Circular RNAs in Cardiovascular Disease: Mechanisms, Biomarkers, and Therapeutic Frontiers
Abstract
1. Introduction
2. circRNAs Involved in Cardiac Hypertrophy
2.1. CircHRCR, Guardian of the Stressed Myocardium
2.2. CircHIPK3, Driver of Maladaptive Hypertrophy
2.3. CircYAP1, Hippo-Pathway Satellite That Guards Against Fibrosis and Hypertrophy
2.4. CircSLC8A1, Calcium-Handling Hub That Fuels Pathological Growth
- •
- SERCA2a (Atp2a2), improving sarcoplasmic reticulum Ca2+ re-uptake and enabling stronger contractions;
- •
- CTGF, a profibrotic matricellular protein;
- •
- RhoA, which remodels the actin cytoskeleton and augments cell size;
- •
- Ccnd2, a G1/S cyclin that extends the cardiomyocyte growth window.
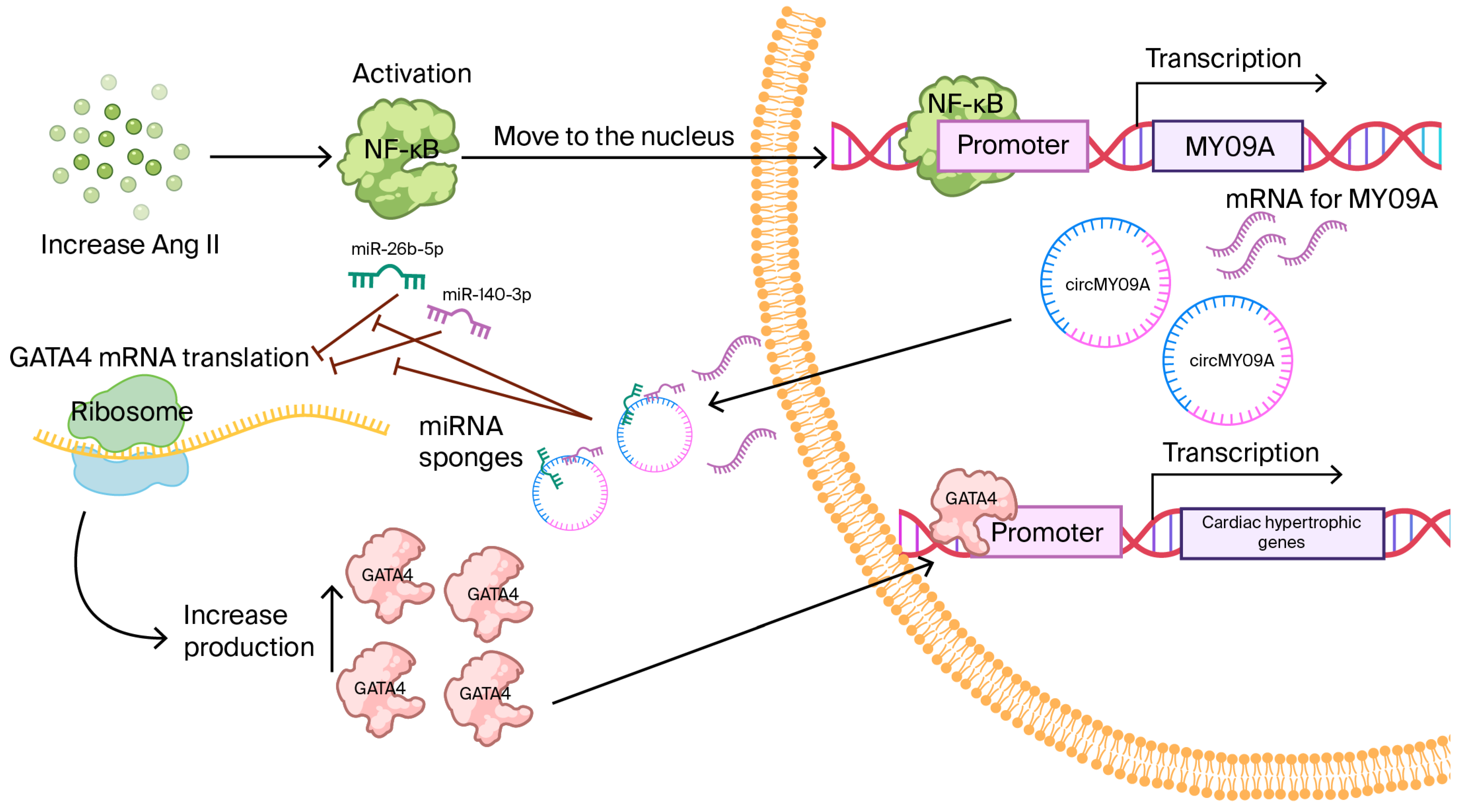
2.5. CircMYO9A, an NF-κB-Responsive Amplifier of the GATA4 Axis
2.6. CircWWP1 Mitigates β-Adrenergic-Driven Hypertrophy
2.7. CircPAN3, an m6A-Sensitive Safeguard That Counteracts Isoproterenol Hypertrophy
3. circRNAs in Atherosclerosis—Orchestrators of Plaque Formation and Vascular Inflammation
3.1. CircANRIL Exhibits Context-Dependent Roles
3.2. CircLRP6—A VSMC-Enriched Sponge That Tilts the Balance Toward the Synthetic Phenotype
3.3. CircGNAQ—An Endothelial Safeguard That Delays Senescence and Slows Plaque Growth
3.4. CircWDR77—A Diazbetes-Linked VSMC Accelerator That Drives Plaque Growth
4. CircRNA Involvement in Coronary Artery Disease (CAD)
4.1. CircYOD1—A Circulating Regulator That Links miRNA Networks with Inflammatory Remodeling in CAD
4.2. CircZNF609—A Dual-Mode Defender: Macrophage Anti-Inflammatory Signaling and Cardiomyocyte Stress Control
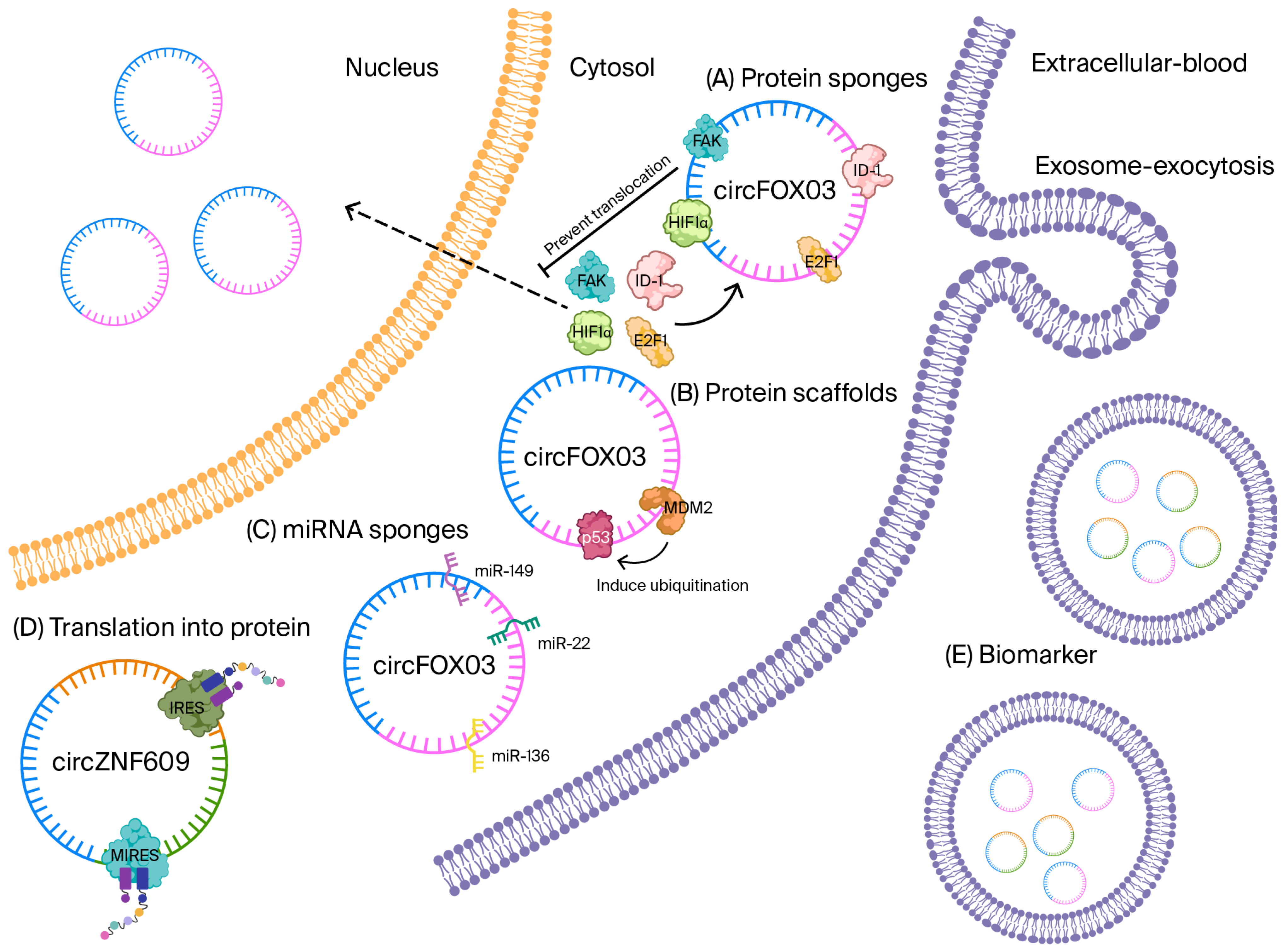
4.3. CircFOXO3—A Senescence-Linked Regulator That Bridges Genetic Susceptibility and Functional Decline in CAD
| Axis | Baseline Role | CircFOXO3 Effect | Cellular Consequence | Clinical Implication | Quantitative Expression (Disease vs. Control) |
|---|---|---|---|---|---|
| Protein scaffolding (ID-1, E2F1, FAK, HIF1α) | Anti-senescence, pro-survival signaling with nuclear translocation | Cytoplasmic sequestration by circFOXO3 | Reduced nuclear protection, ↑ stress susceptibility | Accelerated vascular/cardiac aging, higher CAD burden | 9-fold change, tissue/biofluid, disease, n, p (verify from Refs. [17,56,58]) |
| p21/CDK2 complex | Cell-cycle progression | circFOXO3 scaffold halts CDK2 via p21 | G1 arrest, senescence | Impaired repair capacity in aging myocardium/vasculature | 9-fold change, tissue/biofluid, disease, n, p (verify from Refs. [17,56,58]) |
| p53/MDM2 axis | Apoptosis control | circFOXO3 complex modulates p53 ubiquitination | Context-dependent apoptosis/senescence | Tissue remodeling and functional decline with age | 9-fold change, tissue/biofluid, disease, n, p (verify from Refs. [17,56,58]) |
| miRNA sponging (miR-149/miR-22/ miR-136) | Post-transcriptional repression of stress/cell-cycle genes | Relief of repression via sponging | Gene-expression shifts toward senescence programs | Adds to remodeling in CAD contexts | 9-fold change, tissue/biofluid, disease, n, p (verify from Refs. [17,56,58]) |
4.4. CircROBO2—HASMC-Centric Driver That Links Growth Cues to Inflammatory Signaling in CAD
4.5. CircROBO2 —A Circulatin Classifier for Angiographic CAD
4.6. CircSMARCA5—Diagnostic Adjunct in ACS/CAD
4.7. CircCDR1as/ciRS-7—miR-7 Axis Linking Inflammation and Endothelial Dysfunction
5. Therapeutic Delivery Strategies and Clinical Translation in CircRNA-Based Therapies
6. Methodological Sources of Discrepancy and Study Limitations
7. Conclusions
| CircRNA | Parent Gene | Primary Regulatory Function | Mechanism of Action | Disease Context/Models | Biomarker Potential | Therapeutic Angle | Key Refs. |
|---|---|---|---|---|---|---|---|
| circFOXO3 | FOXO3 | Cellular aging/senescence; stress response | Protein scaffold/decoy (p21–CDK2, p53–MDM2; ID-1/E2F1/FAK/HIF1α sequestration) | Cardiac/vascular aging; CAD models; stress-induced injury | Candidate circulating/tissue marker of cellular aging and disease burden | Modulates scaffolding interactions to restore repair capacity or reduce senescence | [17,56,58] |
| circGNAQ | GNAQ | Endothelial function; vascular homeostasis | miRNA interaction and signaling modulation (reported endothelial pathways) | Atherosclerosis/vascular dysfunction models | Potential marker of vascular health | Augments endothelial protective programs | [49] |
| circLRP6 | LRP6 | Smooth muscle cell phenotype/remodeling | miRNA sponge/signaling node (Wnt/LRP pathways) | Plaque growth; vascular remodeling (mouse/human tissues) | Stage-specific remodeling marker | Inhibits SMC drivers in growth-prone plaques | [48] |
| circROBO2 | ROBO2 | Smooth muscle/endothelial crosstalk | miRNA sponge or RBP interactions (ROBO/Slit axis) | Atherosclerosis models; vascular injury | Remodeling activity marker | Target pathway to limit maladaptive remodeling | [63,64] |
| circZNF609 | ZNF609 | Inflammation and angiogenesis; immune signaling | miRNA sponge; potential translation (cap-independent) reported in other systems | Immune-driven vascular inflammation; ischemic models | Inflammatory activity marker | Attenuates pro-inflammatory signaling | [55,56] |
| circSMARCA5 | SMARCA5 | Endothelial/vascular regulation; anti-angiogenic roles reported | miRNA sponge; RBP interaction | Atherosclerosis; vascular dysfunction | Potential circulating/tissue biomarker | Restores protective endothelial programs | [65] |
| circYOD1 | YOD1 | Vascular/immune modulation | miRNA sponge; pathway modulation | Atherosclerotic burden; vascular injury models | Broad disease-burden signal | Downstream pathway inhibition | [54] |
| ciRS-7/CDR1as | CDR1 | miR-7 sponge (canonical) | High-capacity miRNA sponging | Vascular/neurovascular contexts; broader literature | Context-dependent biomarker relevance | Pathway-level targeting via miR-7 axis | [7,8,9] |
| circHIPK3 | HIPK3 | Cell proliferation/angiogenesis | miRNA sponge (e.g., miR-124 family); RBP interactions | Endothelial dysfunction; diabetic vasculopathy models | Potential diagnostic/prognostic utility (matrix-dependent) | Modulates pro-proliferative/angiogenic signals | [7,8,9] |
| circANRIL | ANRIL | Atheroprotection via nucleolar stress | Protein interaction (PES1) affecting rRNA maturation | Human atherosclerosis genetics; vascular tissue | Protective genetic/functional signal | Pathway reinforcement strategies | [17,18] |
| circHRCR | HRCR (heart-related circRNA; host annotation varies) | Anti-hypertrophic cardioprotection | miR-223 sponge to de-repress ARC (apoptosis repressor with CARD) | Cardiac hypertrophy and heart failure models (pressure overload, TAC) | Potential protective signature in hypertrophy/heart failure | Gene therapy or vector overexpression to reduce hypertrophy | [16,20] |
| circYAP1 | YAP1 | Cardiomyocyte survival; stress response | miRNA sponge (e.g., miR-367-5p/miR-21 axis) modulating Hippo–YAP signaling | Myocardial infarction/ischemia–reperfusion injury models | Injury severity and remodeling marker (context-dependent) | Augments circYAP1 to reduce apoptosis and preserve function | [7,8,9], |
| circSLC8A1 | SLC8A1 (NCX1) | Cardiac hypertrophy/remodeling | miR-133a sponge affecting pro-hypertrophic programs | Pressure overload hypertrophy; heart failure models | Cardiac stress/hypertrophy marker | Therapeutic inhibition/ASO knockdown to limit hypertrophy | [7,8,9,16,17,18] |
| circMYO9A | MYO9A | Vascular remodeling; SMC phenotype | Putative miRNA sponge/RBP interactions (predicted) | Atherosclerotic plaques; vascular tissue datasets | Differential-expression-based biomarker candidate | Target pending functional validation | [26,39] |
| circWWP1 | WWP1 | Inflammation/ubiquitin pathway-linked vascular effects | Likely miRNA sponge; pathway crosstalk via WWP1 signaling | Atherosclerotic vascular tissue/cohort datasets | Expression-based disease activity marker | Pathway-informed targeting after validation | [40] |
| circPAN3 | PAN3 | Endothelial activation/inflammation control | miRNA sponge; post-transcriptional regulation | Endothelial dysfunction; atherosclerosis models | Inflammation/activation activity marker | Attenuate pro-inflammatory signaling | [38,41] |
| circWDR77 | WDR77 | Immune–vascular interface; macrophage signaling | miRNA sponge/RBP interaction (putative) | Plaque macrophage-rich microenvironments | Plaque activity/progression marker | Modulate immune signaling nodes | [50,51] |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Wang, Y.; Cui, Y.; Li, X.; Jin, S.-H.; Wang, H.; Gaipl, U.S.; Ma, H.; Wang, S.; Zhou, J.-G. circRNAs: Functions and emerging roles in cancer and immunotherapy. BMC Med. 2025, 23, 477. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef]
- Piwecka, M.; Glažar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar] [CrossRef]
- Wang, R.; Chen, Y.; Zhou, Q.; Xiao, J. Circular RNAs in Cardiovascular Diseases: Regulation and Translational Potential. Research 2024, 6, 191–204. [Google Scholar]
- Zhang, X.-O.; Wang, H.-B.; Zhang, Y.; Lu, X.; Chen, L.-L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Habara, A. Exploratory Review and In Silico Insights into circRNA and RNA-Binding Protein Roles in γ-Globin to β-Globin Switching. Cells 2025, 14, 312. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E. Translation of circRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Zhao, B.S.; Wang, X.; Beadell, A.V.; Lu, Z.; Shi, H.; Kuuspalu, A.; Ho, R.K.; He, C. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 2017, 542, 475–478. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Chen, Y.; Wu, Z.-K.; Foster, F.S.; Yang, Z.; Li, X.; Yang, B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017, 38, 1402–1412. [Google Scholar] [CrossRef]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef]
- Benjamin, E.J.; Virani, S.S.; Callaway, C.W.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Chiuve, S.E.; Cushman, M.; Delling, F.N.; Deo, R. Heart disease and stroke statistics—2018 update: A report from the American Heart Association. Circulation 2018, 137, e67–e492. [Google Scholar] [CrossRef]
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P. Global burden of cardiovascular diseases and risk factors, 1990–2019: Update from the GBD 2019 study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef]
- Garikipati, V.N.S.; Verma, S.K.; Cheng, Z.; Liang, D.; Truongcao, M.M.; Cimini, M.; Yue, Y.; Huang, G.; Wang, C.; Benedict, C. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 2019, 10, 4317. [Google Scholar] [CrossRef]
- Wang, K.; Gan, T.-Y.; Li, N.; Liu, C.-Y.; Zhou, L.-Y.; Gao, J.-N.; Chen, C.; Yan, K.-W.; Ponnusamy, M.; Zhang, Y.-H. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017, 24, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, J.-D.; Fang, X.-H.; Zhu, J.-N.; Yang, J.; Pan, R.; Yuan, S.-J.; Zeng, N.; Yang, Z.-Z.; Yang, H. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc. Res. 2020, 116, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Koo, C.; Su, W.; You, Q.; Guo, H.; Liu, B. Circular RNAs modulate cell death in cardiovascular diseases. Cell Death Discov. 2025, 11, 214. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Yuan, X.; Yuan, Y. Circular RNAs in coronary heart disease: From molecular mechanism to promising clinical application. J. Mol. Med. 2025, 55, 1–5. [Google Scholar] [CrossRef]
- Martin, T.G.; Juarros, M.A.; Leinwand, L.A. Regression of cardiac hypertrophy in health and disease: Mechanisms and therapeutic potential. Nat. Rev. Cardiol. 2023, 20, 347–363. [Google Scholar] [CrossRef]
- Gajarsa, J.J.; Kloner, R.A. Left ventricular remodeling in the post-infarction heart: A review of cellular, molecular mechanisms, and therapeutic modalities. Heart Fail. Rev. 2011, 16, 13–21. [Google Scholar] [CrossRef]
- Hill, J.A.; Olson, E.N. Cardiac plasticity. N. Engl. J. Med. 2008, 358, 1370–1380. [Google Scholar] [CrossRef]
- Wang, K.; Long, B.; Liu, F.; Wang, J.-X.; Liu, C.-Y.; Zhao, B.; Zhou, L.-Y.; Sun, T.; Wang, M.; Yu, T. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016, 37, 2602–2611. [Google Scholar] [CrossRef]
- Heineke, J.; Molkentin, J.D. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 2006, 7, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Kreutzer, F.P.; Fiedler, J.; Thum, T. Non-coding RNAs: Key players in cardiac disease. J. Physiol. 2020, 598, 2995–3003. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, X.; Zheng, H.; Si, X.; Li, B.; Wei, G.; Li, C.; Chen, Y.; Chen, Y.; Liao, W. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation 2019, 139, 2857–2876. [Google Scholar] [CrossRef] [PubMed]
- Lim, T.B.; Aliwarga, E.; Luu, T.D.A.; Li, Y.P.; Ng, S.L.; Annadoray, L.; Sian, S.; Ackers-Johnson, M.A.; Foo, R.S.-Y. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc. Res. 2019, 115, 1998–2007. [Google Scholar] [CrossRef]
- Zhang, M.-W.; Shen, Y.-J.; Shi, J.; Yu, J.-G. MiR-223-3p in cardiovascular diseases: A biomarker and potential therapeutic target. Front. Cardiovasc. Med. 2021, 7, 610561. [Google Scholar] [CrossRef]
- Xu, X.; Wang, J.; Wang, X. Silencing of circHIPK3 inhibits pressure overload-induced cardiac hypertrophy and dysfunction by sponging miR-185-3p. Drug Des. Dev. Ther. 2020, 14, 5699–5710. [Google Scholar] [CrossRef]
- Wu, N.; Xu, J.; Du, W.W.; Li, X.; Awan, F.M.; Li, F.; Misir, S.; Eshaghi, E.; Lyu, J.; Zhou, L. YAP circular RNA, circYap, attenuates cardiac fibrosis via binding with tropomyosin-4 and gamma-actin decreasing actin polymerization. Mol. Ther. 2021, 29, 1138–1150. [Google Scholar] [CrossRef]
- Jahn, C.; Bär, C.; Thum, T. CircSlc8a1, Breaking a Vicious Circle in Cardiac Hypertrophy; Oxford University Press: Oxford, UK, 2019; pp. 1946–1947. [Google Scholar]
- Han, M.; Yang, Z.; Sayed, D.; He, M.; Gao, S.; Lin, L.; Yoon, S.; Abdellatif, M. GATA4 expression is primarily regulated via a miR-26b-dependent post-transcriptional mechanism during cardiac hypertrophy. Cardiovasc. Res. 2012, 93, 645–654. [Google Scholar] [CrossRef]
- Yang, M.-H.; Wang, H.; Han, S.-N.; Jia, X.; Zhang, S.; Dai, F.-F.; Zhou, M.-J.; Yin, Z.; Wang, T.-Q.; Zang, M.-X. Circular RNA expression in isoproterenol hydrochloride-induced cardiac hypertrophy. Aging 2020, 12, 2530. [Google Scholar] [CrossRef]
- Fang, X.; Ao, X.; Xiao, D.; Wang, Y.; Jia, Y.; Wang, P.; Li, M.; Wang, J. Circular RNA-circPan3 attenuates cardiac hypertrophy via miR-320-3p/HSP20 axis. Cell. Mol. Biol. Lett. 2024, 29, 3. [Google Scholar] [CrossRef]
- Cao, Q.; Guo, Z.; Du, S.; Ling, H.; Song, C. Circular RNAs in the pathogenesis of atherosclerosis. Life Sci. 2020, 255, 117837. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, Y.; Zhou, H.; Li, Y. Circular RNAs in atherosclerosis. Clin. Chim. Acta 2022, 531, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Deng, K.; Zang, Y.; Zhang, Z.; Zhao, B.; Fan, J.; Huang, L. Exploring the regulatory roles of circular RNAs in the pathogenesis of atherosclerosis. Vasc. Pharmacol. 2021, 141, 106898. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.-H.; Zhang, R.-Q.; Huang, X.-S.; Zhou, J.; Guo, Z.; Xu, B.-F.; Liu, R. Transcriptomic and proteomic profiling of human stable and unstable carotid atherosclerotic plaques. Front. Genet. 2021, 12, 755507. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Gaetano, C.; Mazzaccaro, D.; Martelli, F. Circular RNA role in Atherosclerosis Development and Progression. Curr. Atheroscler. Rep. 2025, 27, 60. [Google Scholar] [CrossRef]
- Shi, P.; Ji, H.; Zhang, H.; Yang, J.; Guo, R.; Wang, J. circANRIL reduces vascular endothelial injury, oxidative stress and inflammation in rats with coronary atherosclerosis. Exp. Ther. Med. 2020, 20, 2245–2251. [Google Scholar] [CrossRef]
- Hall, I.F.; Climent, M.; Quintavalle, M.; Farina, F.M.; Schorn, T.; Zani, S.; Carullo, P.; Kunderfranco, P.; Civilini, E.; Condorelli, G. Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ. Res. 2019, 124, 498–510. [Google Scholar] [CrossRef]
- Wu, W.-P.; Zhou, M.-Y.; Liu, D.-L.; Min, X.; Shao, T.; Xu, Z.-Y.; Jing, X.; Cai, M.-Y.; Xu, S.; Liang, X. circGNAQ, a circular RNA enriched in vascular endothelium, inhibits endothelial cell senescence and atherosclerosis progression. Mol. Ther.-Nucleic Acids 2021, 26, 374–387. [Google Scholar] [CrossRef]
- Chen, J.; Cui, L.; Yuan, J.; Zhang, Y.; Sang, H. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem. Biophys. Res. Commun. 2017, 494, 126–132. [Google Scholar] [CrossRef]
- Prandi, F.R.; Lecis, D.; Illuminato, F.; Milite, M.; Celotto, R.; Lerakis, S.; Romeo, F.; Barilla, F. Epigenetic modifications and non-coding RNA in diabetes-mellitus-induced coronary artery disease: Pathophysiological link and new therapeutic frontiers. Int. J. Mol. Sci. 2022, 23, 4589. [Google Scholar] [CrossRef]
- Waterbury, T.M.; Tarantini, G.; Vogel, B.; Mehran, R.; Gersh, B.J.; Gulati, R. Non-atherosclerotic causes of acute coronary syndromes. Nat. Rev. Cardiol. 2020, 17, 229–241. [Google Scholar] [CrossRef]
- Kashyap, A.K.; Duggal, B. CircRNA-miRNA-mRNA network targets ubiquitin-mediated proteolysis pathway in coronary artery disease. Hum. Gene 2023, 35, 201144. [Google Scholar] [CrossRef]
- Miao, L.; Yin, R.-X.; Zhang, Q.-H.; Liao, P.-J.; Wang, Y.; Nie, R.-J.; Li, H. A novel circRNA-miRNA-mRNA network identifies circ-YOD1 as a biomarker for coronary artery disease. Sci. Rep. 2019, 9, 18314. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Zheng, L.; Li, X.; Sun, Y. CircZNF609 sponges miR-135b to up-regulate SEMA3A expression to alleviate ox-LDL-induced atherosclerosis. Mol. Cell. Biochem. 2024, 480, 1105–1120. [Google Scholar] [CrossRef]
- Liang, B.; Li, M.; Deng, Q.; Wang, C.; Rong, J.; He, S.; Xiang, Y.; Zheng, F. CircRNA ZNF609 in peripheral blood leukocytes acts as a protective factor and a potential biomarker for coronary artery disease. Ann. Transl. Med. 2020, 8, 741. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, P.; Wang, J.; Xu, G.; Wang, T.; Feng, J.; Bei, Y.; Xu, J.; Wang, H.; Das, S. Downregulation of circ-ZNF609 promotes heart repair by modulating RNA N6-methyladenosine-modified Yap expression. Research 2022, 2022, 9825916. [Google Scholar] [PubMed]
- Yu, P.; Wang, J.; Xu G-e Zhao, X.; Cui, X.; Feng, J.; Sun, J.; Wang, T.; Spanos, M.; Lehmann, H.I. RNA m6A-regulated circ-ZNF609 suppression ameliorates doxorubicin-induced cardiotoxicity by upregulating FTO. Basic Transl. Sci. 2023, 8, 677–698. [Google Scholar] [CrossRef]
- Zhou, Y.-L.; Wu, W.-P.; Cheng, J.; Liang, L.-L.; Cen, J.-M.; Chen, C.; Liu, X.; Xiong, X.-D. CircFOXO3 rs12196996, a polymorphism at the gene flanking intron, is associated with circFOXO3 levels and the risk of coronary artery disease. Aging 2020, 12, 13076. [Google Scholar] [CrossRef]
- Su, Y.; Zhu, C.; Wang, B.; Zheng, H.; McAlister, V.; Lacefield, J.C.; Quan, D.; Mele, T.; Greasley, A.; Liu, K.; et al. Circular RNA Foxo3 in cardiac ischemia-reperfusion injury in heart transplantation: A new regulator and target. Am. J. Transplant. 2021, 21, 2992–3004. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef]
- Lin, D.-S.; Zhang, C.-Y.; Li, L.; Ye, G.-H.; Jiang, L.-P.; Jin, Q. Circ_robo2/mir-149 axis promotes the proliferation and migration of human aortic smooth muscle cells by activating NF-κB signaling. Cytogenet. Genome Res. 2021, 161, 414–424. [Google Scholar] [CrossRef]
- Xing, A.A.; Zhang, L.; Li, Y.-D.; MuKaiDaiSi, T.; Cao, G.-Q. Association of circular RNA hsa_circ_0124644 and incidence and severity of coronary heart disease in premenopausal women. Int. J. Clin. Exp. Med. 2023, 16, 285–293. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, X.; Gao, C.; Jian, D.; Hao, P.; Rao, L.; Li, M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 2017, 7, 39918. [Google Scholar] [CrossRef] [PubMed]
- Vilades, D.; Martínez-Camblor, P.; Ferrero-Gregori, A.; Bär, C.; Lu, D.; Xiao, K.; Vea, À.; Nasarre, L.; Sanchez Vega, J.; Leta, R. Plasma circular RNA hsa_circ_0001445 and coronary artery disease: Performance as a biomarker. FASEB J. 2020, 34, 4403–4414. [Google Scholar] [CrossRef] [PubMed]
- Xuan, L.; Chen, J.; Yang, H.; Hao, J.; Li, S.; Zhang, Q.; Zhang, H.; Wang, S.; Luo, H.; Guo, J. CircRNA CDR1AS promotes cardiac ischemia–reperfusion injury in mice by triggering cardiomyocyte autosis. J. Mol. Med. 2025, 103, 219–237. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T.; Scharpf, R.B.; Bravo, H.C.; Simcha, D.; Langmead, B.; Johnson, W.E.; Geman, D.; Baggerly, K.; Irizarry, R.A. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010, 11, 733–739. [Google Scholar] [CrossRef]
- Ståhl, P.L.; Salmén, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.O.; Huss, M.; et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353, 78–82. [Google Scholar] [CrossRef]
- Asp, M.; Bergenstråhle, J.; Lundeberg, J. Spatially resolved transcriptomes—Next generation tools for tissue exploration. Nat. Rev. Genet. 2020, 21, 785–802. [Google Scholar] [CrossRef]
- Guo, X.; Huang, Z.; Ju, F.; Zhao, C.; Yu, L. Highly Accurate Estimation of Cell Type Abundance in Bulk Tissues Based on Single-Cell Reference and Domain Adaptive Matching. Adv. Sci. 2024, 11, e2306329. [Google Scholar] [CrossRef]
- Zhang, X.O.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell. 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Hansen, T.B.; Venø, M.T.; Damgaard, C.K.; Kjems, J. Comparison of circular RNA prediction tools reveals differences in sensitivity and precision. Methods 2018, 155, 65–75. [Google Scholar]
- Li, X.; Yang, L.; Chen, L.L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef]
- Szabo, L.; Salzman, J. Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 2016, 17, 679–692. [Google Scholar] [CrossRef]
- Weidle, U.H.; Sela, T.; Brinkmann, U.; Niewoehner, J. Circular RNAs With Efficacy in Preclinical In Vitro and In Vivo Models of Esophageal Squamous Cell Carcinoma. Cancer Genom. Proteom. 2022, 19, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Houseley, J.; LaCava, J.; Tollervey, D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006, 7, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Schlesinger, F.; Davis, C.A.; Zhang, Y.; Li, R.; Salit, M.; Gingeras, T.R.; Oliver, B. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011, 21, 1543–1551. [Google Scholar] [CrossRef] [PubMed]
- Kivioja, T.; Vähärautio, A.; Karlsson, K.; Bonke, M.; Enge, M.; Linnarsson, S.; Taipale, J. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 2011, 9, 72–74. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
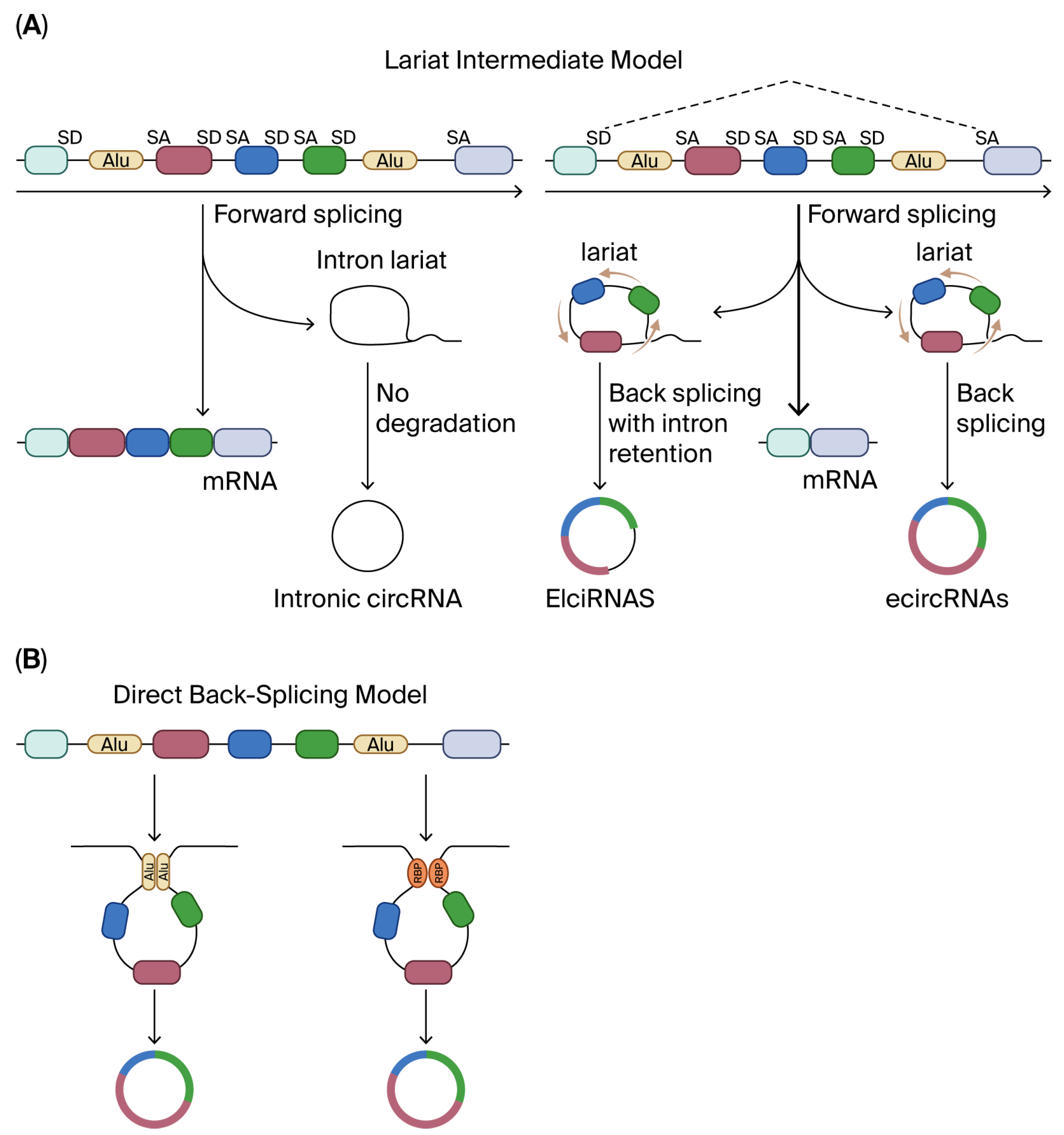
| Mechanism | Representative CircRNA(s) | Molecular Partner(s) | Downstream Consequence | Ref. |
|---|---|---|---|---|
| miRNA sponge | ciRS-7, circHIPK3 | miR-7, miR-124, etc. | De-repression of target mRNAs | [7,8,9] |
| Transcriptional modulator | circEIF3J, circPAIP2 | U1 snRNP, RNA-pol II | Enhanced host-gene transcription | [11,12] |
| Cap-independent translation | circZNF609, circFBXW7 | eIF4G2, eIF3A, YTHDF3 | Peptide generation (e.g., FBXW7-185aa) | [14,15,16,17] |
| Protein scaffold/decoy | circFOXO3, circANRIL | p53/MDM2, PES1 | p53 degradation, nucleolar stress | [18,19] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alali, R.; Almansori, M.; Vatte, C.; Akhtar, M.S.; Abduljabbar, S.S.; Al-Matroud, H.; Alnuwaysir, M.J.; Radhi, H.A.; Keating, B.; Habara, A.; et al. Circular RNAs in Cardiovascular Disease: Mechanisms, Biomarkers, and Therapeutic Frontiers. Biomolecules 2025, 15, 1455. https://doi.org/10.3390/biom15101455
Alali R, Almansori M, Vatte C, Akhtar MS, Abduljabbar SS, Al-Matroud H, Alnuwaysir MJ, Radhi HA, Keating B, Habara A, et al. Circular RNAs in Cardiovascular Disease: Mechanisms, Biomarkers, and Therapeutic Frontiers. Biomolecules. 2025; 15(10):1455. https://doi.org/10.3390/biom15101455
Chicago/Turabian StyleAlali, Rudaynah, Mohammed Almansori, Chittibabu Vatte, Mohammed S. Akhtar, Seba S. Abduljabbar, Hassan Al-Matroud, Mohammed J. Alnuwaysir, Hasan A. Radhi, Brendan Keating, Alawi Habara, and et al. 2025. "Circular RNAs in Cardiovascular Disease: Mechanisms, Biomarkers, and Therapeutic Frontiers" Biomolecules 15, no. 10: 1455. https://doi.org/10.3390/biom15101455
APA StyleAlali, R., Almansori, M., Vatte, C., Akhtar, M. S., Abduljabbar, S. S., Al-Matroud, H., Alnuwaysir, M. J., Radhi, H. A., Keating, B., Habara, A., & Al-Ali, A. K. (2025). Circular RNAs in Cardiovascular Disease: Mechanisms, Biomarkers, and Therapeutic Frontiers. Biomolecules, 15(10), 1455. https://doi.org/10.3390/biom15101455






