Abstract
Collagen VI is an extracellular matrix component encoded by COL6A1, COL6A2 and COL6A3 genes. Causative variants in these genes are associated with the following collagen VI-related myopathies: severe Ullrich congenital muscular dystrophy (UCMD), milder Bethlem myopathy (BM) and intermediate phenotypes (INT). We report the mutation landscape of COL6A genes in 138 Italian patients affected with a collagen VI-related phenotype. The patient cohort included 44 (32%) UCMD, 9 (7%) INT, 61 (44%) BM and 21 (15%) INT/BM patients; 3 patients (2%) with a myosclerosis myopathy (MM) phenotype were also considered. We identified 104 different variants: 26 in COL6A1 (25%), 52 in COL6A2 (50%) and 26 in COL6A3 (25%). The variant spectrum includes missense, splicing, small indel, frameshifting and nonsense variants. Glycine substitutions in the triple helical domain of the collagen VI protein are the commonest variants and occur in all phenotypes. Our genetic profiling disclosed a unique mutation scenario and phenotypic association of the COL6A2 gene with respect to COL6A1 and COL6A3, which may be related to a different evolutive history. Landscape mutation analysis of variants occurring in ultrarare conditions, such as collagen VI-related myopathies, is crucial to better understand the variations’ profile and to gain insight into fundamental knowledge about gene structure and its evolutive origin.
1. Introduction
Collagen VI is an extracellular matrix component encoded by COL6A1, COL6A2 and COL6A3 genes.
The COL6A1 (OMIM *120220) and COL6A2 (OMIM *120240) genes are located on chromosome 21q22.3, whereas COL6A3 (OMIM *120250) is located on chromosome 2q37. The COL6A1 gene is subdivided into 35 exons over an area of 23 kb and is transcribed into a single mRNA of approximately 4.2 Kb [1]. The COL6A2 gene (spanning 28 exons over an area of 35 kb) and the COL6A3 gene (including 44 exons spanning 100 kb) are transcribed in multiple isoforms, undergoing extensive alternative splicing [2,3].
Collagen VI is composed of three main α-chains, the α1 (VI), α2 (VI)—encoded by COL6A1 and COL6A2, respectively—and α3(VI)—encoded by COL6A3—and organized into a network of microfibrils with an essential role in anchoring the basement membrane to the extracellular matrix (ECM) [4].
Each chain contains a central triple helical domain (THD) with repeating Gly-X-Y subunits, flanked by extensive N- and C-terminal domains with homology to the type A domains of von Willebrand factor [5].
Collagen VI is broadly expressed with the highest representation in connective tissues, including bone, skin, tendon, cartilage and interstitial fibroblasts of skeletal muscle; expression in the central and peripheral nervous system, intestine, lung, adipose tissue, pancreatic islets, ovarian follicles, kidney glomeruli, vasculature and cornea has also been reported [6].
Causative variants in COL6A1, COL6A2 and COL6A3 genes result in skeletal muscle disorders, collectively called “collagen VI myopathies” [7], which represent a common type of congenital muscular dystrophy (CMD), identified in 20.24% of Italian CMD patients [8].
Collagen VI-related myopathies constitute a continuum of overlapping clinical phenotypes with Ullrich congenital muscular dystrophy (UCMD; OMIM #254090) at the more severe end and Bethlem myopathy (BM; OMIM #158810) at the milder end; moreover, intermediate phenotypes of various severities have been described and a new category, intermediate collagen VI-related myopathy, has been suggested for patients with a clinical picture between the two extreme ends [9]. An additional, mostly contractural, phenotype is referred to as myosclerosis myopathy (MM; OMIM #255600) [10].
Several nationwide studies have described genetic mutations in COL6A genes. Glycine amino acid substitutions in the THD are the commonest mutations, accounting for about 30% of known pathogenic variations [9,10,11].
Other missense, nonsense, frameshifting and splicing variants, and some rare large genomic deletions have been identified [12,13]. A frequent intronic variant causing pseudoexon recognition and inclusion in COL6A1 has also been reported [14]. Interestingly, variants did not show ethnic specificity [7,15,16,17].
Dominant and recessive mutations occur in COL6A1, COL6A2 and COL6A3 and result in a clinical severity depending on the impact of the mutation on the multimeric complex of collagen VI and its assembly process.
Beside inherited variants, more than 50% of UCMD cases harbor monoallelic, de novo dominant mutations (all types), including those affecting the Gly-X-Y subunits in the N-terminal end of THD. These mutations exert dominant negative effects on the assembly and structure of collagen VI, making the mutant chain unable to perform tetrameric assembly [18,19]. In contrast, mutations in rare recessive cases clustering near the C-terminal of THD or in the C-terminal domain disrupt the initial formation of monomers and consequently prevent their inclusion in the assembly process [11].
The present study reports on an Italian nationwide study describing the COL6A genes mutation scenario in a cohort of 138 patients affected by collagen VI-related myopathies.
Genetic diagnosis is fundamental to establishing genotype/phenotype relationships, facilitating biomarker discovery and patients’ enrollment in personalized therapies.
2. Materials and Methods
- Patient cohort
We studied 138 patients with clinically and genetically confirmed diagnosis of collagen VI-related myopathies referred by 21 Italian diagnostic centers—13 from North Italy, 5 from Central Italy and 3 from South Italy—in a temporal window of 15 years (2007–2022). The study was conducted within the routine diagnostic flowchart and for diagnostic purposes.
Ethical consent was collected in each center as part of the routine diagnostic procedures for COL6A genetic diagnosis and the study was approved by Comitato Etico di Area Vasta Emilia Centro (CE-AVEC) 66/2020/Oss/AOUFe 2020-01-23.
Affected family members of probands were not included in the present study.
The clinical phenotype of patients was defined according to the output of the 166th ENMC International Workshop on Collagen VI Myopathies, where the UCMD phenotype refers to patients who have never walked or have lost the ability to walk by the age of 12, the BM phenotype refers to patients who are able to walk during adulthood (>19 years) and the INT phenotype refers to patients who lose ambulation during their teens (13–19 years) [20]. The myosclerosis myopathy phenotype [10] was also considered, as was an INT/BM phenotype (defined for patients still able to walk at the last clinical evaluation, performed between 12 and 19 years).
- Genetic analysis
Genetic testing was performed by Sanger sequencing [21] and/or next-generation sequencing (NGS) custom panels. In selected cases, a custom oligonucleotide CGH array was used [22].
NGS analysis of COL6A genes was performed through a custom panel called “NEUROMIO”, including 171 neuromuscular genes, of which 80 were for hereditary neuropathies, 46 for congenital myopathies, 42 for muscular dystrophies and 3 for motoneuron iseases.
Library preparation was performed using the Illumina DNA Prep with Enrichment (target enrichment) method before proceeding to paired-end sequencing (150 bp) on the MiSeq™Dx (Illumina Inc., San Diego, CA, USA). The average coverage (Miseq) was 500×.
Coverage achieved from the entire NGS panel was 99.8% at a read depth > 20×; coverage of COL6A genes was 100%. Sequence analysis included the coding exons and flanking intronic regions (±50 bp upstream and downstream of each exon). Specific probes were designed to ensure that intron 11 of COL6A1 was captured.
Standard analysis (alignment, annotation, filtering and prioritization) of the raw data included sequence alignment and variants (e.g., SNP, InDel and copy number variations – CNVs-) calling and annotation were performed using Emedgene v37.0.0 software Illumina (based on the genome assembly Homo sapiens GRCh37-hg19). The integrative genomics viewer [23] was also used to visualize the read depth and the quality of the reads.
Variants with a frequency exceeding 0.1% in population databases, including the Exome Variant Server (ESP), the Exome Aggregation Consortium (ExAC) and gnomAD, were considered common and excluded from further analysis.
All identified variants were classified, according to American College of Medical Genetics and Genomics (ACMG) guidelines, into one of 5 classes: pathogenic (P), likely pathogenic (LP), uncertain significance (VUS), likely benign (LB) and benign (B) [24]. Varsome [25] and/or ClinVar [26] and/or Franklin [27] were used as tools to sum up actual knowledge about the variants. Variants classified as LP or P were considered, as were VUS satisfying the PM2 criterion (“absent from controls”) of the ACMG guidelines. The molecular confirmation of variants was performed by standard Sanger sequencing on an automated analyzer (Applied Biosystems ®3130xl and/or 3500DX, Thermo Fisher Scientific Inc. Waltham, MA, USA).
3. Results

Table 1.
Clinical and genetic data of patients of the COL6A1, COL6A2 and COL6A3 cohorts.
A summary of VUS characteristics identified in COL6A1, COL6A2 and COL6A3 genes is reported in Table S1.
3.1. COL6A Genes-Identified Variants
Twenty-six different COL6A1 variants were identified in 48 patients (35% of the total cohort) (45 heterozygous, 1 compound heterozygous and 2 homozygous), with a predominance of splicing and missense variants.
In the COL6A2 gene, 52 different variants were detected in 56 patients (40% of the total cohort) (32 heterozygous, 12 compound heterozygous and 12 homozygous), encompassing all mutation types, with a predominance of missense variants.
Twenty-six COL6A3 variants, mostly missense, were identified in 34 patients (25%) (31 heterozygous, 1 compound heterozygous and 2 homozygous) (Figure 1a,b).
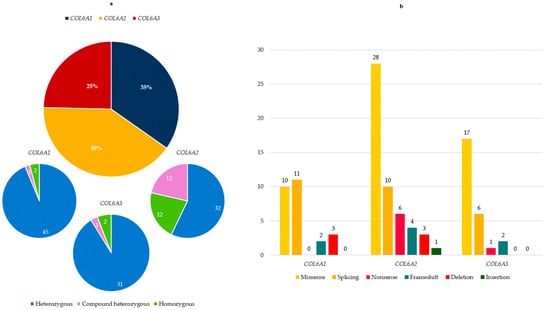
Figure 1.
(a) COL6A gene distribution among patients of our cohort. For each COL6A gene, proportion of heterozygous, compound heterozygous and homozygous is reported; (b) type and number of variants in the three COL6A genes.
For all detected variants, a pathogenicity prediction analysis was performed according to the ACMG criteria, resulting in the following classification:
3.2. COL6A Genes and Phenotypes
Overall, in our study, we identified 44 (32%) UCMD, 9 (7%) INT, 61 (44%) BM and 21 (15%) INT/BM patients. Moreover, three patients (2%) presented with an MM phenotype (Figure 2a).
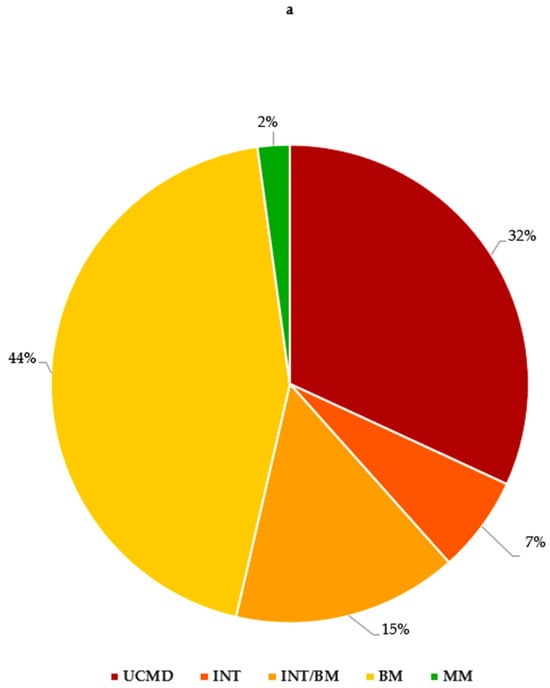
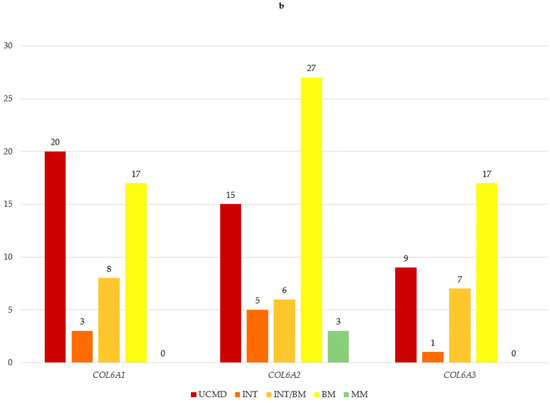
Figure 2.
(a) COL6A gene and phenotypes observed in our cohort; (b) number of variants in COL6A genes for each phenotype. UCMD: Ullrich congenital muscular dystrophy; INT: intermediate collagen VI-related myopathy; INT/BM: intermediate collagen VI-related myopathy/Bethlem myopathy; BM: Bethlem myopathy; MM: myosclerosis myopathy.
COL6A1 variants were associated with all phenotypes except MM (20 UCMD, 3 INT, 8 INT/BM, 17 BM).
The MM phenotype was present only in the COL6A2 cohort, where less severe phenotypes predominated (15 UCMD, 5 INT, 6 INT/BM, 27 BM, 3 MM).
In the COL6A3 gene, a similar prevalence of less severe phenotypes was observed (9 UCMD, 1 INT, 7 INT/BM, 17 BM) (Figure 2b).
3.3. COL6A Genes, Phenotypes and Inheritance
Most patients (108/138—78.3%) presented a monoallelic COL6A gene variant, suggesting an autosomal dominant/de novo inheritance. Multiple (two or more) variants were identified in 30/138 (21.7%) patients, suggesting a recessive inheritance.
Although monoallelic variants were prevalent in all COL6A genes, biallelic variants were significantly represented in the COL6A2 gene; indeed, biallelic variants in the COL6A2 gene were identified in 24 patients, whereas only three patients carried biallelic variants in COL6A1 and COL6A3.
Biallelic variants were exclusively associated with UCMD in the COL6A1 cohort, whereas in the COL6A2 and COL6A3 cohorts, they were linked to different phenotypes, with BM being the most prevalent in the COL6A2 cohort (Figure 3).
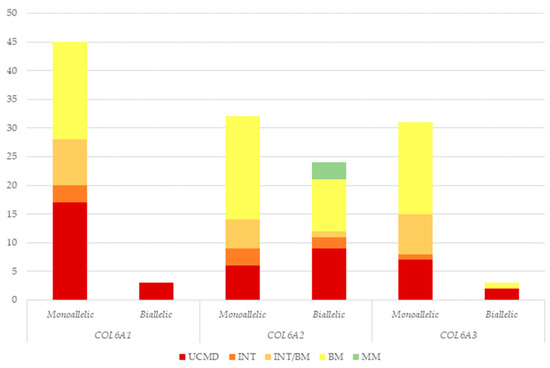
Figure 3.
Number of patients presenting monoallelic or biallelic COL6A gene variants according to each phenotype. UCMD: Ullrich congenital muscular dystrophy; INT: intermediate collagen VI-related myopathy; INT/BM: intermediate collagen VI-related myopathy/Bethlem myopathy; BM: Bethlem myopathy; MM: myosclerosis myopathy.
3.4. COL6A Variants and Protein Domain Distribution
In the COL6A1 and COL6A3 genes, the identified mutations were preferentially located in the N-terminal and THD domains, whereas they were rare in the C-terminal domain. However, in the COL6A2 gene, the mutations were distributed throughout the gene, with a prevalence in the C-terminal domain.
The mutation distribution within different domains of the three genes is reported in Figure 4.
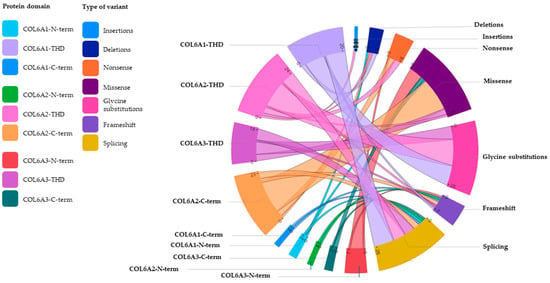
Figure 4.
Schematic representation of the mutation distribution within different domains of the three COL6A genes. The innovative diagram, called Chord (Microsoft Corporation), enables us to intuitively visualize the relationships between several variables, in this case between the three domains (N-terminal, TH and C-terminal) of the COL6A1, COL6A2 and COL6A3 genes and the different types of variants. Each variable, assigned a color code, is placed at a precise point, defined as a ‘node’, along a circular layout and connected to other nodes by arcs (literally ‘strings’). Each connection is assigned a value (in this case, corresponding to the number of variants), which is represented proportionally by the size of each arc, with thicker strings indicating numerically more significant connections and thinner strings representing weaker connections. THD: triple helical domain; N-term: N-terminal; C-term: C-terminal.
In detail, for the α1(VI) chain, THD was the most affected domain, with missense variants (glycine residue substitutions only) and splicing variants being the most common mutations.
For the α2(VI) chain, the variants were equally distributed between the THD and the C-terminal domain; in both regions, missense mutations were the most frequent.
In the α3(VI) chain, variants, mostly missense, were equally distributed in the THD and in the extended N-terminal region.
4. Discussion
We report on the largest study describing a nationwide genetic landscape of unrelated patients with collagen VI-related myopathies.
The cohort of 138 patients came from 21 Italian diagnostic centers, distributed throughout the national territory, diagnosed in a temporal window of 15 years (2007–2022).
Mutation detection was based on sequencing, either traditional Sanger sequencing or the next-generation sequencing approach, due to the evolution of technologies over time. Only in a few cases was a custom oligonucleotide CGH array used. This strategy implies that the occurrence of CNVs in COL6A genes was not fully investigated. Nevertheless, based on the Human Gene Mutation Database (HGMD), large genomic rearrangements account for a small proportion of collagen VI-related myopathy cases (≈5%) [28].
In our cohort, the proportion of COL6A1 (35%), COL6A2 (40%) and COL6A3 (25%) variants substantially overlapped with those already described in Western countries, reported to be 38%, 44% and 18%, respectively [29]. Similar proportions were found in an Egyptian cohort of 23 patients [17], but not in a larger cohort of 119 subjects with collagen VI-related myopathies of Spanish and American origin studied by Natera-de Benito and colleagues [30] or in recent sets of Asian cases in which COL6A1 variants were predominant [7,31,32]; however, the analysis of another Chinese cohort of patients showed a preponderance of COL6A2 variants [16].
This mutational scenario does not seem to support the existence of clear population or ethnic differences in the involvement of COL6A genes. Common to all reported studies, COL6A3 seems to be the least represented in cohorts of patients with collagen VI-related myopathies. Notably, it has been proposed that the α5(VI) and α6(VI) chains, highly homologous with α3(VI), can substitute for α3(VI) in assembling with α1(VI) and α2 (VI), perhaps compensating for the loss of the α3(VI) chain, if mutated.
The identified mutational scenario, including 104 COL6A different variants, further highlights the known considerable allelic heterogeneity of collagen VI-related myopathies [33]. According to the HGMD, more than 2000 different pathogenic variants have been reported in the COL6A genes, mainly point mutations, which account for ≈82% of variants [28].
In our cohort, the variant spectrum encompassed missense, splicing, frameshift/nonsense variants and in-frame deletions/insertion. Interestingly, the COL6A2 gene was the only one in which we observed the entire spectrum of variants; this gene also showed the highest allelic variability, with a total of 52 different variants identified.
Missense variants were the most frequent in all genes (52.9% of the total number of variants detected), followed by splicing variants. This finding is consistent with the HGMD data mentioned above and with the results of other studies [7,16,32].
Almost all variants in the COL6A1 and COL6A3 genes were detected in heterozygosity (monoallelic), whereas variants in compound heterozygosity/homozygosity were commonly observed in the COL6A2 gene, associated with both UCMD and BM phenotypes.
Among the missense variants, heterozygous glycine substitutions interrupting the triple Gly-X-Y repetitive sequence in α(VI) chains and well recognized as pathogenic [11,31] accounted for 49% of the total missense variants; among these, the COL6A1 p.(Gly284Arg) and p.(Gly293Arg) variants recur in our cohort, being detected in eight and six different individuals, respectively and associated with the full phenotypic spectrum.
Phenotypic variability in collagen VI-related myopathies is common and well documented in the literature [34]. It has been suggested that genetic modifiers, epigenetic and/or environmental factors or the occurrence of mosaicism may be involved in the explanation of this variability [35,36,37]. The availability of groups of patients with the same COL6A gene genotype but different degrees of severity could represent a useful tool to further investigate mechanisms underlying phenotypic variability.
Splicing variants were the second most common variant type (25.9% of the total number of variants detected), whereas they were the most common variants identified in other works [30]. Our cohort included four female patients carrying the deep intronic variant in COL6A1 (c.930+189C>T), described as a recurrent mutation that disrupts the Gly-X-Y motif at the N-terminal end of the THD and leads to a dominantly acting in-frame pseudo-exon insertion [14]. All identified patients shared a severe UCMD phenotype and early loss of ambulation, as described in previously reported cases [38]. In fact, previous studies have suggested that this recurrent intronic variant has a more pronounced dominant-negative action than other dominantly acting mutations in the COL6A gene, leading to a distinct phenotype of UCMD [38,39].
This novel and unexpectedly common COL6A1 intronic variant is important to identify, especially considering that it has been shown to be amenable to exon-skipping therapy [39].
Less common in our cohort were nonsense variants (found almost exclusively in the COL6A2 gene), frameshifts, small deletions or in-frame insertions. Variants introducing a premature stop codon (nonsense, frameshift and some splicing variants) were more common in recessive forms and more often associated with severe phenotypes, according to the literature data [30].
As previously discussed, collagen VI myopathies are caused either by recessive loss-of-function mutations or, more commonly, by de novo dominant-negative pathogenic variants in the three major COL6A genes. These dominant-negative variants generally fall into two categories: in-frame deletions/insertions and single nucleotide missense mutations that substitute glycine residues, thereby disrupting the Gly-X-Y motif of the THD. Nevertheless, nonsense or truncating mutations can also exert a dominant-negative effect if nonsense-mediated decay is escaped and the resulting truncated protein fragments are assembled with normal collagen chains, thus altering supramolecular structures [40].
In our cohort, we also identified, through a custom oligonucleotide CGH array, a deletion within intron 1A of the COL6A2 gene occurring in compound heterozygosity with a small deletion in exon 28 in a BM patient [22]. This case highlighted the relevance of array-CGH as a useful complementary diagnostic tool, especially in recessive forms of the disease in which only one mutant allele is detected by standard sequencing.
Unlike published patient cohorts [16,41,42], milder phenotypes (BM and INT/BM) were prevalent in our series, accounting for 59% of the patients. The recognition of mild phenotypes is certainly enhanced by the activity within clinical networks of excellence that bring together highly specialized centers, such as the European Reference Network (ERN) for neuromuscular diseases (ERN EURO-NMD) [43].
The ability to identify milder phenotypes and accurately categorize patients with collagen VI-related myopathies into phenotype subgroups will be crucial in optimizing clinical care, in the design of clinical trials for the therapeutic strategies currently under development, and in the definition of natural history and cohort studies.
Although meaningful genotype–phenotype correlations were not identified in our study, variants in COL6A1 tended to be associated with more severe phenotypes (UCMD), whereas less severe phenotypes (BM) were observed in the COL6A2 and COL6A3 cohorts.
The majority of patients presented with a monoallelic COL6A gene variant, suggesting an autosomal dominant/de novo inheritance (78.3%). Biallelic variants were found in 21.7% of the patients.
Notably, patients carrying dominantly acting variants appear to be more common in several other studies [7,30,41,44]. Rare exceptions are the small French cohorts, in which the number of dominant and recessive cases did not differ significantly [42], and the Egyptian cohort, in which patients with recessive variants predominated (56.5%), probably due to the higher inbreeding rates in this population [17].
In our cohort, biallelic variants in COL6A1 invariably led to a UCMD phenotype, whereas all phenotypes were associated with biallelic COL6A2 and COL6A3 variants. Unlike UCMD, which notoriously displays both autosomal dominant and recessive inheritance, BM has been considered exclusively an autosomal dominant disease for a long time. However, in recent years, few cases of recessive inheritance were described [21,45,46,47,48,49]. Our work significantly contributes to the collection of “milder” autosomal recessive cases because 10 of our BM patients presented a recessive inheritance. Intriguingly, nine of these (90%) displayed biallelic variants in the COL6A2 gene, mostly concentrated in the C-terminal end of the α2(VI) chain.
This finding might indicate that biallelic variants in the COL6A2 gene are more likely to be tolerated than biallelic variants in the other COL6A genes, thus leading to milder phenotypes. However, this hypothesis deserves to be explored with further extensive patient studies.
Concerning the involved protein domain, variants in the COL6A1 and COL6A3 genes were almost exclusively located in the NH2 and THD regions, consistent with data from other works [7,16,31].
In contrast, in the COL6A2 gene, variants were distributed throughout the gene with a prevalence in the carboxy-terminal domain. Interestingly, this finding is consistent with data from a large Chinese multicenter study in which the mutations were observed along the entire length of the COL6A2 gene, from the N-terminus to the THD and C-terminus [33].
On the tetramer structure of collagen VI, N-terminal globular domains are believed to be an attaching site for microfibril formation, but one hypothesis is that the carboxy-terminal domain is also crucial for this [50]. Indeed, after synthesis, the three collagen VI chains associate through the C-terminal globular domain and coil into a triple-helical monomer. The monomers are then assembled in a staggered, antiparallel manner into dimers, which then associate laterally into tetramers. The tetramers are secreted outside of the cells, where they assemble into collagen VI microfibrils [50].
A specific role for the C-terminal globular domain has been proposed by Zhang and colleagues, who suggested that this domain is critical for the proper alignment of the tetramer and for heterotypic interactions with other matrix molecules [40].
In addition, recessive mutations in the carboxy-terminal domain of the COL6A2 gene have been reported in the literature associated with a severe UCMD phenotype, emphasizing the importance of this region [50,51].
The COL6A2 mutation spectrum (all mutation types), together with its capability to be mutated in all collagen VI phenotypes, is interesting. Collagen gene preservation was checked in several species, from fungi to primates, and while COL6A1 and COL6A3 were not present in chimpanzees and rats, COL6A2 was found to be preserved in species ranging from fish to primates [52]. More generally, the COL6A genes were found to be the most ancient of all collagen-encoding genes. This peculiar evolutive aspect might be related to the unique mutation scenario and phenotypic association of COL6A2.
5. Conclusions
To the best of our knowledge, this paper describes the largest cohort of genotyped patients worldwide diagnosed in a temporal window of 15 years in 21 Italian diagnostic centers.
Our data on this large cohort of collagen VI-related myopathy patients reinforces previous reports describing the variability of the clinical spectrum, the considerable allelic heterogeneity and the resulting complexity of diagnosis [53].
Recent innovations in molecular genetics and innovative approaches [54], including the use of artificial intelligence (AI) [55], have provided tremendous support in the diagnostic process for conditions characterized by pronounced heterogeneity, such as collagen VI-related myopathies.
Also extremely significant in this context is the advantageous networking strategy offered by the ERN EURO-NMD [43]. Sharing expertise through ERNs has greatly improved diagnostic accuracy, the ability to identify the milder phenotypes and the categorization of patients into subgroups. Thanks to this network, Health Care Providers (HCPs) can easily interface with each other and use dedicated IT tools, such as the Clinical Patient Management System (CPMS), an innovative digital platform for discussing complex and atypical clinical cases [56].
An important and novel finding that has emerged from our study is how the COL6A2 gene has peculiar characteristics that distinguish it from the COL6A1 and COL6A3 genes. First, the COL6A2 gene was found to be the most frequently involved, with the greatest allelic variability and the full spectrum of variant types. These variants are both dominant and recessive, the latter being much more common than in the other COL6A genes. The distribution of variants along the α2(VI) chain is also characteristic, not limited to the N-terminal end of the THD, but also frequently occurring in the C-terminal domain, which appears to be of primary importance in the complex process of protein assembly. Although the small number of patients reported to date hampers definitive conclusions, it is interesting that COL6A2 is the only gene affected in our myosclerosis myopathy patients, suggesting that this should be the first gene to be investigated when the phenotype is highly suggestive.
This diversity is surprising, considering that COL6A1 and COL6A2 genes have a highly similar sequence and protein structure, cluster on the same 21q22.3 chromosomal region and share an ancestral gene duplication origin. However, evolutionary and structural differences between the two genes exist, as the COL6A2 gene is more ancient than the COL6A1 gene in animal phylogeny, is the unique COL6A gene that is preserved in all vertebrates without gene losses [52]. Additionally, and significantly, the human intron–exon organization of the globular domains of COL6A2 and COL6A1 is different, in contrast with the conservation of the triple-helical exon organization [57]. These differences could be the expression of different functions and deserve further investigations to understand their correlation with the COL6A2 mutation pattern and their implications in the pathophysiology of collagen VI-related myopathies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biom15101426/s1, Table S1: Summary of VUS Characteristics in COL6A1, COL6A2, and COL6A3 genes
Author Contributions
F.F. contributed to data curation, formal analysis, investigation, methodology and manuscript writing—original draft. L.F. contributed to data curation, formal analysis, investigation and methodology. A.F. and F.G. were responsible for conceptualization, formal analysis, supervision, writing—original draft and writing—review & editing. A.M., M.N., A.D., E.B., E.R., E.M.M., M.P. (Marika Pane)., R.M., G.G., A.L.B., C.C., A.P., L.M. (Luciano Merlini)., C.F. (Carlo Fusco)., C.R., S.M., C.F. (Chiara Fiorillo), C.B., M.P. (Marina Pedemonte), M.T., I.M., L.M. (Lorenzo Maggi), S.G., E.P. (Elena Pegoraro), E.P. (Esther Picillo), L.P., M.S., F.V., F.M., M.F., P.G., F.R., T.E.M. and R.S. contributed to patient data collection/investigation. A.F. is the corresponding author. All authors have contributed significantly to the work, and agree to be accountable for all aspects of the work to ensure its accuracy and integrity. All authors have read and agreed to the published version of the manuscript.
Funding
This research received funding the Euro-NMD Project 101156434 (EURO-NMD multi-beneficiary grant agreement 2023–2027; WP4 CPMS).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki. Ethical consent was collected in each center as part of the routine diagnostic procedures for COL6A genetic diagnosis. This study was approved by Comitato Etico di Area Vasta Emilia Centro (CE-AVEC) 66/2020/Oss/AOUFe 2020-01-23.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All variants presented in this study have been submitted to the LOVD gene-specific database for free consultation.
Acknowledgments
The Euro-NMD Project 101156434 (EURO-NMD multi-beneficiary grant agreement 2023–2027; WP4 CPMS, to A.F. and F.F.) is acknowledged. We acknowledge the support of Telethon (grant GSP19003) and Solve-RD (grant agreement No. 779257).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ECM | Extracellular matrix |
| THD | Triple helical domain |
| CMD | Congenital muscular dystrophy |
| UCMD | Ullrich congenital muscular dystrophy |
| BM | Bethlem myopathy |
| MM | Myosclerosis myopathy |
| INT | Intermediate collagen VI-related myopathy |
| INT/BM | Intermediate collagen VI-related myopathy/Bethlem myopathy |
| NGS | Next-generation sequencing |
| CNV | Copy number variation |
| ESP | Exome Variant Server |
| ExAC | Exome Aggregation Consortium |
| ACMG | American College of Medical Genetics and Genomics |
| VUS | Variant of uncertain significance |
| HGMD | Human Gene Mutation Database |
| ERN | European Reference Network |
| HCP | Health care provider |
| CPMS | Clinical Patient Management System |
| AI | Artificial intelligence |
References
- Timpl, R.; Chu, M.L. Collagen type VI. In Extracellular Matrix Assembly and Structure; Yurchenco, P.D., Birk, D.E., Mecham, R.P., Eds.; Academic Press: Orlando, FL, USA, 1994; pp. 207–242. [Google Scholar]
- Saitta, B.; Stokes, D.G.; Vissing, H.; Timpl, R.; Chu, M.L. Alternative splicing of the human alpha 2(VI) collagen gene generates multiple mRNA transcripts which predict three protein variants with distinct carboxyl termini. J. Biol. Chem. 1990, 265, 6473–6480. [Google Scholar] [CrossRef]
- Stokes, D.G.; Saitta, B.; Timpl, R.; Chu, M.L. Human alpha 3(VI) collagen gene. Characterization of exons coding for the amino-terminal globular domain and alternative splicing in normal and tumor cells. J. Biol. Chem. 1991, 266, 8626–8633. [Google Scholar] [CrossRef]
- Kuo, H.J.; Maslen, C.L.; Keene, D.R.; Glanville, R.W. Type VI collagen anchors endothelial basement membranes by interacting with type IV collagen. J. Biol. Chem. 1997, 272, 26522–26529. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.L.; Pan, T.C.; Conway, D.; Saitta, B.; Stokes, D.; Kuo, H.J.; Glanville, R.W.; Timpl, R.; Mann, K.; Deutzmann, R. The structure of type VI collagen. Ann. N. Y Acad. Sci. 1990, 580, 55–63. [Google Scholar] [CrossRef]
- Gregory, C.A.; Ma, J.; Lomeli, S. The coordinated activities of collagen VI and XII in maintenance of tissue structure, function and repair: Evidence for a physical interaction. Front. Mol. Biosci. 2024, 11, 1376091. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Saito, Y.; Yonekawa, T.; Ogawa, M.; Iida, A.; Nishino, I.; Noguchi, S. Causative variant profile of collagen VI-related dystrophy in Japan. Orphanet J. Rare Dis. 2021, 16, 284. [Google Scholar] [CrossRef] [PubMed]
- Graziano, A.; Bianco, F.; D’Amico, A.; Moroni, I.; Messina, S.; Bruno, C.; Pegoraro, E.; Mora, M.; Astrea, G.; Magri, F.; et al. Prevalence of congenital muscular dystrophy in Italy: A population study. Neurology 2015, 84, 904–911. [Google Scholar] [CrossRef]
- Bönnemann, C.G. The collagen VI-related myopathies: Muscle meets its matrix. Nat. Rev. Neurol. 2011, 7, 379–390. [Google Scholar] [CrossRef]
- Merlini, L.; Martoni, E.; Grumati, P.; Sabatelli, P.; Squarzoni, S.; Urciuolo, A.; Ferlini, A.; Gualandi, F.; Bonaldo, P. Autosomal recessive myosclerosis myopathy is a collagen VI disorder. Neurology 2008, 71, 1245–1253. [Google Scholar] [CrossRef]
- Butterfield, R.J.; Foley, A.R.; Dastgir, J.; Asman, S.; Dunn, D.M.; Zou, Y.; Hu, Y.; Donkervoort, S.; Flanigan, K.M.; Swoboda, K.J.; et al. Position of glycine substitutions in the triple helix of COL6A1, COL6A2, and COL6A3 is correlated with severity and mode of inheritance in collagen VI myopathies. Hum. Mutat. 2013, 34, 1558–1567. [Google Scholar] [CrossRef]
- Lampe, A.K.; Bushby, K.M. Collagen VI related muscle disorders. J. Med. Genet. 2005, 42, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Foley, A.R.; Hu, Y.; Zou, Y.; Yang, M.; Medne, L.; Leach, M.; Conlin, L.K.; Spinner, N.; Shaikh, T.H.; Falk, M.; et al. Large genomic deletions: A novel cause of Ullrich congenital muscular dystrophy. Ann. Neurol. 2011, 69, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Cummings, B.B.; Marshall, J.L.; Tukiainen, T.; Lek, M.; Donkervoort, S.; Foley, A.R.; Bolduc, V.; Waddell, L.B.; Sandaradura, S.A.; O’Grady, G.L.; et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med. 2017, 9, eaal5209. [Google Scholar] [CrossRef]
- Okada, M.; Kawahara, G.; Noguchi, S.; Sugie, K.; Murayama, K.; Nonaka, I.; Hayashi, Y.K.; Nishino, I. Primary collagen VI deficiency is the second most common congenital muscular dystrophy in Japan. Neurology 2007, 69, 1035–1042. [Google Scholar] [CrossRef]
- Fan, Y.; Liu, A.; Wei, C.; Yang, H.; Chang, X.; Wang, S.; Yuan, Y.; Bonnemann, C.; Wu, Q.; Wu, X.; et al. Genetic and clinical findings in a Chinese cohort of patients with collagen VI-related myopathies. Clin. Genet. 2018, 93, 1159–1171. [Google Scholar] [CrossRef]
- Sharaf-Eldin, W.E.; Rafat, K.; Issa, M.Y.; Elbendary, H.M.; Eissa, N.R.; Hawaary, B.; Gaboon, N.E.A.; Maroofian, R.; Gleeson, J.G.; Essawi, M.L.; et al. Clinical and Molecular Profiles of a Cohort of Egyptian Patients with Collagen VI-Related Dystrophy. J. Mol. Neurosci. 2024, 74, 93. [Google Scholar] [CrossRef]
- Lamandé, S.R.; Bateman, J.F. Collagen VI disorders: Insights on form and function in the extracellular matrix and beyond. Matrix Biol. 2018, 71–72, 348–367. [Google Scholar] [CrossRef]
- Lampe, A.K.; Zou, Y.; Sudano, D.; O’Brien, K.K.; Hicks, D.; Laval, S.H.; Charlton, R.; Jimenez-Mallebrera, C.; Zhang, R.Z.; Finkel, R.S.; et al. Exon skipping mutations in collagen VI are common and are predictive for severity and inheritance. Hum. Mutat. 2008, 29, 809–822. [Google Scholar] [CrossRef]
- Allamand, V.; Merlini, L.; Bushby, K.; Consortium for Collagen VI-Related Myopathies. 166th ENMC International Workshop on Collagen type VI-related Myopathies, 22–24 May 2009, Naarden, The Netherlands. Neuromuscul Disord. 2010, 20, 346–354. [Google Scholar] [CrossRef]
- Foley, A.R.; Hu, Y.; Zou, Y.; Columbus, A.; Shoffner, J.; Dunn, D.M.; Weiss, R.B.; Bönnemann, C.G. Autosomal recessive inheritance of classic Bethlem myopathy. Neuromuscul. Disord. 2009, 19, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Bovolenta, M.; Neri, M.; Martoni, E.; Urciuolo, A.; Sabatelli, P.; Fabris, M.; Grumati, P.; Mercuri, E.; Bertini, E.; Merlini, L.; et al. Identification of a deep intronic mutation in the COL6A2 gene by a novel custom oligonucleotide CGH array designed to explore allelic and genetic heterogeneity in collagen VI-related myopathies. BMC Med. Genet. 2010, 11, 44. [Google Scholar] [CrossRef]
- IGV. Available online: http://software.broadinstitute.org/software/igv/ (accessed on 11 July 2025).
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Varsome. Available online: https://varsome.com/ (accessed on 11 July 2025).
- Clinvar. Available online: https://www.ncbi.nlm.nih.gov/clinvar/ (accessed on 11 July 2025).
- Franklin. Available online: https://franklin.genoox.com/clinical-db/home (accessed on 11 July 2025).
- Stenson, P.D.; Mort, M.; Ball, E.V.; Shaw, K.; Phillips, A.; Cooper, D.N. The Human Gene Mutation Database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014, 133, 1–9. [Google Scholar] [CrossRef]
- Bushby, K.M.; Collins, J.; Hicks, D. Collagen type VI myopathies. Adv. Exp. Med. Biol. 2014, 802, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Natera-de Benito, D.; Foley, A.R.; Domínguez-González, C.; Ortez, C.; Jain, M.; Mebrahtu, A.; Donkervoort, S.; Hu, Y.; Fink, M.; Yun, P.; et al. Association of Initial Maximal Motor Ability with Long-term Functional Outcome in Patients With COL6-Related Dystrophies. Neurology 2021, 96, e1413–e1424. [Google Scholar] [CrossRef] [PubMed]
- Kwong, A.K.; Zhang, Y.; Ho, R.S.; Gao, Y.; Ling, X.; Tsang, M.H.; Luk, H.M.; Chung, B.H.; Bönnemann, C.G.; Javed, A.; et al. Collagen VI-related myopathies: Clinical variability, phenotype-genotype correlation and exploratory transcriptome study. Neuromuscul. Disord. 2023, 33, 371–381. [Google Scholar] [CrossRef]
- Lee, J.H.; Shin, H.Y.; Park, H.J.; Kim, S.H.; Kim, S.M.; Choi, Y.C. Clinical, Pathologic, and Genetic Features of Collagen VI-Related Myopathy in Korea. J. Clin. Neurol. 2017, 13, 331–339. [Google Scholar] [CrossRef]
- Hu, C.; Shi, Y.; Zhao, L.; Zhu, W.; Jiao, K.; Yu, L.; Li, X.; Wang, Y. Clinical, Pathologic, and Genetic Spectrum of Collagen VI-Related Disorder in China-A Retrospective Observational Multicenter Study. Hum Mutat. 2024, 2024, 3503253. [Google Scholar] [CrossRef]
- Bardakov, S.N.; Deev, R.V.; Magomedova, R.M.; Umakhanova, Z.R.; Allamand, V.; Gartioux, C.; Zulfugarov, K.Z.; Akhmedova, P.G.; Tsargush, V.A.; Titova, A.A.; et al. Intrafamilial Phenotypic Variability of Collagen VI-Related Myopathy Due to a New Mutation in the COL6A1 Gene. J. Neuromuscul. Dis. 2021, 8, 273–285. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, A.; Fattori, F.; Tasca, G.; Petrini, S.; Gualandi, F.; Bruselles, A.; D’Oria, V.; Verardo, M.; Carrozzo, R.; Niceta, M.; et al. Somatic mosaicism represents an underestimated event underlying collagen 6-related disorders. Eur. J. Paediatr. Neurol. 2017, 21, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, A.; Kinali, M.; Main, M.; Jimenez-Mallebrera, C.; Aloysius, A.; Clement, E.; North, B.; Manzur, A.Y.; Robb, S.A.; Mercuri, E.; et al. Natural history of Ullrich congenital muscular dystrophy. Neurology 2009, 73, 25–31. [Google Scholar] [CrossRef]
- Silverstein, R.S.; Wang, D.D.; Haruno, L.S.; Lotze, T.E.; Scott, A.C.; Rosenfeld, S.B. Bethlem Myopathy (Collagen VI-Related Dystrophies): A Retrospective Cohort Study on Musculoskeletal Pathologies and Clinical Course. J. Pediatr. Orthop. 2023, 43, e163–e167. [Google Scholar] [CrossRef]
- Foley, A.R.; Bolduc, V.; Guirguis, F.; Donkervoort, S.; Hu, Y.; Orbach, R.; McCarty, R.M.; Sarathy, A.; Norato, G.; Cummings, B.B.; et al. The recurrent deep intronic pseudoexon-inducing variant COL6A1 c.930+189C>T results in a consistently severe phenotype of COL6-related dystrophy: Towards clinical trial readiness for splice-modulating therapy. medRxiv 2024. [Google Scholar] [CrossRef]
- Aguti, S.; Bolduc, V.; Ala, P.; Turmaine, M.; Bönnemann, C.G.; Muntoni, F.; Zhou, H. Exon-Skipping Oligonucleotides Restore Functional Collagen VI by Correcting a Common COL6A1 Mutation in Ullrich CMD. Mol. Ther. Nucleic Acids. 2020, 21, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.-Z.; Sabatelli, P.; Pan, T.-C.; Squarzoni, S.; Mattioli, E.; Bertini, E.; Pepe, G.; Chu, M.-L. Effects on collagen VI mRNA stability and microfibrillar assembly of three COL6A2 mutations in two families with Ullrich congenital muscular dystrophy. J. Biol. Chem. 2002, 277, 43557–43564. [Google Scholar] [CrossRef] [PubMed]
- Zanoteli, E.; Soares, P.S.; Silva, A.M.S.D.; Camelo, C.G.; Fonseca, A.T.Q.S.M.; Albuquerque, M.A.V.; Moreno, C.A.M.; Lopes Abath Neto, O.; Novo Filho, G.M.; Kulikowski, L.D.; et al. Clinical features of collagen VI-related dystrophies: A large Brazilian cohort. Clin. Neurol. Neurosurg. 2020, 192, 105734. [Google Scholar] [CrossRef]
- Morel, V.; Audic, F.; Tardy, C.; Verschueren, A.; Attarian, S.; Nguyen, K.; Salort-Campana, E.; Krahn, M.; Chabrol, B.; Gorokhova, S. Retrospective clinical and genetic analysis of COL6-RD patients with a long-term follow-up at a single French center. Front. Genet. 2023, 14, 1242277. [Google Scholar] [CrossRef]
- ERN EURO-NMD. Available online: https://ern-euro-nmd.eu/ (accessed on 11 July 2025).
- Foley, A.R.; Mohassel, P.; Donkervoort, S.; Bolduc, V.; Bönnemann, C.G. Collagen VI-Related Dystrophies; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; GeneReviews®; University of Washington: Seattle, WA, USA, 2004. [Google Scholar]
- Aldharee, H.; Hamdan, H.Z. Segregation of the COL6A2 Variant (c.1817-3C>G) in a Consanguineous Saudi Family with Bethlem Myopathy. Genes 2024, 15, 1405. [Google Scholar] [CrossRef]
- Zamurs, L.K.; Idoate, M.A.; Hanssen, E.; Gomez-Ibañez, A.; Pastor, P.; Lamandé, S.R. Aberrant mitochondria in a Bethlem myopathy patient with a homozygous amino acid substitution that destabilizes the collagen VI α2(VI) chain. J. Biol. Chem. 2015, 290, 4272–4281. [Google Scholar] [CrossRef]
- Deconinck, N.; Richard, P.; Allamand, V.; Behin, A.; Lafôret, P.; Ferreiro, A.; de Becdelievre, A.; Ledeuil, C.; Gartioux, C.; Nelson, I.; et al. Bethlem myopathy: Long-term follow-up identifies COL6 mutations predicting severe clinical evolution. J. Neurol. Neurosurg. Psychiatry 2015, 86, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Sframeli, M.; Sarkozy, A.; Bertoli, M.; Astrea, G.; Hudson, J.; Scoto, M.; Mein, R.; Yau, M.; Phadke, R.; Feng, L.; et al. Congenital muscular dystrophies in the UK population: Clinical and molecular spectrum of a large cohort diagnosed over a 12-year period. Neuromuscul. Disord. 2017, 27, 793–803. [Google Scholar] [CrossRef]
- Annunziata, A.; Langella, G.; Cauteruccio, R.; Fiorentino, L.; Fiorentino, G. Severe progressive respiratory involvement requiring ventilator support in autosomal recessive Bethlem myopathy. A case report. Acta Myol. 2024, 43, 149–152. [Google Scholar] [CrossRef]
- Zhang, R.Z.; Zou, Y.; Pan, T.C.; Markova, D.; Fertala, A.; Hu, Y.; Squarzoni, S.; Reed, U.C.; Marie, S.K.N.; Bönnemann, C.G.; et al. Recessive COL6A2 C-globular missense mutations in Ullrich congenital muscular dystrophy: Role of the C2a splice variant. J. Biol. Chem. 2010, 285, 10005–10015. [Google Scholar] [CrossRef]
- Baker, N.L.; Mörgelin, M.; Peat, R.; Goemans, N.; North, K.N.; Bateman, J.F.; Lamandé, S.R. Dominant collagen VI mutations are a common cause of Ullrich congenital muscular dystrophy. Hum. Mol. Genet. 2005, 14, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Haq, F.; Ahmed, N.; Qasim, M. Comparative genomic analysis of collagen gene diversity. 3 Biotech 2019, 9, 83. [Google Scholar] [CrossRef]
- Merlini, L.; Sabatelli, P.; Gualandi, F.; Redivo, E.; Di Martino, A.; Faldini, C. New Clinical and Immunofluoresence Data of Collagen VI-Related Myopathy: A Single Center Cohort of 69 Patients. Int. J. Mol. Sci. 2023, 24, 12474. [Google Scholar] [CrossRef]
- Godwin, A.R.F.; Becker, M.H.; Dajani, R.; Snee, M.; Roseman, A.M.; Baldock, C. Collagen VI microfibril structure reveals mechanism for molecular assembly and clustering of inherited pathogenic mutations. Nat. Commun. 2025, 16, 7549. [Google Scholar] [CrossRef]
- Frías, M.; Badosa, C.; Jimenez-Mallebrera, C.; Porta, J.M.; Roldán, M. The artificial intelligence challenge in rare disease diagnosis: A case study on collagen VI muscular dystrophy. Comput. Biol. Med. 2025, 196, 110610. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, F.; Bianchi, F.; Ricci, G.; Torri, F.; Gualandi, F.; Neri, M.; Farnè, M.; Giannini, F.; Malandrini, A.; Volpi, N.; et al. Digital health and Clinical Patient Management System (CPMS) platform utility for data sharing of neuromuscular patients: The Italian EURO-NMD experience. Orphanet J. Rare Dis. 2023, 18, 196. [Google Scholar] [CrossRef]
- Trikka, D.; Davis, T.; Lapenta, V.; Brahe, C.; Kessling, A.M. Human COL6A1, Genomic characterization of the globular domains structural and evolutionary comparison with COL6A2. Mamm. Genome 1997, 8, 342–345. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).