underlies replicative asymmetry in a bacterium. Proc. Natl. Acad. Sci. USA 2011, 108, 1052–1057. [Google Scholar] [CrossRef] [PubMed]
- Tsokos, C.G.; Perchuk, B.S.; Laub, M.T. A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev. Cell 2011, 20, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Zeng, W.; Findlay, K.C.; Buttner, M.J.; Brennan, R.G.; Tschowri, N. The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex. Nucleic Acids Res. 2017, 45, 6923–6933. [Google Scholar] [CrossRef]
- Tao, F.; He, Y.W.; Wu, D.H.; Swarup, S.; Zhang, L.H. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J. Bacteriol. 2010, 192, 1020–1029. [Google Scholar] [CrossRef]
- Wilksch, J.J.; Yang, J.; Clements, A.; Gabbe, J.L.; Short, K.R.; Cao, H.; Cavaliere, R.; James, C.E.; Whitchurch, C.B.; Schembri, M.A.; et al. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 2011, 7, e1002204. [Google Scholar] [CrossRef]
- Chambers, J.R.; Liao, J.; Schurr, M.J.; Sauer, K. BrlR from Pseudomonas aeruginosa is a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2014, 92, 471–487. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Wu, Y.; Yuan, Z.H.; Cai, Z.; Qian, W.; Ge, X.; Wang, F.F. Dual Regulatory Role Exerted by Cyclic Dimeric GMP To Control FsnR-Mediated Bacterial Swimming. mBio 2022, 13, e0141422. [Google Scholar] [CrossRef] [PubMed]
- Skotnicka, D.; Steinchen, W.; Szadkowski, D.; Cadby, I.T.; Lovering, A.L.; Bange, G.; Søgaard-Andersen, L. CdbA is a DNA-binding protein and c-di-GMP receptor important for nucleoid organization and segregation in Myxococcus xanthus. Nat. Commun. 2020, 11, 1791. [Google Scholar] [CrossRef]
- Russell, M.H.; Bible, A.N.; Fang, X.; Gooding, J.R.; Campagna, S.R.; Gomelsky, M.; Alexandre, G. Integration of the second messenger c-di-GMP into the chemotactic signaling pathway. mBio 2013, 4, e00001-13. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xie, Z.; Sui, F.; Liu, X.; Cheng, W. Identification of Cbp1, a c-di-GMP Binding Chemoreceptor in Azorhizobium caulinodans ORS571 Involved in Chemotaxis and Nodulation of the Host Plant. Front. Microbiol. 2019, 10, 638. [Google Scholar] [CrossRef] [PubMed]
- Seidel, M.; Skotnicka, D.; Glatter, T.; Søgaard-Andersen, L. During heat stress in Myxococcus xanthus, the CdbS PilZ domain protein, in concert with two PilZ-DnaK chaperones, perturbs chromosome organization and accelerates cell death. PLoS Genet. 2023, 19, e1010819. [Google Scholar] [CrossRef] [PubMed]
- Pratt, J.T.; Tamayo, R.; Tischler, A.D.; Camilli, A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 2007, 282, 12860–12870. [Google Scholar] [CrossRef]
- Benach, J.; Swaminathan, S.S.; Tamayo, R.; Handelman, S.K.; Folta-Stogniew, E.; Ramos, J.E.; Forouhar, F.; Neely, H.; Seetharaman, J.; Camilli, A.; et al. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007, 26, 5153–5166. [Google Scholar] [CrossRef]
- Lee, V.T.; Matewish, J.M.; Kessler, J.L.; Hyodo, M.; Hayakawa, Y.; Lory, S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 2007, 65, 1474–1484. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, H.; Zhang, X.; Jiang, H.; Liu, C.; Wu, F.; Qian, L.; Hao, B.; Czajkowsky, D.M.; Guo, S.; et al. Interplay between the bacterial protein deacetylase CobB and the second messenger c-di-GMP. EMBO J. 2019, 38, e100948. [Google Scholar] [CrossRef]
- Liu, C.; Shi, R.; Jensen, M.S.; Zhu, J.; Liu, J.; Liu, X.; Sun, D.; Liu, W. The global regulation of c-di-GMP and cAMP in bacteria. mLife 2024, 3, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Wörmann, M.E.; Henderson, M.; Salinas, R.; Latoscha, A.; Al-Bassam, M.M.; Singh, K.S.; Barclay, E.; Gunka, K.; Tschowri, N. Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP. Nat. Commun. 2022, 13, 5834. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.; Mahmoud, S.A.; Kim, S.K.; Ogdahl, J.L.; Lee, V.T.; Chien, P.; Yildiz, F.H. c-di-GMP inhibits LonA-dependent proteolysis of TfoY in Vibrio cholerae. PLoS Genet. 2020, 16, e1008897. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, J.; Yun, G.; Liu, B.; Li, X.; Jiang, M.; Yu, X.; Zhang, J.; Han, Y.; Liu, D.; Zhao, J.; et al. Dynamic Executors of Bacterial Signals: Functional Versatility and Regulatory Networks of c-di-GMP Effectors. Biomolecules 2025, 15, 1471. https://doi.org/10.3390/biom15101471
Jia J, Yun G, Liu B, Li X, Jiang M, Yu X, Zhang J, Han Y, Liu D, Zhao J, et al. Dynamic Executors of Bacterial Signals: Functional Versatility and Regulatory Networks of c-di-GMP Effectors. Biomolecules. 2025; 15(10):1471. https://doi.org/10.3390/biom15101471
Chicago/Turabian StyleJia, Jia, Ge Yun, Bingxin Liu, Xinxin Li, Meiling Jiang, Xinlu Yu, Jing Zhang, Yufei Han, Dan Liu, Junlong Zhao, and et al. 2025. "Dynamic Executors of Bacterial Signals: Functional Versatility and Regulatory Networks of c-di-GMP Effectors" Biomolecules 15, no. 10: 1471. https://doi.org/10.3390/biom15101471
APA StyleJia, J., Yun, G., Liu, B., Li, X., Jiang, M., Yu, X., Zhang, J., Han, Y., Liu, D., Zhao, J., Wang, Y., & Chen, G. (2025). Dynamic Executors of Bacterial Signals: Functional Versatility and Regulatory Networks of c-di-GMP Effectors. Biomolecules, 15(10), 1471. https://doi.org/10.3390/biom15101471






