Identification of Novel Quantitative Trait Loci and Candidate Genes Associated with Grain Yield and Related Traits Under Low-Light Stress Conditions in Rice
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
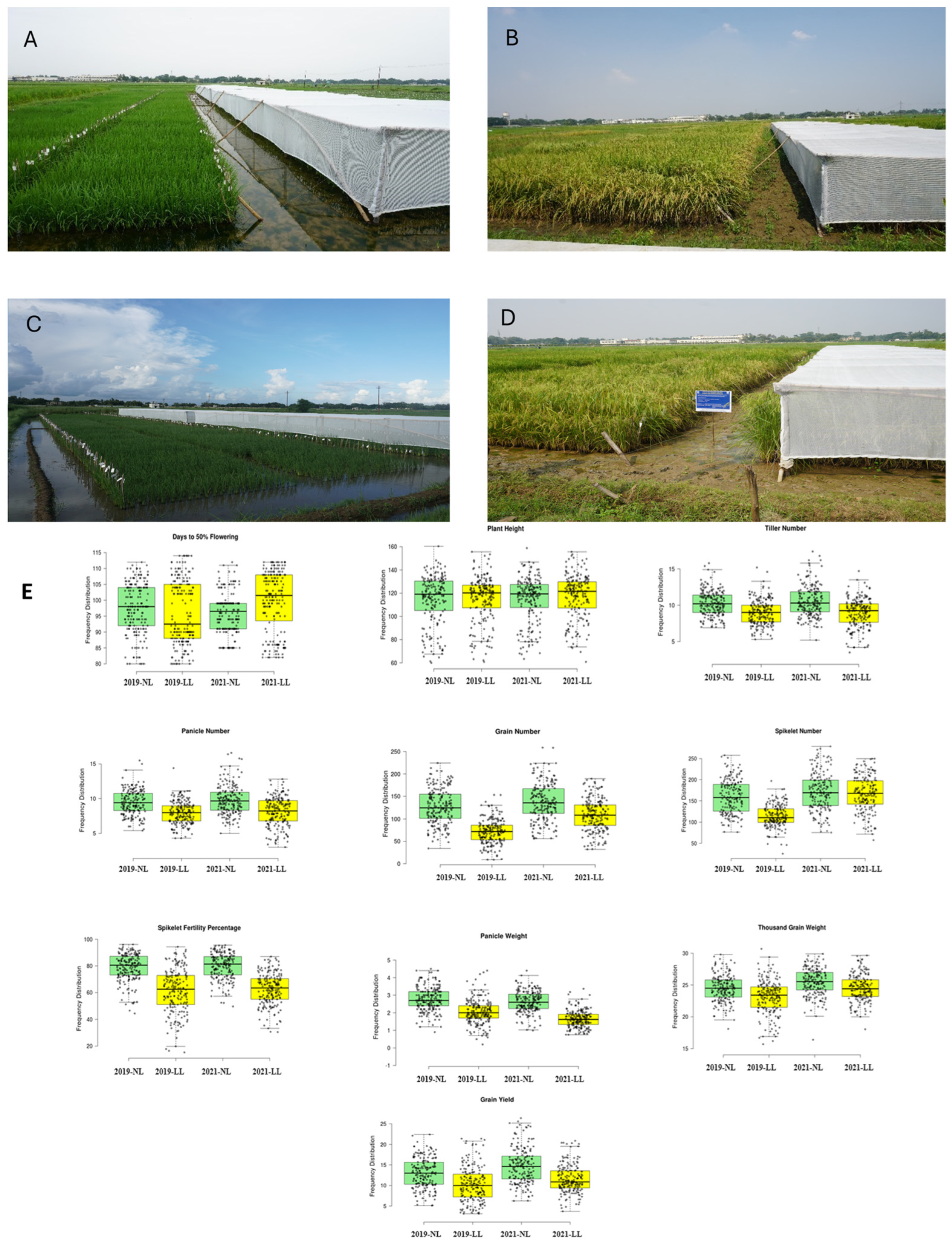
2.2. Phenotypic Evaluation of RIL Mapping Population for Grain Yield and Related Traits Under Low-Light Stress Conditions
2.3. Genotyping of Parents and RIL Mapping Population
2.4. Construction of Linkage Map and Identification of QTLs
2.5. In Silico Analysis to Identify Candidate GENES Associated with QTLs
2.6. Expression Analysis of Candidate Genes
2.7. Pathway and Network Analysis of Selected Candidate Genes
2.8. Statistical Analysis
3. Results
3.1. Phenotypic Variations and Correlations Among Traits in the RIL Population
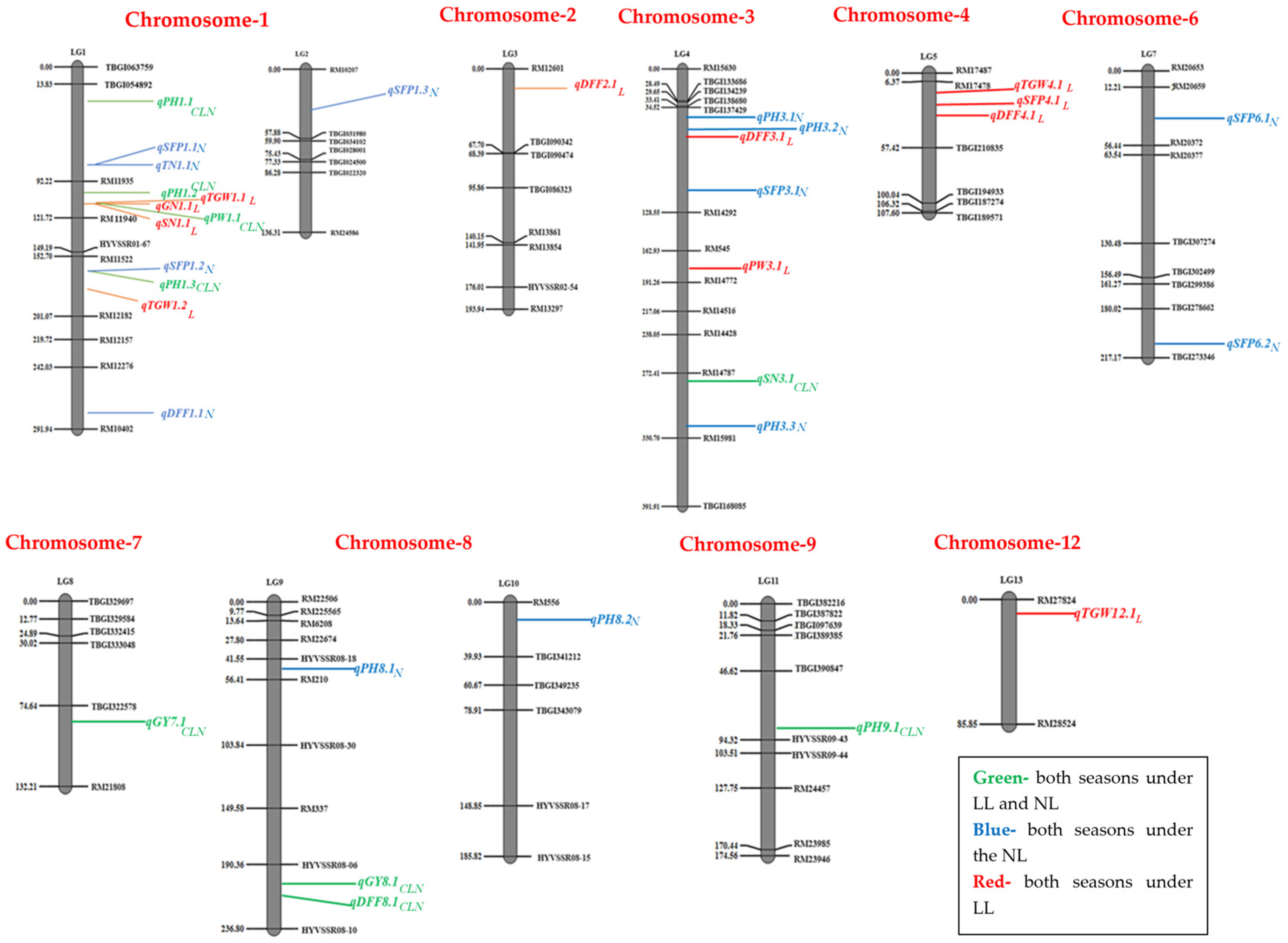
3.2. Genotyping and Linkage Map Construction
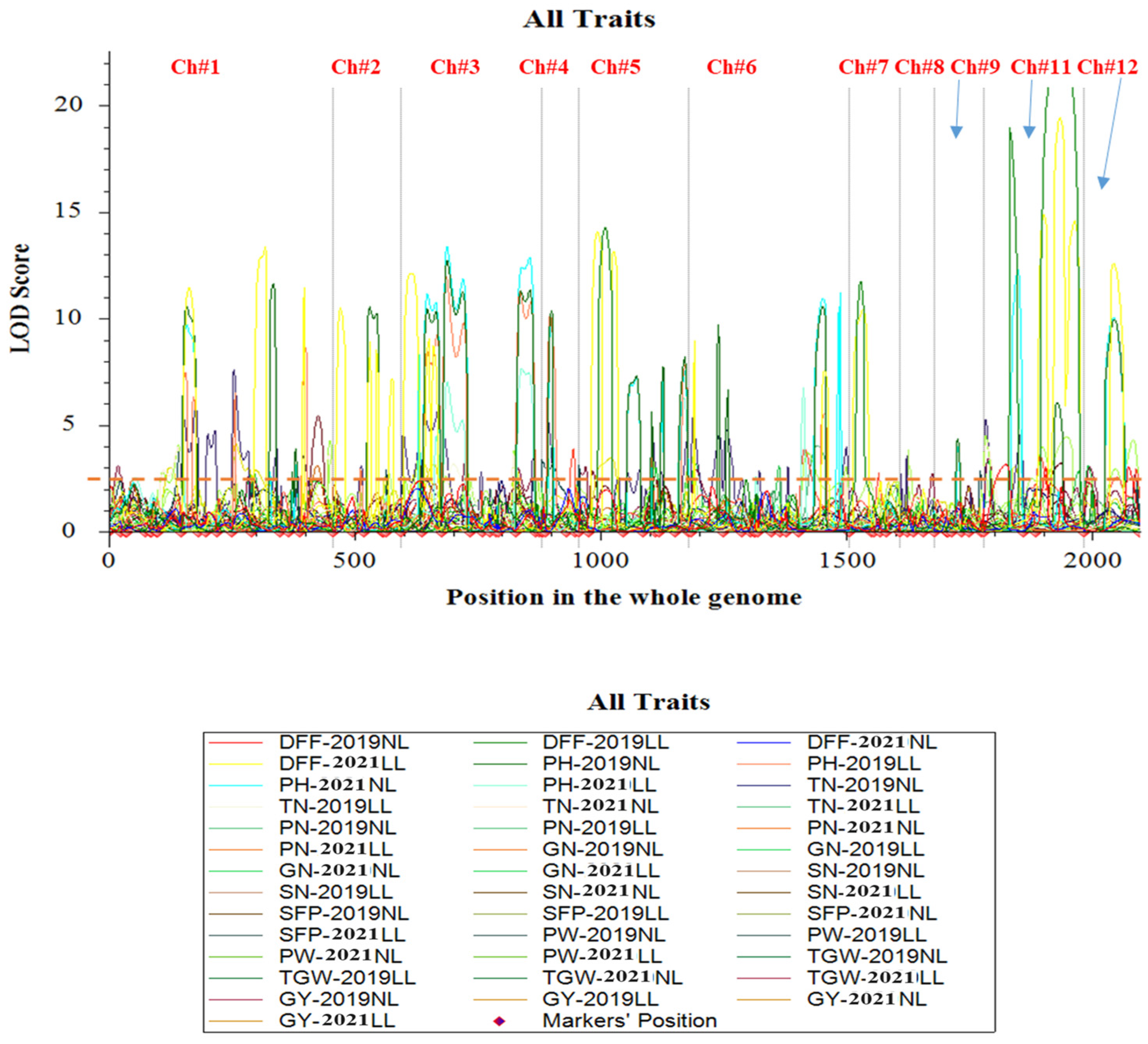
3.3. Composite Interval Mapping and Identification of QTLs
3.4. Novel QTLs
3.5. QTL Hotspots
3.6. In Silico Analysis for the Identification of Candidate Genes in QTL Hotspot Genomic Regions
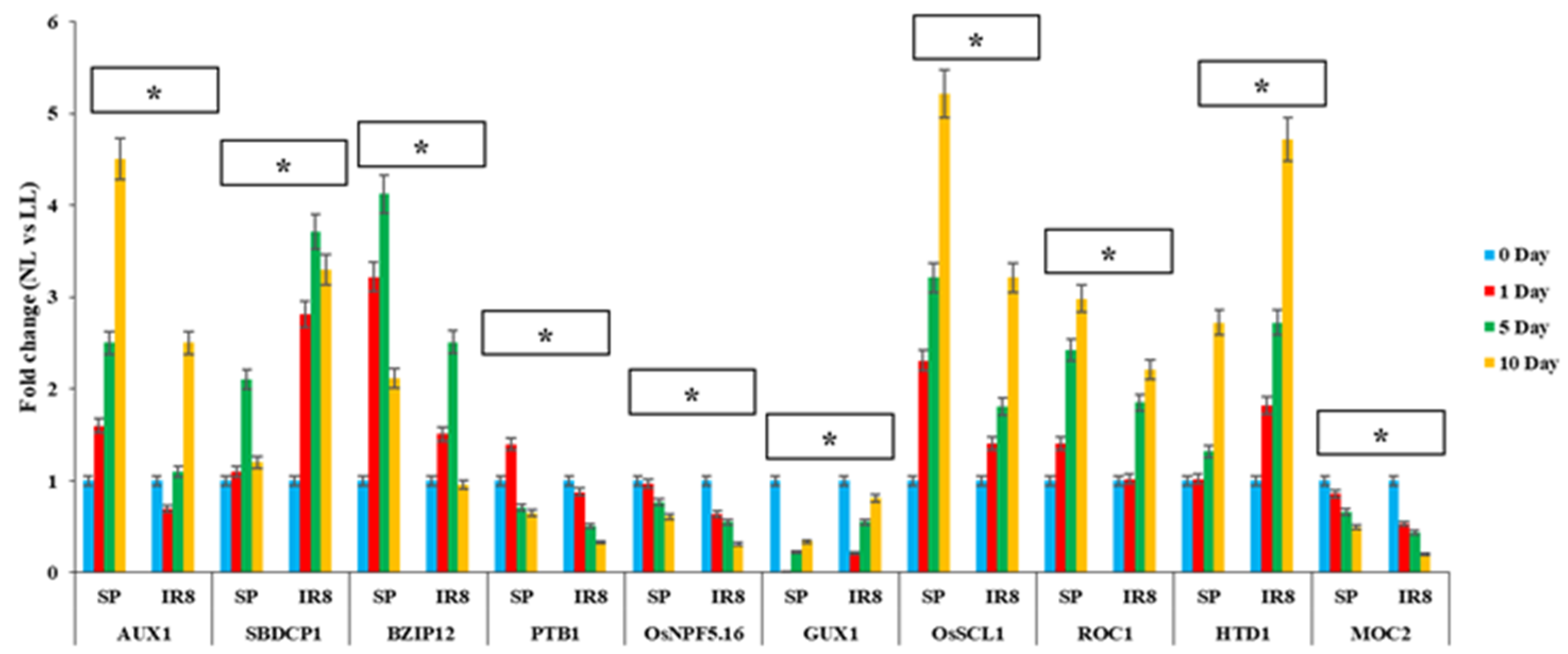
3.7. Expression Analysis of Candidate Genes
3.8. Pathway and Network Analysis of Hub Genes
4. Discussion
4.1. Phenotypic Diversity Among Parents and RILs
4.2. Correlations Among Grain Yield and Related Traits
4.3. Identification of QTLs Associated with Grain Yield and Related Traits Under Low-Light and Normal-Light Conditions
4.4. Identification of QTL Hotspots
4.5. Identification and Expression Analysis of Candidate Genes Associated with QTL Hotspots
4.6. Pathway and Network Analysis of Selected Candidate Genes to Identify Hub Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luo, Y.; Schuur, E.A. Model parameterization to represent processes at unresolved scales and changing properties of evolving systems. Glob. Change Biol. 2020, 26, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Panda, D.; Mohanty, S.; Das, S.; Sah, R.P.; Kumar, A.; Behera, L.; Tripathy, B.C. The role of phytochrome-mediated gibberellic acid signaling in the modulation of seed germination under low light stress in rice (O. sativa L.). Physiol. Mol. Biol. Plants. 2022, 28, 585–605. [Google Scholar] [CrossRef] [PubMed]
- Panda, D.; Dash, G.K.; Mohanty, S.; Sekhar, S.; Roy, A.; Tudu, C.; Baig, M.J. Phytochrome A mediated modulation of photosynthesis, development and yield in rice (Oryza sativa L.) in fluctuating light environment. Environ. Exp. Bot. 2023, 206, 105183. [Google Scholar] [CrossRef]
- Assuero, S.G.; Tognetti, J.A. Tillering regulation by endogenous and environmental factors and its agricultural management. Am. J. Plant Sci. 2010, 4, 35–48. [Google Scholar]
- Roeber, V.M.; Bajaj, I.; Rohde, M.; Schmülling, T.; Cortleven, A. Light acts as a stressor and influences abiotic and biotic stress responses in plants. Plant Cell Environ. 2021, 44, 645–664. [Google Scholar] [CrossRef]
- Praba, M.L.; Vanangamudi, M.; Thandapani, V. Effect of low light on yield and physiological attributes of rice. Int. Rice Res. 2004, 29, 71–73. [Google Scholar]
- Kumar, A.; Panda, D.; Biswal, M.; Dey, P.; Behera, L.; Baig, M.J.; Sharma, S. Low light stress influences resistant starch content and glycemic index of rice (O. sativa L). Starch Stärke 2019, 71, 1800216. [Google Scholar] [CrossRef]
- Murty, K.S.; Sahu, G. Impact of low-light stress on growth and yield of rice. In Weather and Rice, Proceedings of the International Workshop on the Impact of Weather Parameters on Growth and Yield of Rice, Manila, Philippines, 7–10 April 1986; International Rice Research Institute: Manila, Philippines, 1987; pp. 93–101. [Google Scholar]
- Ramachandran, N. Climate change, seasonality and hunger: The South Asian experience. In Global Food Insecurity, Rethinking Agricultural and Rural Development Paradigm and Policy; Behnassi, M., Draggan, S., Yaya, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 201–215. [Google Scholar]
- Wassmann, R.; Jagadish, S.V.K.; Sumfleth, K.; Pathak, H.; Howell, G.; Ismail, A.; Heuer, S. Regional vulnerability of climate change impacts on Asian rice production and scope for adaptation. Adv. Agron. 2009, 102, 91–133. [Google Scholar]
- Mandal, G.; Joshi, S.P. Eco-physiology and habitat invasibility of an invasive, tropical shrub (Lantana camara) in western Himalayan forests of India. For. Sci. Technol. 2015, 11, 182–196. [Google Scholar] [CrossRef]
- Fageria, N.K. Yield physiology of rice. J. Plant Nutr. 2007, 30, 843–879. [Google Scholar] [CrossRef]
- Fujita, K.; Yoshida, S. Partitioning of photosynthates between panicle and vegetative organs of rice under different planting densities. J. Soil Sci. Plant Nutr. 1984, 30, 519–525. [Google Scholar] [CrossRef]
- Panda, D.; Biswal, M.; Behera, L.; Baig, M.J.; Dey, P.; Nayak, L.; Kumar, A. Impact of low light stress on physiological, biochemical and agronomic attributes of rice. J. Pharmacogn. Phytochem. 2019, 8, 1814–1821. [Google Scholar]
- Wang, R.; Yu, G.; He, N.; Wang, Q.; Zhao, N.; Xu, Z.; Ge, J. Latitudinal variation of leaf stomatal traits from species to community level in forests, linkage with ecosystem productivity. Sci. Rep. 2015, 5, 14454. [Google Scholar] [CrossRef]
- Lawlor, D.W. Photosynthesis, productivity and environment. J. Exp. Bot. 1995, 46, 1449–1461. [Google Scholar] [CrossRef]
- Umesh, M.R.; Angadi, S.; Begna, S.; Gowda, P.; Prasad, P.V. Shade tolerance response of legumes in terms of biomass accumulation, leaf photosynthesis, and chlorophyll pigment under reduced sunlight. Crop Sci. 2023, 63, 278–292. [Google Scholar] [CrossRef]
- Liu, Q.H.; Xiu, W.U.; Chen, B.C.; Jie, G.A.O. Effects of low light on agronomic and physiological characteristics of rice including grain yield and quality. Rice Sci. 2014, 21, 243–251. [Google Scholar] [CrossRef]
- Panda, D.; Biswal, M.; Mohanty, S.; Dey, P.; Swain, A.; Behera, D.; Behera, L. Contribution of phytochromea in the regulation of sink capacity starch biosynthesis, grain quality, grain yield and related traits in rice. Plant Archiv. 2020, 20, 1179–1194. [Google Scholar]
- Taiz, L.; Zeiger, E. (Eds.) Responses and adaptations to abiotic stress. In Plant Physiology, 5th ed.; Sinauer Associates Inc.: Sunderland, MA, USA, 2010; pp. 755–778. [Google Scholar]
- Radha, B.; Sunitha, N.C.; Sah, R.P.; TP, M.A.; Krishna, G.K.; Umesh, D.K.; Siddique, K.H. Physiological and molecular implications of multiple abiotic stresses on yield and quality of rice. Front. Plant Sci. 2023, 13, 996514. [Google Scholar] [CrossRef]
- Ubierna, N.; Sun, W.; Cousins, A.B. The efficiency of C4 photosynthesis under low light conditions, assumptions and calculations with CO2 isotope discrimination. J. Exp. Bot. 2011, 62, 3119–3134. [Google Scholar] [CrossRef]
- Sheibani, F.; Bourget, M.; Morrow, R.C.; Mitchell, C.A. Close-canopy lighting, an effective energy-saving strategy for overhead sole-source LED lighting in indoor farming. Front. Plant Sci. 2023, 14, 1215919. [Google Scholar] [CrossRef]
- Lichtenthaler, H.; Wenzel, O.; Buschmann, C.; Gitelson, A. Plant Stress Detection by Reflectance and Fluorescence. Ann. N. Y. Acad. Sci. 1999, 851, 271–285. [Google Scholar] [CrossRef]
- Mathur, S.; Jain, L.; Jajoo, A. Photosynthetic efficiency in sun and shade plants. Photosynthetica 2018, 56, 354–365. [Google Scholar] [CrossRef]
- Hussain, S.; Iqbal, N.; Pang, T.; Khan, M.N.; Liu, W.-G.; Yang, W.-Y. Weak stem under shade reveals the lignin reduction behavior. J. Integr. Agric. 2019, 18, 496–505. [Google Scholar] [CrossRef]
- Murchie, E.H.; Hubbart, S.; Peng, S.; Horton, P. Acclimation of photosynthesis to high irradiance in rice, gene expression and interactions with leaf development. J. Exp. Bot. 2005, 56, 449–460. [Google Scholar] [CrossRef]
- Achkar, N.P.; Cho, S.K.; Poulsen, C.; Arce, A.L.; Re, D.A.; Giudicatti, A.J.; Manavella, P.A. A quick HYL1-dependent reactivation of microRNA production is required for a proper developmental response after extended periods of light deprivation. Dev. Cell 2018, 46, 236–247. [Google Scholar] [CrossRef]
- Sekhar, S.; Panda, D.; Kumar, J.; Mohanty, N.; Biswal, M.; Baig, M.J.; Behera, L. Comparative transcriptome profiling of low light tolerant and sensitive rice varieties induced by low light stress at active tillering stage. Sci. Rep. 2019, 9, 5753. [Google Scholar] [CrossRef] [PubMed]
- Panigrahy, M.; Panigrahi, K.C.S.; Poli, Y.; Ranga, A.; Majeed, N. Integrated expression analysis of small RNA, degradome and microarray reveals complex regulatory action of miRNA during prolonged shade in Swarnaprabha rice. Biology 2022, 11, 798. [Google Scholar] [CrossRef]
- Sekhar, S.; Das, S.; Panda, D.; Mohanty, S.; Mishra, B.; Kumar, A.; Mohapatra, T. Identification of microRNAs that provide a low light stress tolerance-mediated signaling pathway during vegetative growth in rice. Plants 2022, 11, 2558. [Google Scholar] [CrossRef]
- Wang, Z.W.; Zhang, T.Q.; Xing, Y.D.; Zeng, X.Q.; Ling, W.; Liu, Z.X.; He, G.H. YGL9, encoding the putative chloroplast signal recognition particle 43 kDa protein in rice, is involved in chloroplast development. J. Integr. Agric. 2016, 15, 944–953. [Google Scholar] [CrossRef]
- Dutta, S.S.; Tyagi, W.; Pale, G.; Pohlong, J.; Aochen, C.; Pandey, A.; Pattanayak, A.; Rai, M. Marker–Trait association for low-light intensity tolerance in rice genotypes from Eastern India. Mol. Genet. Genom. 2018, 293, 1493–1506. [Google Scholar] [CrossRef]
- Ganguly, S.; Nimitha, K.; Saha, S.; Sinha Mahapatra, N.; Bhattacharya, K.; Kundu, R.; Bhattacharyya, S. Identification and analysis of low light responsive yield enhancing QTLs in rice. Sci. Rep. 2024, 14, 21011. [Google Scholar] [CrossRef]
- Saha, S.; Mahapatra, N.S.; Bhattacharya, K.; Kundu, R.; Nimitha, K.; Ganguly, S.; Bhattacharyya, S. The Ratio of A400/A1800 Mapping Identifies Chromosomal Regions Containing Known Photoprotection Recovery-Related Genes in Rice. Rice 2024, 17, 62. [Google Scholar] [CrossRef] [PubMed]
- Khumaida, N.; Kisman, K.; Sopandie, D. Cloning and characterization of partial chlorophyll a oxygenase (CAO) gene involved in soybean shade tolerance mechanism. J. Trop. Crop Sci. 2015, 2, 1–4. [Google Scholar] [CrossRef]
- Zhao, J.; Shi, X.; Chen, L.; Chen, Q.; Tian, X.; Ai, L.; Zhao, H.; Yang, C.; Yan, L.; Zhang, M. Genetic and transcriptome analyses reveal the candidate genes and pathways involved in the inactive shade-avoidance response enabling high-density planting of soybean. Front. Plant Sci. 2022, 13, 973643. [Google Scholar] [CrossRef]
- Su, Y.; Yang, H.; Wu, Y.; Gong, W.; Gul, H.; Yan, Y.; Yang, W. Photosynthetic acclimation of shade-grown soybean seedlings to a high-light environment. Plants 2023, 12, 2324. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.; Gupta, P.; Gowtham, T.P.; Yogesh, K.S.; Sanjay, T.D.; Singh, A.; Duong, H.V.; Pradhan, S.K.; Bisht, D.S.; Singh, N.K.; et al. Generation of High-Value Genomic Resource in Rice, A “Subgenomic Library” of Low-Light Tolerant Rice Cultivar Swarnaprabha. Biology 2023, 12, 428. [Google Scholar] [CrossRef]
- Nayak, S.K.; Murty, K.S. Effect of varying light intensities on yield and growth parameters in rice. Ind. J. Plant Physiol. 1980, 23, 51–54. [Google Scholar]
- Murray, M.G.; Thompson, W. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping, Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1944, 12, 172–175. [Google Scholar] [CrossRef]
- Li, H.; Hearne, S.; Bänziger, M.; Li, Z.; Wang, J. Statistical properties of QTL linkage mapping in biparental genetic populations. Heredity 2010, 105, 257–267. [Google Scholar] [CrossRef]
- Li, J.; Cui, F.; Ding, A.M.; Zhao, C.H.; Wang, X.Q.; Wang, L.; Wang, H.G. QTL detection of seven quality traits in wheat using two related recombinant inbred line populations. Euphytica 2012, 183, 207–226. [Google Scholar] [CrossRef]
- Lander, E.; Kruglyak, L. Genetic dissection of complex traits, guidelines for interpreting and reporting linkage results. Nat. Genet. 1995, 11, 241–247. [Google Scholar] [CrossRef]
- McCouch, S.R.; Chen, X.; Panaud, O.; Temnykh, S.; Xu, Y.; Cho, Y.G.; Blair, M. Microsatellite marker development, mapping and applications in rice genetics and breeding. Plant Mol. Biol. 1997, 35, 89–99. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Namiki, N.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Ikawa, H.; Nagamura, Y. Rice FREND, a platform for retrieving coexpressed gene networks in rice. Nucleic Acids Res. 2013, 41, D1214–D1221. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Ideker, T. Cytoscape, a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG, new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Fisher, R.A. Statistical Methods for Research Workers, 4th ed.; Oliver and Boyd: Edinburgh, UK; London, UK, 1936; p. 352. [Google Scholar]
- Addinsoft. XLSTAT Statistical and Data Analysis Solution; Addinsoft: Long Island, NY, USA, 2020; Available online: https://www.xlstat.com (accessed on 1 January 2014).
- Terashima, I.; Hikosaka, K. Comparative ecophysiology of leaf and canopy photosynthesis. Plant Cell Environ. 1995, 18, 1111–1128. [Google Scholar] [CrossRef]
- Mohammed, R.; Scholz, M. Adaptation strategy to mitigate the impact of climate change on water resources in arid and semi-arid regions, a case study. Water Resour. Manag. 2017, 31, 3557–3573. [Google Scholar] [CrossRef]
- Casal, J.J. Photoreceptor signaling networks in plant responses to shade. Ann. Rev. Plant Biol. 2013, 64, 403–427. [Google Scholar] [CrossRef] [PubMed]
- Collard, B.C.; Mackill, D.J. Marker-assisted selection, an approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef]
- Burr, B.; Burr, F.A.; Thompson, K.H.; Albertson, M.C.; Stuber, C.W. Gene mapping with recombinant inbreds in maize. Genetics 1988, 118, 519–526. [Google Scholar] [CrossRef]
- Burr, B.; Burr, F.A. Recombinant inbreds for molecular mapping in maize, theoretical and practical considerations. Trends Genet. 1991, 7, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, L.; Rao, M.B.; Raju, C.S.; Reddy, S.N. Variability, correlation and path analysis in advanced generation of aromatic rice. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 1798–1806. [Google Scholar] [CrossRef]
- Abhilash, R.; Thirumurugan, T.; Sassikumar, D.; Chitra, S. Genetic studies in F2 for biometrical traits in Rice (Oryza sativa L.). Electron. J. Plant Breed. 2018, 9, 1067–1076. [Google Scholar] [CrossRef]
- Abigail, M.E.A. Biochar-based nanocarriers: Fabrication, characterization, and application as 2,4-dichlorophenoxyacetic acid nanoformulation for sustained release. 3 Biotech 2019, 9, 317. [Google Scholar]
- Priyanka, A.R.; Gnanamalar, R.P.; Banumathy, S.; Senthil, N.; Hemalatha, G. Genetic variability and frequency distribution studies in F2 segregating generation of rice. Electron. J. Plant Breed. 2019, 10, 988–994. [Google Scholar] [CrossRef]
- Shahid, M.; Saleem, M.F.; Saleem, A.; Raza, M.A.S.; Kashif, M.; Shakoor, A.; Sarwar, M. Exogenous potassium–Instigated biochemical regulations confer terminal heat tolerance in wheat. J. Soil Sci. Plant Nutr. 2019, 19, 137–147. [Google Scholar] [CrossRef]
- Renuprasath, P.; Ganesan, N.M.; Bama, K.S.; Boominathan, P.; Suresh, R. Variability and association analysis for yield and yield contributing traits in early segregating backcross population in Rice (Oryza sativa L.). J. Pharm. Innov. 2023, 12, 3218–3222. [Google Scholar]
- Rani, C.S.; Anandakumar, C.R.; Raveendran, M.; Subramanian, K.S.; Robin, S. Genetic variability studies and multivariate analysis in F2 segregating populations involving medicinal rice (Oryza sativa L.) cultivar Kavuni. Int. J. Agril. Sci. 2016, 8, 1733–1735. [Google Scholar]
- Vijaya, I.; Shailaja, H. Assessment of genetic parameters for yield and its related traits in F2 population of KRH-4 hybrid rice (Oryza sativa L.). Int. J. Agril. Sci. Res. 2016, 6, 87–96. [Google Scholar]
- Khan, I.; Muhammad, A.; Chattha, M.U.; Skalicky, M.; BilalChattha, M.; AhsinAyub, M.; El Sabagh, A. Mitigation of salinity-induced oxidative damage, growth, and yield reduction in fine rice by sugarcane press mud application. Front. Plant Sci. 2022, 13, 840900. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.R.; Mahmud, A.; Ghosh, U.K.; Hossain, M.S.; Siddiqui, M.N.; Islam, A.A.; Tran, L.S.P. Exploring the Phenotypic and Genetic Variabilities in Yield and Yield-Related Traits of the Diallel-Crossed F5 Population of Aus Rice. Plants 2023, 12, 3601. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z. Genomic variation in 3010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Babar, M.; Khan, A.A.; Arif, A.; Zafar, Y.; Arif, M. Path analysis of some leaf and panicle traits affecting grain yield in doubled haploid lines of rice (Oryza sativa L.). Pak. J. Agric. 2007, 45, 245–252. [Google Scholar]
- Krishnan, V.; Sivaranjani, V.; Tamilzharasi, M.; Anandhan, T. Characterization of morpho-phenological traits in the traditional landraces of rice. Electron. J. Plant Breed. 2023, 14, 234–245. [Google Scholar]
- Kumar, A.; Rangare, N.R.; Vidyakar, V. Study of genetic variability of Indian and exotic rice germplasm in Allahabad agroclimate. Bioscan 2013, 8, 1445–1451. [Google Scholar]
- Mohanty, S.; Mohanty, N.; Sulakshana, S.; Pradhan, S.K.; Dash, S.K.; Behera, L. Assessment of polymorphism at molecular level, association studies, multivariate analysis and genetic diversity among recombinant inbred lines of rice (Oryza sativa L.). Oryza 2017, 54, 174–185. [Google Scholar] [CrossRef]
- Biswas, A.; Adhikari, A.; Adhikari, S.; Paul, A.; Ghosh, P. Assessment of variability and interrelationship between yield and yield related traits towards divergence in rice (Oryza sativa L.) landraces. Nucleus 2024, 67, 467–482. [Google Scholar] [CrossRef]
- Agalya, J.S.; Hari, P.P.; Ramchander, S.; Kumar, P.; Devesena, N.; Naveenkumar, R.; John, K.N. Assessment of variability parameters and diversity of panicle architectural traits associated with yield in rice (Oryza sativa L.). Plant Sci. Today 2024, 11, 109–118. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, Y.; Guo, X. Low-light image enhancement via feature restoration. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Singapore, 22–27 May 2022; pp. 2440–2444. [Google Scholar]
- Mohanty, S.K.; Panda, R.S.; Mohapatra, S.L.; Nanda, A.; Behera, L.; Jena, M.; Mohapatra, T. Identification of novel quantitative trait loci associated with brown plant hopper resistance in the rice landrace Salkathi. Euphytica 2017, 213, 38. [Google Scholar] [CrossRef]
- Verma, R.K.; Chetia, S.K.; Rahman, A.; Dey, P.C.; Sen, P.; Modi, M.K. Study on genetic diversity and population structure of upland rice accessions using SSR markers associated with grain yield under drought. Crop Res. 2017, 52, 180–187. [Google Scholar] [CrossRef]
- Chen, X.; Cho, Y.; McCouch, S. Sequence divergence of rice microsatellites in Oryza and other plant species. Mol. Genet. Genom. 2002, 268, 331–343. [Google Scholar] [CrossRef]
- Sekhar, S.; Kumar, J.; Mohanty, S.; Mohanty, N.; Panda, R.S.; Das, S.; Behera, L. Identification of novel QTLs for grain fertility and associated traits to decipher poor grain filling of basal spikelets in dense panicle rice. Sci. Rep. 2021, 11, 13617. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Kumar, A.; Ellur, R.K.; Gopala Krishnan, S.; Vinod, K.K.; Dixit, D.; Singh, A.K. Novel quantitative trait loci for yield and yield related traits identified in Basmati rice (Oryza sativa). Plant Breed. 2023, 142, 327–337. [Google Scholar] [CrossRef]
- Li, C.; Lu, C.; Yang, M.; Wu, G.; Nyasulu, M.; He, H.; He, X.; Bian, J. Uncovering Novel QTLs and Candidate Genes for Salt Tolerance at the Bud Burst Stage in Rice through Genome-Wide Association Study. Plants 2024, 13, 174. [Google Scholar] [CrossRef] [PubMed]
- Donde, R.; Mohapatra, S.; Baksh, S.Y.; Padhy, B.; Mukherjee, M.; Roy, S.; Dash, S.K. Identification of QTLs for high grain yield and component traits in new plant types of rice. PLoS ONE 2020, 15, e0227785. [Google Scholar] [CrossRef]
- Marathi, B.; Guleria, S.; Mohapatra, T.; Parsad, R.; Mariappan, N.; Kurungara, V.K.; Singh, A.K. QTL analysis of novel genomic regions associated with yield and yield related traits in new plant type based recombinant inbred lines of rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 137. [Google Scholar] [CrossRef]
- Kulkarni, S.R.; Balachandran, S.M.; Ulaganathan, K.; Balakrishnan, D.; Praveen, M.; Prasad, A.H.; Sundaram, R.M. Molecular mapping of QTLs for yield related traits in recombinant inbred line (RIL) population derived from the popular rice hybrid KRH-2 and their validation through SNP genotyping. Sci. Rep. 2020, 10, 13695. [Google Scholar] [CrossRef]
- Yang, J.; Yuan, Z.; Meng, Q.; Huang, G.; Périn, C.; Bureau, C.; Zhang, D. Dynamic regulation of auxin response during rice development revealed by newly established hormone biosensor markers. Front. Plant Sci. 2017, 8, 242614. [Google Scholar] [CrossRef] [PubMed]
- Cakir, B.; Tian, L.; Crofts, N.; Chou, H.L.; Koper, K.; Ng, C.Y.; Okita, T.W. Re-programming of gene expression in the CS 8 rice line over-expressing ADP glucose pyrophosphorylase induces a suppressor of starch biosynthesis. Plant J. 2019, 97, 1073–1088. [Google Scholar] [CrossRef]
- Koumoto, T.; Shimada, H.; Kusano, H.; She, K.C.; Iwamoto, M.; Takano, M. Rice monoculm mutation moc2, which inhibits outgrowth of the second tillers, is ascribed to lack of a fructose-1, 6-bisphosphatase. Plant Biotechnol. 2013, 30, 47–56. [Google Scholar] [CrossRef]
- Lee, D.J.; Kim, S.; Ha, Y.M.; Kim, J. Phosphorylation of Arabidopsis response regulator 7 (ARR7) at the putative phospho-accepting site is required for ARR7 to act as a negative regulator of cytokininsignaling. Planta 2008, 227, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Mosa, K.A.; Ismail, A.; Helmy, M. Introduction to Plant Stresses. In Plant Stress Tolerance; Springer Briefs in Systems Biology; Springer: Cham, Switzerland, 2017; pp. 1–19. [Google Scholar]
- Zhang, C.; Liu, J.; Zhao, T.; Gomez, A.; Li, C.; Yu, C.; Lin, C.A. Drought-inducible transcription factor delays reproductive timing in rice. Plant Physiol. 2016, 171, 334–343. [Google Scholar] [CrossRef]
- Ruhl, C.; Stauffer, E.; Kahles, A.; Wagner, G.; Drechsel, G.; Rätsch, G.; Wachter, A. Polypyrimidine tract binding protein homologs from Arabidopsis are key regulators of alternative splicing with implications in fundamental developmental processes. Plant Cell 2012, 24, 4360–4375. [Google Scholar] [CrossRef]
- Evans, J.R. Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 1989, 78, 9–19. [Google Scholar] [CrossRef]
- Mae, T. Physiological nitrogen efficiency in rice, nitrogen utilization, photosynthesis, and yield potential. Plant Soil 1997, 196, 201–210. [Google Scholar] [CrossRef]
- Wang, J.; Wan, R.; Nie, H.; Xue, S.; Fang, Z. OsNPF5.16, a nitrate transporter gene with natural variation, is essential for rice growth and yield. Crop J. 2022, 10, 397–406. [Google Scholar] [CrossRef]
- Gao, D.; Sun, W.; Wang, D.; Dong, H.; Zhang, R.; Yu, S. A xylanglucuronosyl transferase gene exhibits pleiotropic effects on cellular composition and leaf development in rice. Sci. Rep. 2020, 10, 3726. [Google Scholar]
- Kovi, M.R.; Zhang, Y.; Yu, S.; Yang, G.; Yan, W.; Xing, Y. Candidacy of a chitin-inducible gibberellin-responsive gene for a major locus affecting plant height in rice that is closely linked to Green Revolution gene sd1. Theor. Appl. Genet. 2011, 123, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Zhang, S.; Zhang, W.; Li, G.; Chen, Z.; Zhai, W.; Zhu, L. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 2006, 48, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Sentoku, N.; Nishimura, A.; Hong, S.; Sato, Y.; Matsuoka, M. Position dependent expression of GL2-type homeobox gene, Roc1, significance for protoderm differentiation and radial pattern formation in early rice embryogenesis. J. Mol. Cell Biol. 2002, 29, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Zhang, S.; Zhang, W.; Li, G.; Chen, Z.; Zhai, W.; Zhu, L. OsSBDCP1 and its role in carbohydrate metabolism under low light conditions. Plant J. 2020, 97, 1073–1088. [Google Scholar]

| Chrom# | No. of SSR Markers Used | No. of SNP Markers Used | No. of Polymorphic SSR Markers | No. of Polymorphic SNP Markers Used | Total Number of Polymorphic Markers | Polymorphism % |
|---|---|---|---|---|---|---|
| 1 | 153 | 50 | 11 | 9 | 20 | 9.85 |
| 2 | 151 | 20 | 6 | 4 | 10 | 6.43 |
| 3 | 170 | 21 | 12 | 5 | 17 | 7.85 |
| 4 | 111 | 24 | 2 | 4 | 6 | 4.44 |
| 5 | 105 | 20 | 4 | 5 | 9 | 6.45 |
| 6 | 98 | 22 | 7 | 5 | 12 | 10.83 |
| 7 | 76 | 15 | 5 | 5 | 10 | 9.89 |
| 8 | 92 | 12 | 14 | 3 | 17 | 16.35 |
| 9 | 67 | 14 | 5 | 5 | 10 | 12.35 |
| 10 | 40 | 9 | 0 | 2 | 2 | 2.04 |
| 11 | 61 | 9 | 5 | 1 | 6 | 11.43 |
| 12 | 59 | 8 | 4 | 0 | 4 | 7.46 |
| Total | 1183 | 224 | 75 | 48 | 123 | 8.74 |
| Sl. No. | Trait Name | QTL Name | Chromosome | Position (cM) | Flanking Markers | Physical Position (Mb) | LOD | PVE (%) | Additive | Condition | Season (Kharif) | Parental Contribution |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | DFF | qDFF1.1N | 1 | 289–291 | RM12276-RM10402 | 83.81–84.39 | 3.16–3.23 | 6.67–11.37 | 1.41–1.45 | NL | 2019, 2021 | SP |
| 2 | qDFF2.1L | 2 | 19–35 | RM12601-TBGI090342 | 5.51–10.15 | 14.08–18.39 | 10.69–11.32 | (−1.3–2.18) | LL | 2019, 2021 | IR8 | |
| 3 | qDFF3.1L | 3 | 80–81 | TBGI137429-RM14292 | 23.2–23.49 | 9.24–18.56 | 9.96–10.24 | 0.72–10.21 | LL | 2019, 2021 | SP | |
| 4 | qDFF4.1L | 4 | 21–32 | RM17478-TBGI210835 | 27.6–30.8 | 10.32–12.66 | 6.63–10.21 | (−0.96–0.17) | LL | 2019, 2021 | IR8 | |
| 5 | qDFF8.1CLN | 8 | 170–214 | HYVSSR8-06-HYVSSR8-10 | 4.6–5.2 | 3.37–13.24 | 7.74–11.33 | (−0.75-2.16) | NL, LL | 2019, 2021 | IR8 | |
| 6 | PH | qPH1.1CLN | 1 | 27–32 | TBGI054892-RM11935 | 7.83–37.7 | 3.34–12.57 | 5–10.82 | 0.35–0.55 | NL, LL | 2019, 2021 | SP |
| 7 | qPH1.2CLN | 1 | 100–104 | RM11935-RM11940 | 37.7–37.8 | 4.06–5.85 | 4.76–9.58 | 8.50–10.51 | NL, LL | 2019, 2021 | SP | |
| 8 | qPH1.3CLN | 1 | 158–164 | RM11522-RM12182 | 45.82–47.56 | 5.01–13.18 | 4.16–11.22 | 2.60–3.94 | NL, LL | 2019, 2021 | SP | |
| 9 | qPH3.1N | 3 | 6–7 | RM15630-TBGI133686 | 1.74–2.03 | 7.88–8.43 | 8.86–11.25 | 0.87–1.13 | NL | 2019, 2021 | SP | |
| 10 | qPH3.2N | 3 | 76–78 | TBGI137429-RM14292 | 22.04–22.62 | 8.06–9.55 | 10.95–11.19 | 17.88–18.47 | NL | 2019, 2021 | SP | |
| 11 | qPH3.3N | 3 | 320 | RM14787-RM15981 | 92.8–92.9 | 14.13–14.64 | 6.95–9.21 | 2.14–3.05 | NL | 2019, 2021 | SP | |
| 12 | qPH8.1N | 8 | 50–68 | HYVSSR8-18-RM210 | 14.5–19.72 | 3.27–13.02 | 5.14–8.52 | 0.28–0.85 | NL | 2019, 2021 | SP | |
| 13 | qPH8.2N | 8 | 12 | RM556-TBGI341212 | 3.48–4.01 | 11.94–12.51 | 10.92–11.01 | (−0.33–0.84) | NL | 2019, 2021 | IR8 | |
| 14 | qPH9.1CLN | 9 | 83–102 | TBGI390847-HYVSSR9-43 | 24.07–29.58 | 3.93–14.24 | 10.91–11.94 | 0.67–6.37 | NL, LL | 2019, 2021 | SP | |
| 15 | TN | qTN1.1N | 1 | 79–85 | TBGI054892-RM11935 | 22.91–37.7 | 3.69–3.98 | 12.47–13.75 | (−0.39–1.39) | NL | 2019, 2021 | IR8 |
| 16 | GN | qGN1.1L | 1 | 100–107 | RM11935-RM11940 | 37.7–37.8 | 3.01–4.89 | 11.83–12.69 | 0.51–9.78 | LL | 2019, 2021 | SP |
| 17 | SN | qSN1.1L | 1 | 101–103 | RM11935-RM11940 | 37.7–37.8 | 3.68–4.01 | 11.20–15.21 | 2.23–8.73 | LL | 2019, 2021 | SP |
| 18 | qSN3.1CLN | 3 | 278–292 | RM14787-RM15981 | 80.62–84.68 | 3.05–3.95 | 11.28–17.70 | 0.08–10.34 | NL, LL | 2019, 2021 | SP | |
| 19 | SFP | qSFP1.1N | 1 | 80–85 | TBGI054892-RM11935 | 37.2–37 | 3.46–4.39 | 10.08–11.65 | (−0.03–1.20) | NL | 2019, 2021 | IR8 |
| 20 | qSFP1.2N | 1 | 157–159 | RM11522-RM12182 | 45.53–46.11 | 3.72–4.39 | 6.05–10.09 | 0.25–1.005 | NL | 2019, 2021 | SP | |
| 21 | qSFP1.3N | 1 | 53–54 | RM10207-TBGI031980 | 15.37–15.66 | 4.33–7.43 | 7.19–8.25 | (−0.4–0.16) | NL | 2019, 2021 | IR8 | |
| 22 | qSFP3.1N | 3 | 111–120 | TBGI137429-RM14292 | 32.19–34.8 | 2.73–2.80 | 5.21–9.29 | 1.29–2.84 | NL | 2019, 2021 | SP | |
| 23 | qSFP4.1L | 4 | 13–72 | RM17487-RM17478 | 31.1–30.8 | 3.18–5.28 | 7.14–8.34 | 0.26–1.27 | LL | 2019, 2021 | SP | |
| 24 | qSFP6.1N | 6 | 32–53 | RM20659-RM20372 | 9.28–15.37 | 3.13–3.23 | 5.92–7.32 | (−0.45–1.03) | NL | 2019, 2021 | IR8 | |
| 25 | qSFP6.2N | 6 | 205–207 | TBGI278662-TBGI273346 | 59.45–60.03 | 2.99–3.94 | 10.11–10.24 | 0.54–0.55 | NL | 2019, 2021 | SP | |
| 26 | PW | qPW1.1CLN | 1 | 85–108 | RM11935-RM11940 | 37.7–37.8 | 3.10–3.99 | 6.50–14.52 | 0.19–0.24 | NL, LL | 2019, 2021 | SP |
| 27 | qPW3.1L | 3 | 188–215 | RM545-RM14772 | 54.52–62.35 | 3.48–3.58 | 11.74–12.11 | 0.04–0.15 | LL | 2019, 2021 | SP | |
| 28 | TGW | qTGW1.1L | 1 | 102–103 | RM11935-RM11940 | 37.7–37.8 | 3.50–4.17 | 6.44–7.55 | 0.45–0.96 | LL | 2019, 2021 | SP |
| 29 | qTGW1.2L | 1 | 158–207 | RM11522-RM12182 | 45.82–60.03 | 3.40–6.12 | 10.17–10.21 | 0.09–0.13 | LL | 2019, 2021 | SP | |
| 30 | qTGW4.1L | 4 | 12–49 | RM17478-TBGI210835 | 27.6–30.8 | 2.76–4.89 | 10.59–11.22 | (−0.30–0.06) | LL | 2019, 2021 | IR8 | |
| 31 | qTGW12.1L | 12 | 8–10 | RM27824-RM28524 | 2.32–2.9 | 3.06–5.65 | 7.91–8.56 | 0.56–0.58 | LL | 2019, 2021 | SP | |
| 32 | GY | qGY7.1CLN | 7 | 81–125 | TBGI322578-RM21808 | 23.49–36.25 | 2.89–3.35 | 15.08–17.06 | 1.16–1.84 | NL, LL | 2019, 2021 | SP |
| 33 | qGY8.1CLN | 8 | 161–201 | HYVSSR8-06-HYVSSR8-10 | 4.6–5.2 | 2.52–4.18 | 11.27–18.78 | 0.15–0.24 | NL, LL | 2019, 2021 | SP |
| Scheme | QTL Cluster No. | Chrom# | Marker Interval | Position (Mb) for Flanking Markers | Peak Interval (cM) | Window Size (Mb) | No. of QTLs | Name of the QTLs | Traits | No. of Genes | No. of Candidate Genes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | I | 1 | RM11935-RM11940 | 37.7–37.8 | 85–108 | 0.16 | 5 | qPH1.2CLN, qGN1.1L, qSN1.1L, qPW1.1CLN, qTGW1.1L | PH, GN, SN, PW, TGW | 239 | 17 |
| 2 | II | 4 | RM17478-TBGI210835 | 27.6–30.8 | 12–49 | 3.2 | 3 | qDFF4.1L, qSFP4.1L, qTGW4.1L | DFF, SFP, TGW | 516 | 1 |
| 3 | III | 8 | HYVSSR8-06-HYVSSR8-10 | 4.6–5.2 | 161–204 | 0.6 | 2 | qDFF8.1CLN, qGY8.1CLN | DFF, GY | 171 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohanty, S.; Das, S.; Panda, D.; Choudhury, N.K.; Mishra, B.; Jena, R.K.; Sah, R.P.; Chandrappa, A.K.; Basavantraya Navadagi, D.; Raj K.R., R.; et al. Identification of Novel Quantitative Trait Loci and Candidate Genes Associated with Grain Yield and Related Traits Under Low-Light Stress Conditions in Rice. Biomolecules 2025, 15, 1388. https://doi.org/10.3390/biom15101388
Mohanty S, Das S, Panda D, Choudhury NK, Mishra B, Jena RK, Sah RP, Chandrappa AK, Basavantraya Navadagi D, Raj K.R. R, et al. Identification of Novel Quantitative Trait Loci and Candidate Genes Associated with Grain Yield and Related Traits Under Low-Light Stress Conditions in Rice. Biomolecules. 2025; 15(10):1388. https://doi.org/10.3390/biom15101388
Chicago/Turabian StyleMohanty, Soumya, Swagatika Das, Darshan Panda, Nalini Kanta Choudhury, Baneeta Mishra, Ranjan Kumar Jena, Rameswar Prasad Sah, Anil Kumar Chandrappa, Devanna Basavantraya Navadagi, Reshmi Raj K.R., and et al. 2025. "Identification of Novel Quantitative Trait Loci and Candidate Genes Associated with Grain Yield and Related Traits Under Low-Light Stress Conditions in Rice" Biomolecules 15, no. 10: 1388. https://doi.org/10.3390/biom15101388
APA StyleMohanty, S., Das, S., Panda, D., Choudhury, N. K., Mishra, B., Jena, R. K., Sah, R. P., Chandrappa, A. K., Basavantraya Navadagi, D., Raj K.R., R., Kumar, A., Pradhan, S. K., Samantaray, S., Baig, M. J., & Behera, L. (2025). Identification of Novel Quantitative Trait Loci and Candidate Genes Associated with Grain Yield and Related Traits Under Low-Light Stress Conditions in Rice. Biomolecules, 15(10), 1388. https://doi.org/10.3390/biom15101388










