A Mesophilic Argonaute from Cohnella algarum Mediates Programmable DNA/RNA Cleavage with Distinctive Guide Specificity
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Sequence Analysis
2.3. CalAgo Expression and Purification
2.4. Enzymatic Activity Assay
2.5. Electrophoretic Mobility Shift Assay (EMSA)
3. Results
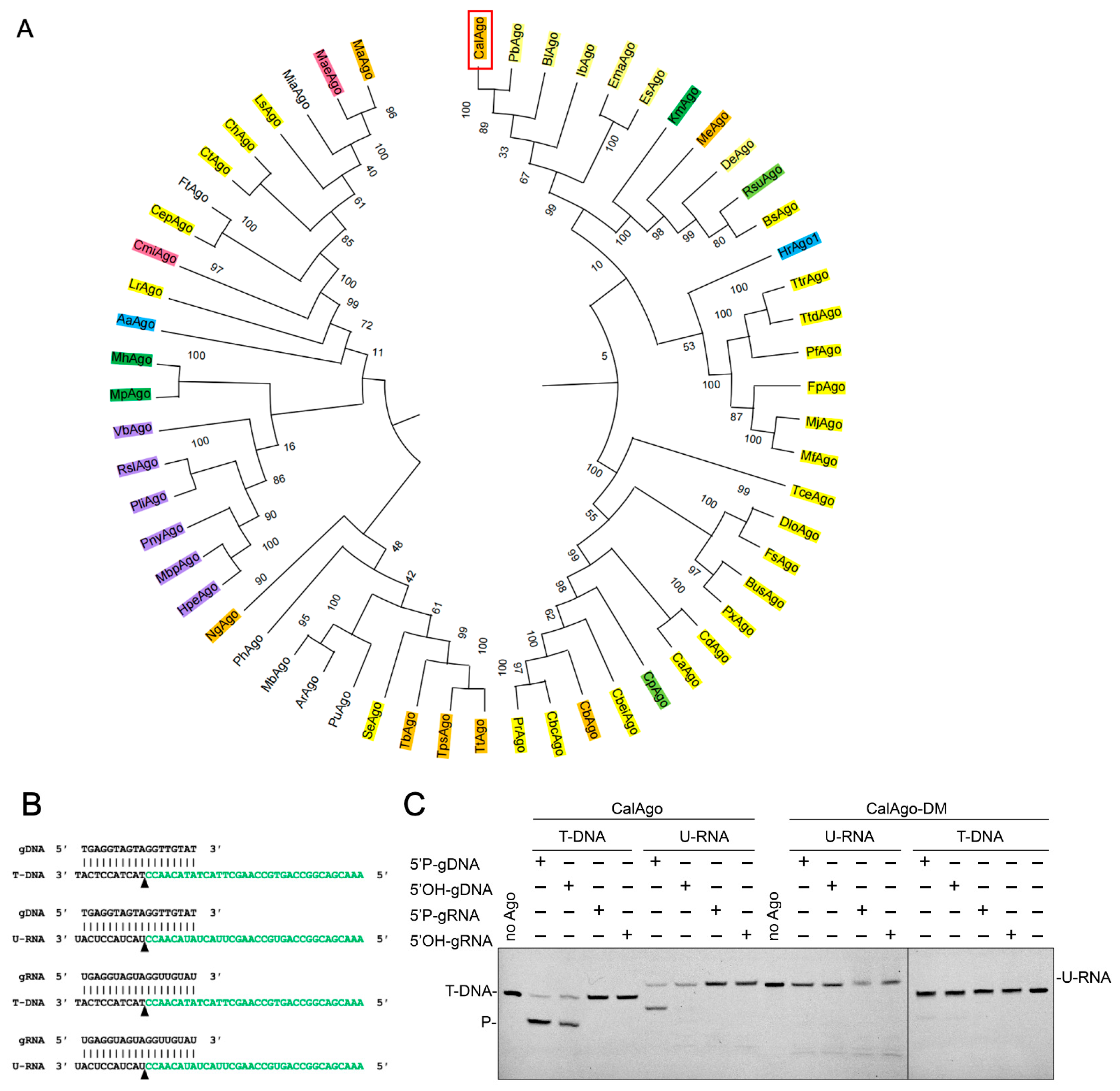
3.1. Sequence Analysis of CalAgo
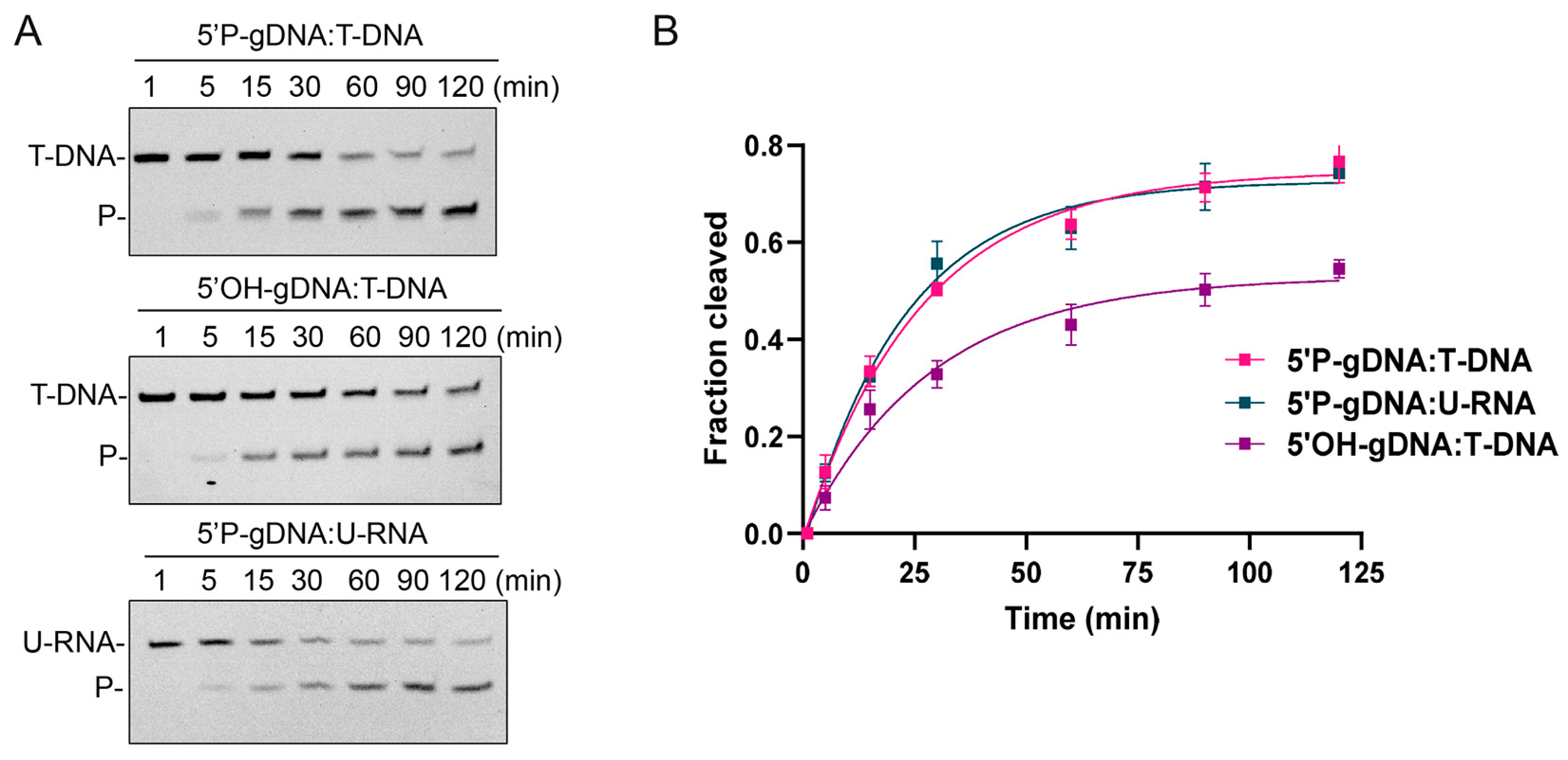
3.2. CalAgo Mediates DNA-Guided DNA and RNA Cleavage at 37 °C
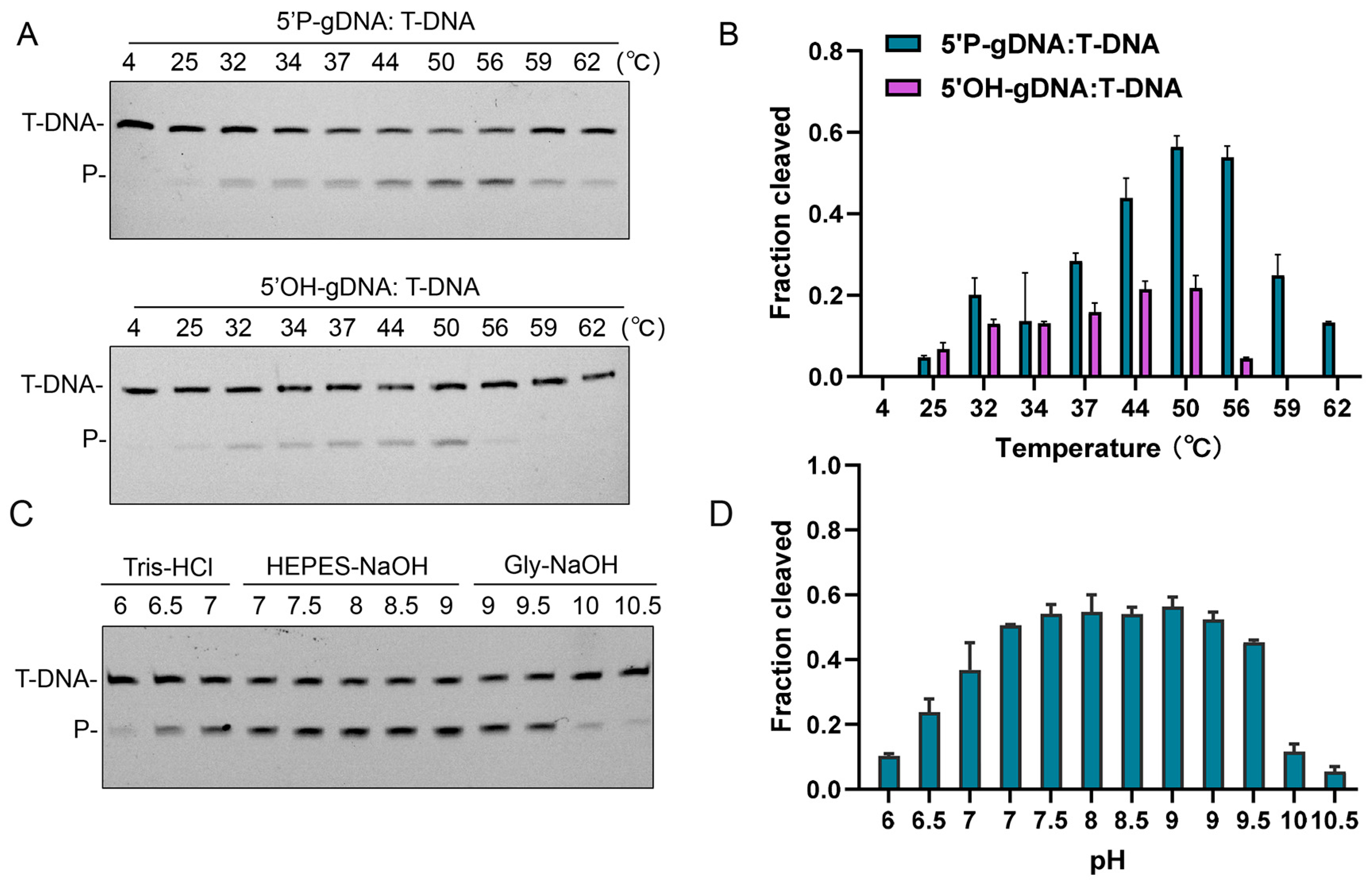
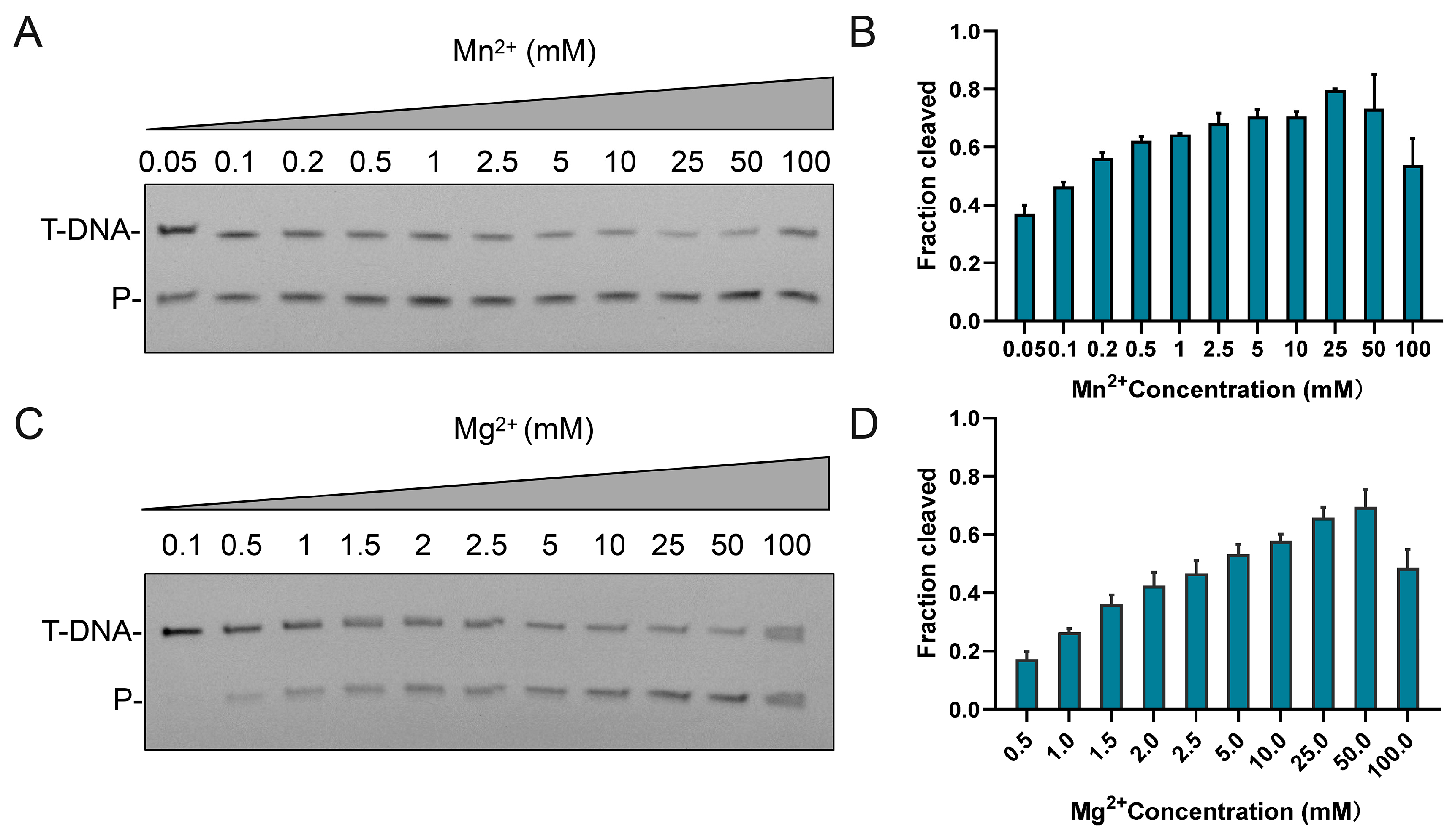
3.3. Effects of Temperature, pH, and Metal Ion on CalAgo Cleavage Activity
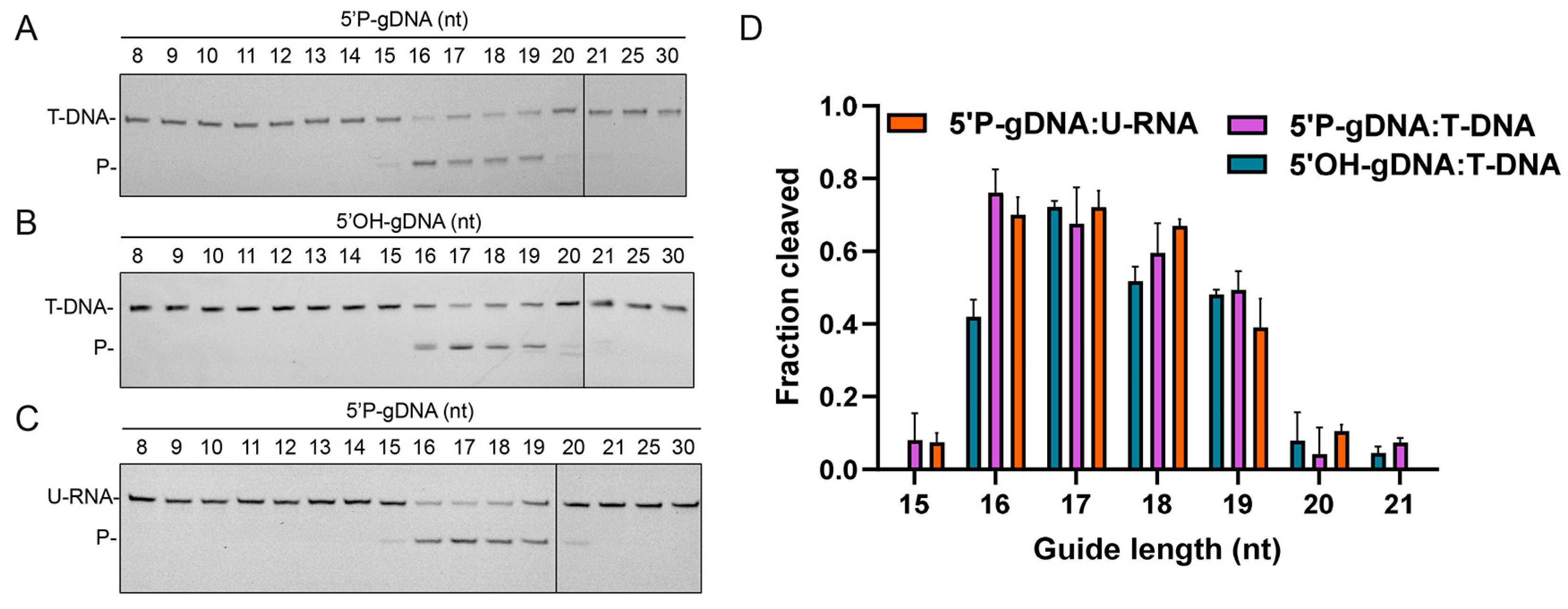
3.4. Effects of gDNA Length on Cleavage Activity
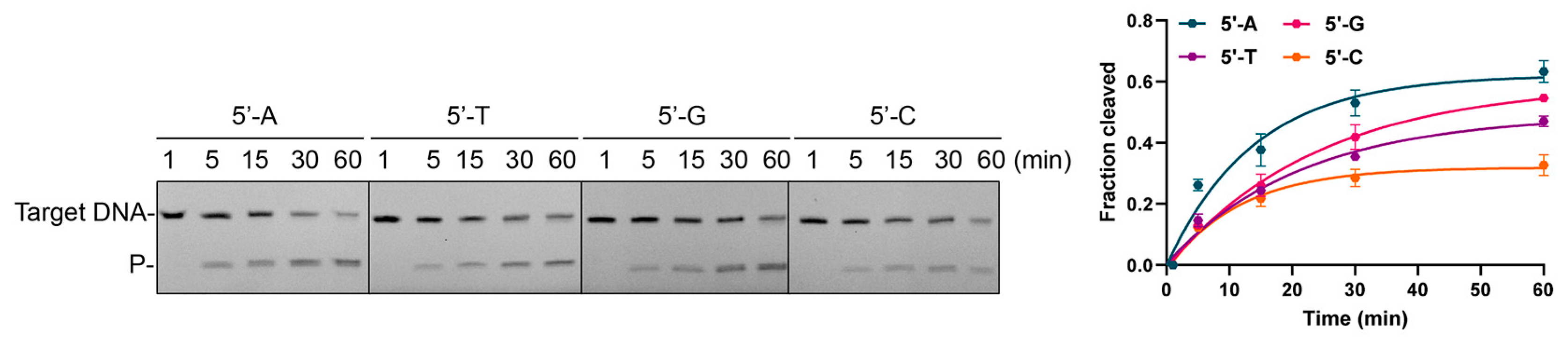
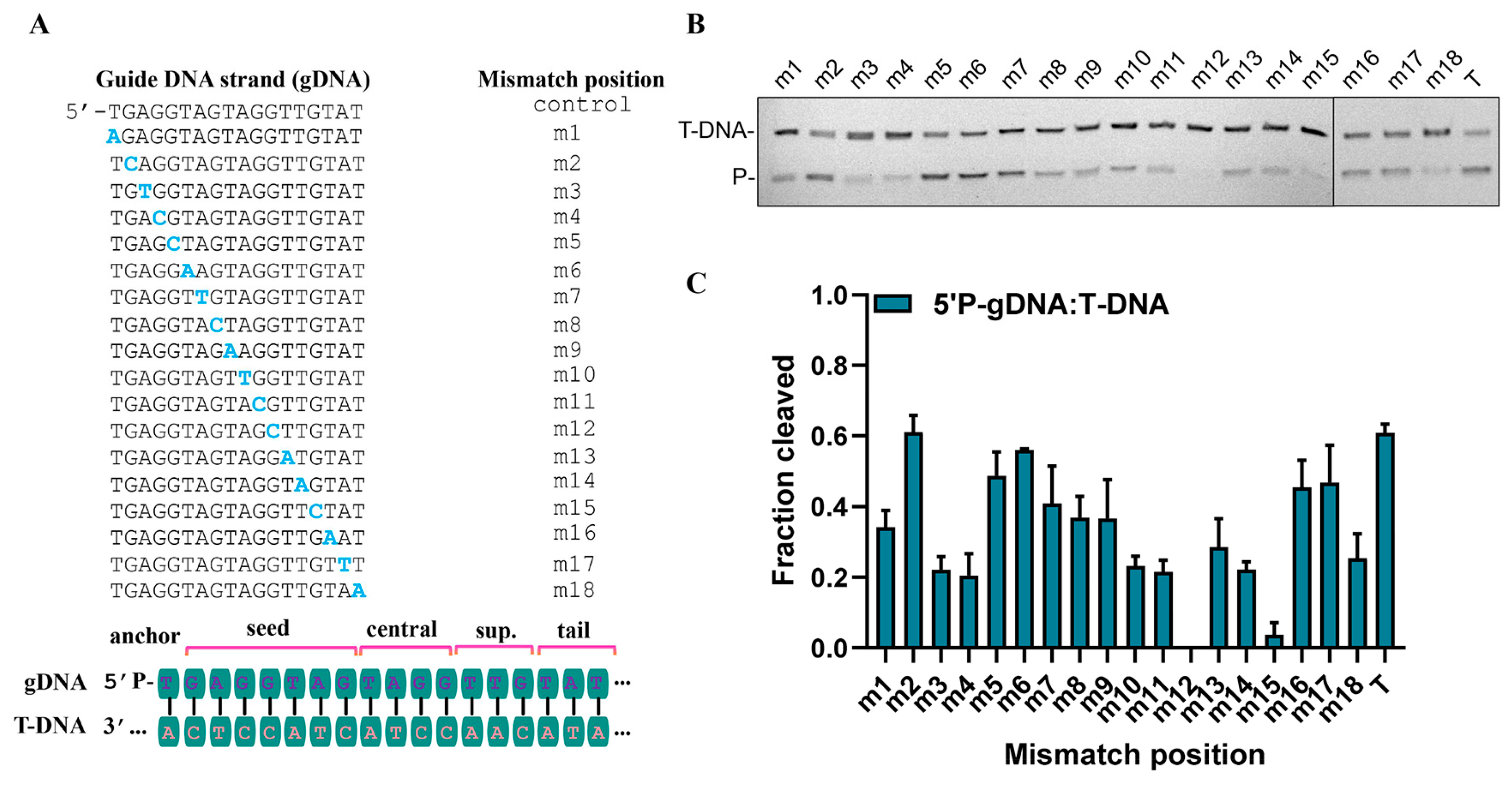
3.5. Effects of 5′-End Nucleotide and Mismatches of gDNA on Cleavage Activity
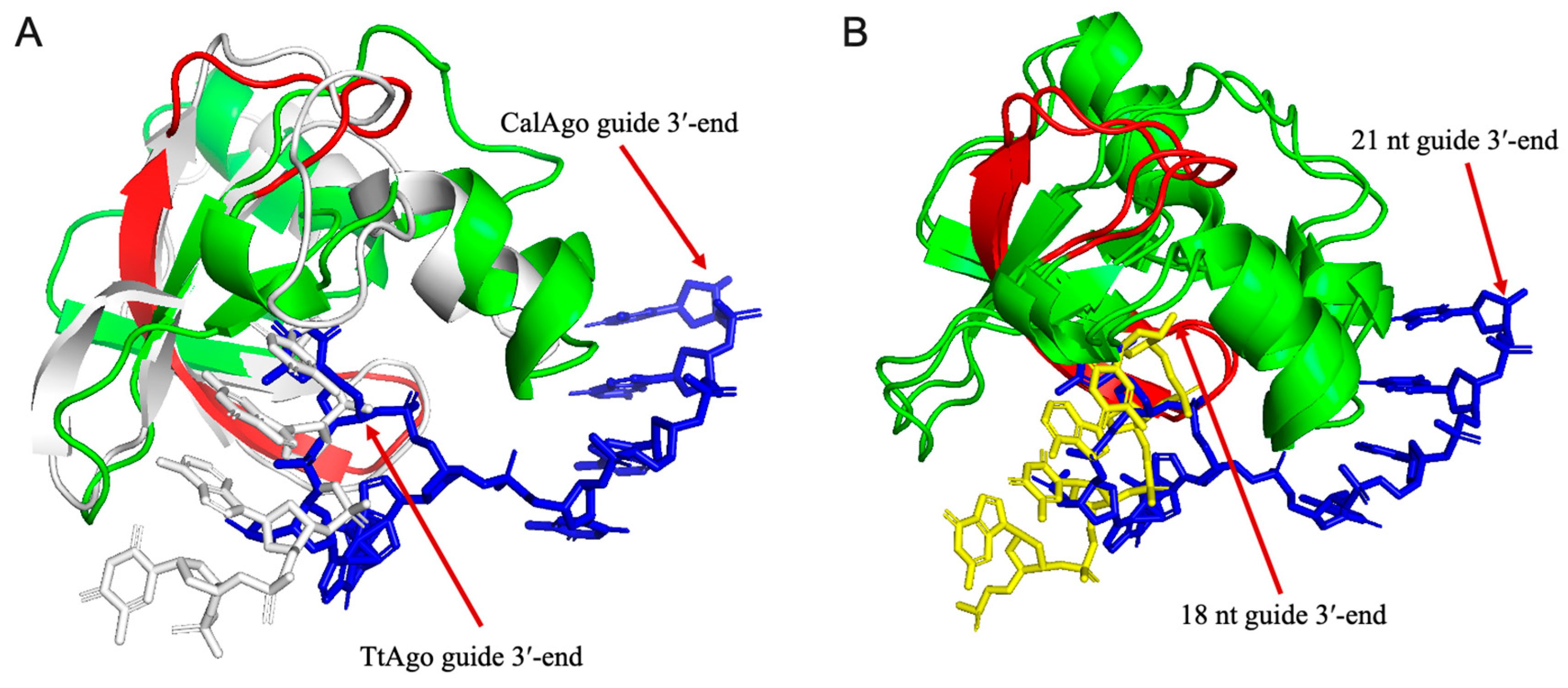
3.6. Structure Prediction and Analysis of the PAZ Domain of CalAgo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Swarts, D.C.; Makarova, K.; Wang, Y.; Nakanishi, K.; Ketting, R.F.; Koonin, E.V.; Patel, D.J.; van der Oost, J. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 2014, 21, 743–753. [Google Scholar] [CrossRef]
- Koonin, E.V. Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: Common ancestry vs convergence. Biol. Direct 2017, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Shabalina, S.A.; Koonin, E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008, 23, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Kuzmenko, A.; Oguienko, A.; Esyunina, D.; Yudin, D.; Petrova, M.; Kudinova, A.; Maslova, O.; Ninova, M.; Ryazansky, S.; Leach, D.; et al. DNA targeting and interference by a bacterial Argonaute nuclease. Nature 2020, 587, 632–637. [Google Scholar] [CrossRef]
- Ryazansky, S.; Kulbachinskiy, A.; Aravin, A.A. The Expanded Universe of Prokaryotic Argonaute Proteins. mBio 2018, 9, e01935-18. [Google Scholar] [CrossRef]
- Lisitskaya, L.; Aravin, A.A.; Kulbachinskiy, A. DNA interference and beyond: Structure and functions of prokaryotic Argonaute proteins. Nat. Commun. 2018, 9, 5165. [Google Scholar] [CrossRef]
- Loeff, L.; Adams, D.W.; Chanez, C.; Stutzmann, S.; Righi, L.; Blokesch, M.; Jinek, M. Molecular mechanism of plasmid elimination by the DdmDE defense system. Science 2024, 385, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Hegge, J.W.; Swarts, D.C.; van der Oost, J. Prokaryotic Argonaute proteins: Novel genome-editing tools? Nat. Rev. Microbiol. 2018, 16, 5–11. [Google Scholar] [CrossRef]
- Jin, S.; Zhan, J.; Zhou, Y. Argonaute proteins: Structures and their endonuclease activity. Mol. Biol. Rep. 2021, 48, 4837–4849. [Google Scholar] [CrossRef]
- Koopal, B.; Mutte, S.K.; Swarts, D.C. A long look at short prokaryotic Argonautes. Trends Cell Biol. 2023, 33, 605–618. [Google Scholar] [CrossRef]
- Tao, X.; Ding, H.; Wu, S.; Wang, F.; Xu, H.; Li, J.; Zhai, C.; Li, S.; Chen, K.; Wu, S.; et al. Structural and mechanistic insights into a mesophilic prokaryotic Argonaute. Nucleic Acids Res. 2024, 52, 11895–11910. [Google Scholar] [CrossRef]
- Nakanishi, K.; Weinberg, D.E.; Bartel, D.P.; Patel, D.J. Structure of yeast Argonaute with guide RNA. Nature 2012, 486, 368–374. [Google Scholar] [CrossRef]
- Sheng, G.; Zhao, H.; Wang, J.; Rao, Y.; Tian, W.; Swarts, D.C.; van der Oost, J.; Patel, D.J.; Wang, Y. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. USA 2014, 111, 652–657. [Google Scholar] [CrossRef]
- Miyoshi, T.; Ito, K.; Murakami, R.; Uchiumi, T. Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nat. Commun. 2016, 7, 11846. [Google Scholar] [CrossRef] [PubMed]
- Swarts, D.C.; Hegge, J.W.; Hinojo, I.; Shiimori, M.; Ellis, M.A.; Dumrongkulraksa, J.; Terns, R.M.; Terns, M.P.; van der Oost, J. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 2015, 43, 5120–5129. [Google Scholar] [CrossRef]
- Yuan, Y.R.; Pei, Y.; Ma, J.B.; Kuryavyi, V.; Zhadina, M.; Meister, G.; Chen, H.Y.; Dauter, Z.; Tuschl, T.; Patel, D.J. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell 2005, 19, 405–419. [Google Scholar] [CrossRef]
- Swarts, D.C.; Jore, M.M.; Westra, E.R.; Zhu, Y.; Janssen, J.H.; Snijders, A.P.; Wang, Y.; Patel, D.J.; Berenguer, J.; Brouns, S.J.J.; et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature 2014, 507, 258–261. [Google Scholar] [CrossRef]
- Zander, A.; Willkomm, S.; Ofer, S.; van Wolferen, M.; Egert, L.; Buchmeier, S.; Stockl, S.; Tinnefeld, P.; Schneider, S.; Klingl, A.; et al. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2017, 2, 17034. [Google Scholar] [CrossRef] [PubMed]
- Graver, B.A.; Chakravarty, N.; Solomon, K.V. Prokaryotic Argonautes for in vivo biotechnology and molecular diagnostics. Trends Biotechnol. 2024, 42, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Kuzmenko, A.; Yudin, D.; Ryazansky, S.; Kulbachinskiy, A.; Aravin, A.A. Programmable DNA cleavage by Ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea. Nucleic Acids Res. 2019, 47, 5822–5836. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, Y.; Liu, Q.; Ma, L. Characterization of a Programmable Argonaute Nuclease from the Mesophilic Bacterium Rummeliibacillus suwonensis. Biomolecules 2022, 12, 355. [Google Scholar] [CrossRef]
- Kaya, E.; Doxzen, K.W.; Knoll, K.R.; Wilson, R.C.; Strutt, S.C.; Kranzusch, P.J.; Doudna, J.A. A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl. Acad. Sci. USA 2016, 113, 4057–4062. [Google Scholar] [CrossRef]
- Bastiaanssen, C.; Bobadilla Ugarte, P.; Kim, K.; Finocchio, G.; Feng, Y.; Anzelon, T.A.; Kostlbacher, S.; Tamarit, D.; Ettema, T.J.G.; Jinek, M.; et al. RNA-guided RNA silencing by an Asgard archaeal Argonaute. Nat. Commun. 2024, 15, 5499. [Google Scholar] [CrossRef]
- Li, W.; Liu, Y.; He, R.; Wang, L.; Wang, Y.; Zeng, W.; Zhang, Z.; Wang, F.; Ma, L. A programmable pAgo nuclease with RNA target preference from the psychrotolerant bacterium Mucilaginibacter paludis. Nucleic Acids Res. 2022, 50, 5226–5238. [Google Scholar] [CrossRef] [PubMed]
- Lisitskaya, L.; Shin, Y.; Agapov, A.; Olina, A.; Kropocheva, E.; Ryazansky, S.; Aravin, A.A.; Esyunina, D.; Murakami, K.S.; Kulbachinskiy, A. Programmable RNA targeting by bacterial Argonaute nucleases with unconventional guide binding and cleavage specificity. Nat. Commun. 2022, 13, 4624. [Google Scholar] [CrossRef]
- Liu, Y.; Li, W.; Jiang, X.; Wang, Y.; Zhang, Z.; Liu, Q.; He, R.; Chen, Q.; Yang, J.; Wang, L.; et al. A programmable omnipotent Argonaute nuclease from mesophilic bacteria Kurthia massiliensis. Nucleic Acids Res. 2021, 49, 1597–1608. [Google Scholar] [CrossRef] [PubMed]
- Kropocheva, E.; Kuzmenko, A.; Aravin, A.A.; Esyunina, D.; Kulbachinskiy, A. A programmable pAgo nuclease with universal guide and target specificity from the mesophilic bacterium Kurthia massiliensis. Nucleic Acids Res. 2021, 49, 4054–4065. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, L.; Wang, J.; Ma, L.; Bai, Y.; Feng, F. Argonaute-Based Nucleic Acid Detection Technology: Advantages, Current Status, Challenges, and Perspectives. ACS Sens. 2024, 9, 5665–5682. [Google Scholar] [CrossRef]
- Enghiad, B.; Xue, P.; Singh, N.; Boob, A.G.; Shi, C.; Petrov, V.A.; Liu, R.; Peri, S.S.; Lane, S.T.; Gaither, E.D.; et al. PlasmidMaker is a versatile, automated, and high throughput end-to-end platform for plasmid construction. Nat. Commun. 2022, 13, 2697. [Google Scholar] [CrossRef]
- Esyunina, D.; Okhtienko, A.; Olina, A.; Panteleev, V.; Prostova, M.; Aravin, A.A.; Kulbachinskiy, A. Specific targeting of plasmids with Argonaute enables genome editing. Nucleic Acids Res. 2023, 51, 4086–4099. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Vaiskunaite, R.; Vainauskas, J.; Morris, J.J.L.; Potapov, V.; Bitinaite, J. Programmable cleavage of linear double-stranded DNA by combined action of Argonaute CbAgo from Clostridium butyricum and nuclease deficient RecBC helicase from E. coli. Nucleic Acids Res. 2022, 50, 4616–4629. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Liu, J.; Zhang, S.; An, L.; Xie, W.; Zhang, K.; Li, S. The PAZ pocket and dimerization drive CpAgo’s guide-independent and DNA-guided dual catalysis. Nat. Commun. 2025, 16, 6599. [Google Scholar] [CrossRef] [PubMed]
- Swarts, D.C.; Szczepaniak, M.; Sheng, G.; Chandradoss, S.D.; Zhu, Y.; Timmers, E.M.; Zhang, Y.; Zhao, H.; Lou, J.; Wang, Y.; et al. Autonomous Generation and Loading of DNA Guides by Bacterial Argonaute. Mol. Cell 2017, 65, 985–998 e986. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Li, X.; Wang, F.; Liu, Y.; Zhai, C.; Li, W.; Ma, L.; Chen, W. TLTC, a T5 exonuclease-mediated low-temperature DNA cloning method. Front. Bioeng. Biotechnol. 2023, 11, 1167534. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Wang, Y.; Juranek, S.; Li, H.; Sheng, G.; Tuschl, T.; Patel, D.J. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 2008, 456, 921–926. [Google Scholar] [CrossRef]
- Willkomm, S.; Oellig, C.A.; Zander, A.; Restle, T.; Keegan, R.; Grohmann, D.; Schneider, S. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat. Microbiol. 2017, 2, 17035. [Google Scholar] [CrossRef]
- Guo, X.; Sun, Y.; Chen, L.; Huang, F.; Liu, Q.; Feng, Y. A Hyperthermophilic Argonaute from Ferroglobus placidus with Specificity on Guide Binding Pattern. Front. Microbiol. 2021, 12, 654345. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Y.; Pan, W.; Wang, Y.; Liu, Y.; Ma, L. A Mesophilic Argonaute from Cohnella algarum Mediates Programmable DNA/RNA Cleavage with Distinctive Guide Specificity. Biomolecules 2025, 15, 1459. https://doi.org/10.3390/biom15101459
Peng Y, Pan W, Wang Y, Liu Y, Ma L. A Mesophilic Argonaute from Cohnella algarum Mediates Programmable DNA/RNA Cleavage with Distinctive Guide Specificity. Biomolecules. 2025; 15(10):1459. https://doi.org/10.3390/biom15101459
Chicago/Turabian StylePeng, Yanhong, Wang Pan, Yang Wang, Yang Liu, and Lixin Ma. 2025. "A Mesophilic Argonaute from Cohnella algarum Mediates Programmable DNA/RNA Cleavage with Distinctive Guide Specificity" Biomolecules 15, no. 10: 1459. https://doi.org/10.3390/biom15101459
APA StylePeng, Y., Pan, W., Wang, Y., Liu, Y., & Ma, L. (2025). A Mesophilic Argonaute from Cohnella algarum Mediates Programmable DNA/RNA Cleavage with Distinctive Guide Specificity. Biomolecules, 15(10), 1459. https://doi.org/10.3390/biom15101459





