Abstract
Cancer’s heterogeneity presents significant challenges in accurate diagnosis and effective treatment, including the complexity of identifying tumor subtypes and their diverse biological behaviors. This review examines how feature selection techniques address these challenges by improving the interpretability and performance of machine learning (ML) models in high-dimensional datasets. Feature selection methods—such as filter, wrapper, and embedded techniques—play a critical role in enhancing the precision of cancer diagnostics by identifying relevant biomarkers. The integration of multi-omics data and ML algorithms facilitates a more comprehensive understanding of tumor heterogeneity, advancing both diagnostics and personalized therapies. However, challenges such as ensuring data quality, mitigating overfitting, and addressing scalability remain critical limitations of these methods. Artificial intelligence (AI)-powered feature selection offers promising solutions to these issues by automating and refining the feature extraction process. This review highlights the transformative potential of these approaches while emphasizing future directions, including the incorporation of deep learning (DL) models and integrative multi-omics strategies for more robust and reproducible findings.
1. Introduction
Cancer remains one of the leading causes of morbidity and mortality worldwide, characterized by its remarkable complexity and heterogeneity [1,2,3]. Tumor heterogeneity manifests at various levels, including genetic, epigenetic, transcriptomic, and proteomic variations, which contribute to differing tumor behaviors, responses to treatment, and patient prognoses [4,5]. Understanding and accurately classifying tumor subtypes is critical for the advancement of personalized medicine, enabling tailored therapeutic strategies that can significantly improve patient outcomes [6,7].
The classification of tumors into distinct subtypes has evolved from a purely histopathological approach to a more integrated methodology that incorporates molecular and genomic information. The advent of high-throughput technologies, such as next-generation sequencing (NGS) and various omics approaches, has generated vast amounts of biological data [8,9], offering unprecedented insights into the molecular underpinnings of cancer. However, this wealth of data also presents a challenge: how to distill complex datasets into meaningful insights that facilitate effective tumor subtype classification [10]. Feature selection techniques emerge as essential tools in this context, serving to identify and prioritize the most informative variables from high-dimensional datasets [11,12]. Feature selection refers to the process of identifying the most informative variables from high-dimensional datasets to reduce dimensionality while retaining essential information. By reducing the dimensionality of data, feature selection not only enhances the interpretability of models but also improves the accuracy of classification algorithms [13,14,15]. This is particularly important in the field of oncology, where the identification of relevant biomarkers can lead to improved diagnostics, prognostics, and treatment strategies [16,17]. With the rise of artificial intelligence (AI), particularly machine learning (ML) and deep learning (DL) algorithms, the integration of AI-driven feature selection techniques has further enhanced the ability to manage and analyze large-scale datasets, thereby refining cancer subtype classification models [18,19]. ML is a branch of AI that involves training algorithms to recognize patterns and make predictions based on data. DL, a subset of ML, uses multi-layered neural networks to automatically learn hierarchical feature representations from raw data. These AI approaches have proven crucial in automating the extraction of relevant features, significantly improving diagnostic accuracy and personalized treatment planning.
In essence, feature selection acts as a bridge between raw data and actionable clinical insights, highlighting its indispensable role in cancer research. Feature selection techniques can be broadly categorized into three main types: filter methods, wrapper methods, and embedded methods [20,21,22]. Filter methods assess the relevance of features based on intrinsic properties of the data, often utilizing statistical tests to evaluate the relationship between features and the target variable [23]. Wrapper methods, on the other hand, involve evaluating the performance of a predictive model using different subsets of features, allowing for a more tailored selection process [24]. Embedded methods integrate feature selection into the model training process itself, identifying the most relevant features as part of model optimization [25]. Each of these approaches offers unique advantages and limitations, and the choice of technique can significantly influence classification outcomes [13,26].
The application of feature selection in tumor subtype classification has been demonstrated in various studies, revealing its potential to enhance the accuracy of predictions and inform clinical decision-making [27,28]. For instance, research has shown that specific gene expression profiles can serve as reliable indicators for distinguishing between subtypes of breast cancer, thereby guiding targeted therapy selection [29,30]. Similar advancements have been observed in other malignancies, such as lung cancer and colorectal cancer, where feature selection has played a pivotal role in improving diagnostic precision and therapeutic strategies [27,31,32,33]. However, the integration of feature selection techniques is not without its challenges. Data quality remains a pressing concern in cancer research, as inconsistencies, missing values, and variations in data generation methods can impact the reliability of feature selection outcomes [20,22,34]. Moreover, interindividual variability introduces additional complexity, as genetic and environmental factors can lead to diverse responses among patients with the same tumor subtype [13,35]. These challenges underscore the importance of adopting robust and standardized methodologies for feature selection to enhance the reproducibility and generalizability of findings [36]. Leveraging AI for data cleaning and preprocessing can address some of these challenges, allowing for more consistent feature extraction across diverse datasets. Furthermore, the risk of overfitting poses a significant concern when utilizing high-dimensional datasets [37,38]. Effective feature selection can reduce this risk by ensuring that only the most relevant features are retained for analysis, thereby improving the robustness of the model, which is critical for the effectiveness of subtype classification [4,39].
Previous reviews have addressed feature selection in various contexts. For example, one review highlights its role in dimensionality reduction and disease understanding through real-world medical case studies [40]. Another explores how feature selection mitigates the curse of dimensionality in disease risk prediction using genetic data [41]. A third survey examines feature selection alongside dimensionality reduction and classification techniques for chronic disease diagnosis, emphasizing their impact on predictive accuracy and computational efficiency [42]. Building upon these studies, our review focuses specifically on the role of feature selection techniques in cancer subtype classification. In this review, we provide a comprehensive exploration of feature selection techniques, their role in unraveling tumor complexity, and their contributions to improving subtype classification. We begin by highlighting the significance of accurately classifying tumors to advance personalized medicine, emphasizing the challenges posed by the heterogeneity of cancer. We then provide an overview of various feature selection methods, detailing their unique characteristics and applications in cancer research. The discussion on practical applications illustrates how these techniques enhance predictive accuracy and inform clinical decision-making across different tumor types. Additionally, we emphasize the transformative impact of AI integration in refining feature selection, demonstrating its potential in streamlining cancer diagnostics and advancing therapeutic strategies. Furthermore, we address the challenges and limitations inherent in feature selection, including data quality and the risks of overfitting. Finally, we outline future directions, emphasizing the need for integration of multi-omics data and advancements in computational methods.
2. Overview of Feature Selection Techniques
2.1. Fundamentals and Workflow of Feature Selection Techniques in High-Dimensional Data Analysis
Feature selection is a critical step in the data analysis process, especially in high-dimensional settings such as genomics, transcriptomics, and proteomics [38,39]. The goal of feature selection is to identify the most relevant variables from a dataset, thereby reducing dimensionality while preserving essential information. This not only enhances the performance of classification algorithms but also aids in interpreting the underlying biological significance of selected features [36,40,41]. For instance, one study presents a machine learning framework that integrates somatic mutation and gene expression data to classify breast cancer subtypes. The authors employ feature selection techniques to identify key genes associated with specific subtypes. These selected features improved model accuracy by reducing noise and overfitting while also providing biologically meaningful insights into the molecular mechanisms of breast cancer [43]. Similar approaches have been applied in other cancer types, emphasizing the versatility and effectiveness of feature selection in handling high-dimensional datasets and improving clinical decision-making. By focusing on the most informative features, these techniques enhance both the reliability and interpretability of predictive models, ultimately supporting advancements in precision medicine. Recent advancements in AI, particularly ML and DL, have further optimized feature selection techniques by automating the identification of relevant features, thereby improving the accuracy and interpretability of cancer subtype classification models. In general, the basic workflow of feature selection methods consists of four fundamental processes: generating feature subsets, evaluating feature subsets, determining stopping criteria, and validating results, as shown in Figure 1. In the subset generation phase, the search strategy is crucial; an effective search strategy should achieve optimal solutions, have local search capabilities, and ensure computational efficiency. The evaluation function guides the feature selection process by deciding whether a feature should be added to or removed from the feature subset. When the stopping criteria are met, the feature selection process concludes, and the selected feature subset is applied to the validation dataset for verification. AI-enhanced methods, such as reinforcement learning, have introduced new possibilities in optimizing the search strategy by learning from iterative feedback, enabling more precise feature subset generation and refinement.
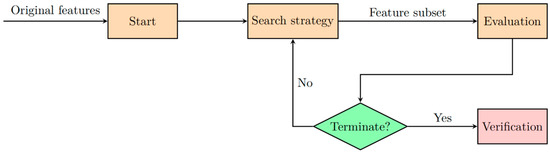
Figure 1.
The workflow diagram of feature selection techniques.
2.2. Categorization of Feature Selection Techniques and Their Operational Mechanisms
In this review, we categorize feature selection into four primary approaches: filter methods [23,27], wrapper methods [37,44], embedded methods [25,45], and swarm intelligence methods [46,47], and each of these approaches has its unique advantages, limitations, and applications in cancer research, as shown in Figure 2. Here, we introduce swarm intelligence methods to reflect their increasing importance in modern applications [48]. Swarm intelligence algorithms are often viewed as an organic combination of the three conventional feature selection methods—filter, wrapper, and embedded techniques. These algorithms leverage the strengths of each approach by combining the statistical evaluation of features (from filter methods), the model performance optimization (from wrapper methods), and the integration of feature selection within the model training process (from embedded methods). The integration of AI has allowed for more dynamic, hybrid approaches that combine the strengths of different feature selection techniques. For example, the combination of wrapper methods with DL models can lead to more robust feature selection by identifying nonlinear patterns in complex biological data.
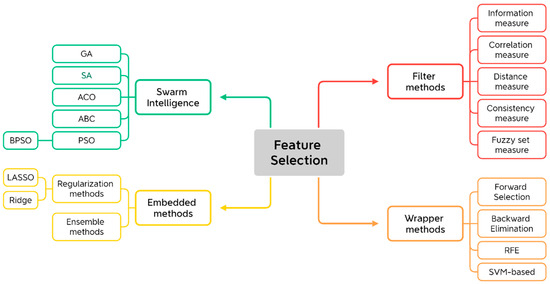
Figure 2.
Overall categories of feature selection techniques. RFE: recursive feature elimination; SVM: support vector machines; LASSO: least absolute shrinkage and selection operator; GA: genetic algorithms; SA: simulated annealing; ACO: ant colony optimization; ABC: artificial bee colony; PSO: particle swarm optimization; BPSO: binary particle swarm optimization.
2.2.1. Filter Methods
Filter methods operate independently of any ML algorithm and rely on statistical measures to evaluate the relevance of features. These methods assess the relationship between each feature and the target variable (e.g., tumor subtype) before any modeling occurs. Common filter techniques include the following:
(1) Correlation Coefficients
Techniques like Pearson [49] or Spearman correlation [50] coefficients are used to quantify the strength and direction of the relationship between each feature and the target. Features with high correlation to the target are typically retained, while those with low correlation are discarded.
(2) Mutual Information
This measure quantifies the amount of information that one variable contains about another. It is particularly useful for assessing nonlinear relationships and can help identify features that are highly informative for classification [51]. Table 1 summarizes various filter-based feature selection algorithms that utilize mutual information. It highlights whether each algorithm considers factors such as relevance, redundancy, complementarity, and interactions among features. Relevance refers to the statistical relationship between features, where highly correlated features may introduce redundancy. Redundancy occurs when multiple features provide overlapping information, which can negatively impact model efficiency. Complementarity describes how different features can collectively contribute unique information to improve predictive performance. Interactions among features capture complex dependencies, such as nonlinear relationships, that are crucial for understanding the underlying patterns in the data. By addressing these factors, feature selection algorithms aim to identify the most informative subset of features while minimizing noise and redundancy. The table also outlines the objective functions of these algorithms, providing researchers with a comparative framework to select appropriate feature selection methods.

Table 1.
Overview of filter-based feature selection algorithms based on mutual information.
Filter methods are computationally efficient and straightforward to implement. However, they have limitations, as they do not consider the interactions between features [61]. As a result, a feature that might be less significant in isolation could be critical in conjunction with other features, potentially leading to suboptimal selections.
2.2.2. Wrapper Methods
Wrapper methods evaluate feature subsets based on the performance of a specific ML algorithm. These techniques involve a more iterative process, where various combinations of features are tested, and the performance of the model is assessed for each subset. Common wrapper methods include the following:
(1) Forward Selection
This approach starts with no features and adds them one by one based on their contribution to model performance. The process continues until adding more features no longer improves the model significantly [46,62].
(2) Backward Elimination
In contrast to forward selection, backward elimination begins with all available features and systematically removes the least significant ones until the model’s performance begins to degrade [63,64].
(3) RFE
Recursive feature elimination (RFE) employs a recursive approach to identify and remove the least important features based on model performance. It continues this process iteratively until the desired number of features is achieved [65,66].
(4) SVM-based
Support vector machines (SVMs) are often favored as an evaluation strategy due to their ability to deliver robust and high-accuracy results, especially in high-dimensional feature spaces. SVMs excel in finding the optimal decision boundary by maximizing the margin between classes, making them particularly effective for feature selection tasks where the goal is to identify the most relevant features for classification. Other popular algorithms such as decision trees (DTs), naïve Bayes (NB), K-nearest neighbors (KNN), artificial neural networks (ANNs), and linear discriminant analysis (LDA) are also commonly used in wrapper approaches. Table 2 displays the feature subset search strategies and evaluation criteria used by several wrapper-based algorithms. These methods utilize swarm intelligence algorithms as search strategies, such as particle swarm optimization (PSO) and genetic algorithms (GAs), which are particularly effective in exploring large, complex search spaces and are widely used in wrapper-based feature selection.

Table 2.
Search strategies and evaluation criteria of some wrapper-based methods.
2.2.3. Embedded Methods
Embedded methods integrate feature selection into the model training process, combining the advantages of both filter and wrapper methods. In these approaches, feature selection is performed as part of the model optimization. Embedded methods strike a balance between computational efficiency and classification performance, as they consider feature interactions while training the model. However, the choice of the underlying model can influence feature selection outcomes, necessitating careful consideration when applying these techniques.
(1) Regularization Techniques
Methods such as least absolute shrinkage and selection operator (LASSO, L1 regularization) [82,83] and ridge regression (L2 regularization) [84,85] penalize the complexity of the model by adding a penalty term. LASSO can shrink some feature coefficients to zero, effectively performing feature selection while fitting the model. Table 3 provides an overview of several classic LASSO-based feature selection algorithms, highlighting their key advantages and limitations, and offering insights into their applicability for different types of data and classification tasks.
(2) Decision Trees and Ensemble Methods
Algorithms like random forests (RFs) and gradient boosting inherently perform feature selection by evaluating the importance of features during the model-building process [25,86,87,88,89]. RF feature selection is an embedded method widely used in high-dimensional data analysis. Ranking features based on their contribution to model accuracy helps in identifying the most significant predictors. This method is particularly effective for data with many features [90,91]. Features that contribute more significantly to the model’s predictive power receive higher importance scores.

Table 3.
Summary of classic LASSO-based feature selection algorithms with key advantages and limitations.
Table 3.
Summary of classic LASSO-based feature selection algorithms with key advantages and limitations.
| Name | Advantages | Limitations | Refs. |
|---|---|---|---|
| L1-regularized LASSO |
|
| [92,93] |
| Elastic Net |
|
| [94,95] |
| Sparse LASSO |
|
| [96,97] |
| Group LASSO |
|
| [98,99] |
| Fused LASSO |
|
| [100,101] |
| Adaptive LASSO |
|
| [102,103] |
2.2.4. Swarm Intelligence
Swarm intelligence refers to the collective behavior of decentralized, self-organized systems, often observed in nature, such as the flocking behavior of birds, the schooling of fish, or the foraging behavior of ants [46,104]. These algorithms are widely used in feature selection due to their ability to effectively explore large search spaces and find optimal or near-optimal solutions [105]. The most used swarm intelligence algorithms in feature selection are PSO, GA, ant colony optimization (ACO), artificial bee colony (ABC), and simulated annealing (SA) algorithms. These swarm intelligence-based methods are particularly advantageous in wrapper-based feature selection approaches, where performance is evaluated by training a model on different feature subsets. Swarm intelligence algorithms help search the vast space of feature combinations more efficiently than exhaustive methods, making them valuable tools for high-dimensional data.
(1) PSO
PSO is inspired by the social behavior of birds flocking or fish schooling. Each particle (representing a potential solution) moves through the search space, adjusting its position based on both its own experience and the experiences of neighboring particles [73]. The algorithm is especially useful for optimizing complex, high-dimensional problems like feature selection, where traditional search methods might struggle.
(2) GA
GA is an evolutionary algorithm inspired by the process of natural selection and genetics [80]. In the context of feature selection, GA treats the feature subsets as “individuals” in a population, with everyone being a set of selected features. These individuals undergo processes like selection, crossover, and mutation to generate new feature subsets. The fitness of each subset is evaluated based on model performance (e.g., accuracy or error rate). Through generations, GA evolves better feature subsets that improve model performance.
(3) ACO
ACO is based on the foraging behavior of ants, where ants deposit pheromones that influence the decisions of other ants [106]. This technique has been applied to feature selection by simulating an ant colony that explores different feature subsets, guided by pheromone intensity. Over time, the colony converges on an optimal or near-optimal feature subset.
(4) ABC
ABC is a swarm intelligence optimization technique inspired by the foraging behavior of honeybees [106,107]. In this algorithm, artificial bees are divided into employed, onlooker, and scout bees, each performing different tasks to explore and exploit the search space. Employed bees search for food sources (solutions), onlooker bees choose food sources based on probability distribution, and scout bees randomly search for new solutions. The algorithm is particularly useful in solving optimization problems, such as feature selection, due to its ability to efficiently explore large solution spaces and converge towards optimal or near-optimal solutions.
(5) SA
SA is a probabilistic optimization algorithm inspired by the annealing process in metallurgy, where materials are heated and then slowly cooled to remove defects and reach a stable configuration [108,109]. SA mimics this process by starting with a random solution and iteratively exploring neighboring solutions. At each step, a new solution is accepted based on a temperature-dependent probability, allowing for occasional acceptance of worse solutions to escape local minima. Over time, the temperature decreases, and the algorithm converges to an optimal or near-optimal solution. SA is widely used for problems with large search spaces and complex objective functions, such as feature selection in ML.
2.3. Comparative Evaluation of Feature Selection Techniques: Advantages and Limitations
Selecting the most effective feature selection method requires a careful balance between efficiency, accuracy, and computational resources. Filter methods are valued for their swiftness and ability to handle high-dimensional data without relying on specific ML models. However, their primary shortcoming lies in ignoring the complex interplay between features, which could lead to suboptimal predictive accuracy. In contrast, wrapper methods, despite their high computational cost due to the need for repeated model training, provide a customized approach that accounts for feature interactions, yet they are prone to overfitting and are less effective with large datasets. Embedded methods strike a balance by integrating feature selection into the model training process, which boosts efficiency and performance, but their reliance on certain models and the complexity of their implementation can be restrictive. Swarm intelligence methods excel in their flexibility and ability to conduct global searches in high-dimensional spaces, but they are sensitive to parameter tuning and have higher computational expenses. Therefore, when aiming to optimize feature selection for cancer subtype classification, it is imperative to weigh the advantages and limitations of filter, wrapper, embedded, and swarm intelligence methods against the analytical objectives and the nature of the dataset in question. Table 4 contrasts these methods, highlighting their key benefits and constraints, to guide researchers in choosing the most suitable approach for their specific needs.

Table 4.
Key advantages and limitations of four types of feature selection methods.
It is also worth noting that certain feature selection methods are particularly well-suited to specific machine learning algorithms. For example, LASSO is highly effective in regression problems due to its ability to shrink coefficients and perform feature selection through regularization, making it ideal for linear models. Similarly, RF excels in handling high-dimensional datasets by inherently ranking feature importance during model training. Methods like RFE work well with SVM because they iteratively refine feature subsets, optimizing for model performance. Furthermore, mutual information-based methods are particularly effective in capturing nonlinear relationships, which are beneficial for models like decision trees or kernel-based algorithms. These alignments highlight the importance of selecting feature selection methods that align with the characteristics of the machine learning algorithm used.
3. Applications of Feature Selection in Tumor Subtype Classification
3.1. Application Cases of Feature Selection Techniques in Tumor Subtype Classification
The application of feature selection techniques in tumor subtype classification is crucial for enhancing diagnostic accuracy, personalizing treatment strategies, and improving patient outcomes. Specifically, feature selection helps refine diagnostic models by identifying the most relevant biomarkers associated with specific tumor subtypes, reducing noise and enhancing model performance. This not only aids in accurate subtype classification but also enables the development of personalized prognostic models. These models support clinicians in tailoring treatment strategies to individual patients, ultimately leading to improved therapeutic efficacy and patient outcomes. By focusing on the most relevant features within high-dimensional datasets, researchers can better understand the molecular underpinnings of different tumor types and their associated behaviors [4,16]. The effectiveness of feature selection techniques has been underscored by various studies across different cancer types, as detailed in Table 5. For example, this approach has been successfully applied in the classification of breast cancer, a heterogeneous disease with distinct molecular subtypes such as Luminal A (LUMA), Luminal B (LUMB), HER2-Enriched, and basal-like (similar to the triple-negative breast cancer, TNBC), each with different biological characteristics and clinical implications. In addition, feature selection has been instrumental in lung cancer classification, the leading cause of cancer-related deaths globally, which is primarily divided into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC, accounting for approximately 85% of all lung cancers, includes subtypes like lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). On the other hand, SCLC is known for its rapid growth and aggressive behavior, necessitating distinct therapeutic approaches. Moreover, renal cell carcinoma (RCC), the most common type of kidney cancer in adults, exhibits heterogeneity and can be subtyped into kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), and kidney chromophobe cell carcinoma (KICH) based on pathological and molecular features, each with differing genetic mutations, prognoses, and treatment strategies. As summarized in Table 5, the studies provide information on the cancer subtypes, the feature selection methods employed, the data types used, and the key biomarkers identified. These findings underscore the impact of feature selection on clinical outcomes, demonstrating its utility in improving the accuracy of tumor subtype classification and, by extension, the personalization of treatment and enhancement of patient care.

Table 5.
Summary of studies utilizing feature selection techniques for cancer subtype classification.
3.2. Impact of Tumor Subtype Classification on Clinical Outcomes
The impact of cancer subtype classification on clinical outcomes is profound, as it directly influences diagnostic precision, treatment decisions, and patient prognosis. By accurately identifying tumor subtypes, clinicians can tailor therapies to specific molecular profiles, improving treatment efficacy. For instance, certain subtypes of breast cancer, such as HER2-enriched tumors, are known to respond positively to targeted therapies like trastuzumab [135,136]. Conversely, subtypes like basal-like and TNBC require alternative treatment strategies due to their distinct molecular characteristics. This personalized treatment strategy has the potential to reduce adverse effects and enhance overall survival rates. Recent advancements in AI algorithms have further enhanced the precision of subtype classifications by integrating multi-dimensional datasets. These algorithms enable more accurate subtype identification, facilitating the selection of therapies that align with the molecular features of the tumor [137,138].
Moreover, robust subtype classification plays a pivotal role in predicting disease progression and guiding follow-up care. By distinguishing between aggressive and indolent subtypes, clinicians can make informed decisions about surveillance and treatment intensity. The incorporation of AI-driven models, particularly those utilizing ML and DL techniques, has proven effective in analyzing complex patterns within high-dimensional biomedical data, facilitating early detection and risk stratification [139,140]. In cases where early detection and subtype identification are pivotal—such as lung cancer—subtype classification can help identify high-risk patients who may benefit from more aggressive surveillance or early intervention, ultimately improving survival rates [141,142]. In oral diseases, subtype classification supported by ML techniques has provided novel insights into diagnosis and prognosis. These approaches have optimized treatment planning and reduced the potential for human error, ensuring that patients receive data-driven, personalized care [143]. Furthermore, AI applications have demonstrated promise in automating the identification of subtle molecular markers, supporting clinicians in making precise, evidence-based decisions that enhance patient outcomes.
4. Challenges and Limitations
Cancer subtype classification using feature selection techniques faces significant challenges, primarily stemming from the inherent complexity and heterogeneity of cancer. For instance, the high dimensionality and sparsity of omics data, where datasets often contain thousands of features (e.g., genes, proteins) but relatively few samples, increase the risk of overfitting. This imbalance can lead to models capturing noise rather than meaningful biological signals, reducing their generalizability [144]. Additionally, integrating multi-omics data, such as genomic, transcriptomic, and clinical information, poses difficulties due to variations in data quality, measurement platforms, and scales. Inappropriate data fusion may obscure meaningful relationships, compromising the accuracy and interpretability of classification models [4].
On the other hand, existing feature selection methodologies exhibit notable limitations that hinder their application. For example, while filter, wrapper, and embedded methods are widely used, each has its drawbacks—filter methods may overlook interaction effects between features, wrapper methods are computationally expensive and prone to overfitting in high-dimensional data, and traditional methods often struggle with feature correlation, resulting in unstable feature selection. This instability reduces confidence in the selected features, as multiple equally optimal signatures are frequently produced [20,145,146]. Moreover, the lack of standardized best practices reduces consistency and reproducibility across studies. Finally, a limitation of this review is its primary focus on structured/tabular data, which does not fully encompass the application of feature selection techniques to multimodal datasets. These datasets integrate structured (e.g., tabular), semi-structured (e.g., time series), and unstructured data (e.g., images, clinical notes) [147,148,149]. Future research should explore feature selection methods tailored to multimodal data, leveraging the complementary nature of diverse data types to uncover novel insights.
5. Future Directions
Future directions in cancer subtype classification using feature selection techniques can focus on several key areas. First, integrating multi-omics data holds significant promise. While most studies rely on a single data type (e.g., gene expression), combining genomic, transcriptomic, proteomic, and even clinical data can provide a more comprehensive understanding of tumor heterogeneity. This integrative approach could lead to more accurate and robust classification models, better capturing the complexity of cancer subtypes and their underlying biology. Second, addressing the challenge of imbalanced datasets is crucial for enhancing classification performance. Many cancer datasets suffer from small sample sizes and class imbalance, which can lead to overfitting and unreliable results. Advanced feature selection methods, such as cost-sensitive algorithms or hybrid models, could be developed to better handle these issues, ensuring that subtype classification remains accurate and applicable in clinical settings. Third, combining feature selection methods offers a promising avenue for future research. Hybrid strategies that integrate filter, wrapper, and embedded methods can capitalize on the strengths of each approach. For example, filter methods can efficiently reduce the dimensionality of large datasets, while wrapper methods can refine the selection process by evaluating subsets of features in relation to specific ML models. Such combinations may help improve the robustness, accuracy, and interpretability of predictive models. Moreover, there is a critical need to explore feature selection techniques capable of handling multimodal datasets. Integrating diverse data types could significantly enhance the interpretability and clinical utility of cancer subtype classification models. Finally, the incorporation of DL and AI-based models represents a frontier for improving cancer subtype classification. While traditional ML methods have proven effective, DL models, particularly those using convolutional neural networks (CNNs) or recurrent neural networks (RNNs), have the potential to automatically learn complex features from raw data. These models could offer improved performance in identifying subtle patterns and biomarkers, enabling more personalized cancer treatment strategies. By focusing on these areas, future research can further enhance the precision and applicability of cancer subtype classification, ultimately advancing personalized medicine.
6. Conclusions
In this review, we explored various feature selection methodologies, including filter, wrapper, and embedded techniques, each with its strengths and limitations. Case studies across diverse cancer types illustrate the practical application of these techniques, showcasing their potential to uncover critical biomarkers that can inform clinical decision-making. The integration of AI into biomedicine has been a cornerstone, underscoring its transformative role in enhancing feature selection and tumor subtype classification across various biomedical applications.
However, the journey toward optimized feature selection is fraught with challenges, including issues related to data quality, interindividual variability, and the risk of overfitting. Addressing these challenges is crucial for ensuring that feature selection outcomes are reliable, reproducible, and clinically relevant, which is of paramount importance in the field of AI in biomedicine. Looking ahead, the integration of multi-omics data and advancements in ML offer promising avenues for enhancing feature selection and tumor subtype classification. These innovations can lead to more comprehensive biomarker discovery and improved model performance, which are key areas of focus in AI applications in bioinformatics and molecular biology. As we continue to unravel the complexities of tumor biology, the role of feature selection will remain pivotal in advancing personalized medicine, improving patient outcomes, and enhancing the overall landscape of cancer care.
Author Contributions
Conceptualization, J.W. and Y.W.; software, Y.W. and Z.Z.; validation, Z.Z.; writing—original draft preparation, J.W. and Y.W.; writing—review and editing, J.W. and Y.W.; visualization, Y.W.; funding acquisition, J.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Open Funds for Shaanxi Provincial Key Laboratory of Infection and Immune Diseases (No. 2023-KFMS-1).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Dagogo-Jack, I.; Shaw, A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
- Zhu, L.; Jiang, M.; Wang, H.; Sun, H.; Zhu, J.; Zhao, W.; Fang, Q.; Yu, J.; Chen, P.; Wu, S.; et al. A Narrative Review of Tumor Heterogeneity and Challenges to Tumor Drug Therapy. Ann. Transl. Med. 2021, 9, 1351. [Google Scholar] [CrossRef] [PubMed]
- Proietto, M.; Crippa, M.; Damiani, C.; Pasquale, V.; Sacco, E.; Vanoni, M.; Gilardi, M. Tumor Heterogeneity: Preclinical Models, Emerging Technologies, and Future Applications. Front. Oncol. 2023, 13, 1164535. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, J.W.; Park, M. Comparison of Cancer Subtype Identification Methods Combined with Feature Selection Methods in Omics Data Analysis. BioData Min. 2023, 16, 18. [Google Scholar] [CrossRef] [PubMed]
- Källberg, D.; Vidman, L.; Rydén, P. Comparison of Methods for Feature Selection in Clustering of High-Dimensional RNA-Sequencing Data to Identify Cancer Subtypes. Front. Genet. 2021, 12, 632620. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.; Sanz-Pamplona, R.; Nadal, E.; Grasselli, J.; Pernas, S.; Dienstmann, R.; Moreno, V.; Tabernero, J.; Salazar, R. Intrinsic Cancer Subtypes-next Steps into Personalized Medicine. Cell Oncol. 2015, 38, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.P.; Rai, S.; Pandey, A.; Singh, N.K.; Srivastava, S. Molecular Subtypes of Colorectal Cancer: An Emerging Therapeutic Opportunity for Personalized Medicine. Genes Dis. 2021, 8, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Misra, B.B.; Langefeld, C.; Olivier, M.; Cox, L.A. Integrated Omics: Tools, Advances and Future Approaches. J. Mol. Endocrinol. 2019, 62, R21–R45. [Google Scholar] [CrossRef] [PubMed]
- Graw, S.; Chappell, K.; Washam, C.L.; Gies, A.; Bird, J.; Robeson, M.S.; Byrum, S.D. Multi-Omics Data Integration Considerations and Study Design for Biological Systems and Disease. Mol. Omics 2021, 17, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Gligorijević, V.; Pržulj, N. Methods for Biological Data Integration: Perspectives and Challenges. J. R. Soc. Interface 2015, 12, 20150571. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Sánchez-Maroño, N.; Alonso-Betanzos, A. Feature Selection for High-Dimensional Data. Prog. Artif. Intell. 2016, 5, 65–75. [Google Scholar] [CrossRef]
- Ghaddar, B.; Naoum-Sawaya, J. High Dimensional Data Classification and Feature Selection Using Support Vector Machines. Eur. J. Oper. Res. 2018, 265, 993–1004. [Google Scholar] [CrossRef]
- Jović, A.; Brkić, K.; Bogunović, N. A Review of Feature Selection Methods with Applications. In Proceedings of the 2015 38th International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO), Opatija, Croatia, 25–29 May 2015; pp. 1200–1205. [Google Scholar]
- Zebari, R.; Abdulazeez, A.; Zeebaree, D.; Zebari, D.; Saeed, J. A Comprehensive Review of Dimensionality Reduction Techniques for Feature Selection and Feature Extraction. J. Appl. Sci. Technol. Trends 2020, 1, 56–70. [Google Scholar] [CrossRef]
- Mladenić, D. Feature Selection for Dimensionality Reduction. In Proceedings of the Subspace, Latent Structure and Feature Selection, Bohinj, Slovenia, 23–25 February 2005; Springer: Berlin/Heidelberg, Germany, 2006; pp. 84–102. [Google Scholar]
- Huda, S.; Yearwood, J.; Jelinek, H.F.; Hassan, M.M.; Fortino, G.; Buckland, M. A Hybrid Feature Selection with Ensemble Classification for Imbalanced Healthcare Data: A Case Study for Brain Tumor Diagnosis. IEEE Access 2016, 4, 9145–9154. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, X.; Ru, X.; Sun, P.; Wang, J. A Hybrid Feature Selection Algorithm and Its Application in Bioinformatics. PeerJ Comput. Sci. 2022, 8, e933. [Google Scholar] [CrossRef]
- Ahn, J.S.; Shin, S.; Yang, S.-A.; Park, E.K.; Kim, K.H.; Cho, S.I.; Ock, C.-Y.; Kim, S. Artificial Intelligence in Breast Cancer Diagnosis and Personalized Medicine. J. Breast Cancer 2023, 26, 405–435. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, G.; Mahmoud, H.; Abd El-Hafeez, T.; E ElAraby, M. The Power of Deep Learning in Simplifying Feature Selection for Hepatocellular Carcinoma: A Review. BMC Med. Inf. Decis. Mak. 2024, 24, 287. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cheng, K.; Wang, S.; Morstatter, F.; Trevino, R.P.; Tang, J.; Liu, H. Feature Selection: A Data Perspective. ACM Comput. Surv. 2018, 50, 1–45. [Google Scholar] [CrossRef]
- Cai, J.; Luo, J.; Wang, S.; Yang, S. Feature Selection in Machine Learning: A New Perspective. Neurocomputing 2018, 300, 70–79. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Chang, Q. Feature Selection Methods for Big Data Bioinformatics: A Survey from the Search Perspective. Methods 2016, 111, 21–31. [Google Scholar] [CrossRef]
- Sánchez-Maroño, N.; Alonso-Betanzos, A.; Tombilla-Sanromán, M. Filter Methods for Feature Selection—A Comparative Study. In Proceedings of the Intelligent Data Engineering and Automated Learning—IDEAL 2007, Birmingham, UK, 16–19 December 2007; Springer: Berlin/Heidelberg, Germany, 2007; pp. 178–187. [Google Scholar]
- Maldonado, S.; Weber, R. A Wrapper Method for Feature Selection Using Support Vector Machines. Inf. Sci. 2009, 179, 2208–2217. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, M.; Liu, Q. An Embedded Feature Selection Method for Imbalanced Data Classification. IEEE/CAA J. Autom. Sin. 2019, 6, 703–715. [Google Scholar] [CrossRef]
- Chandrashekar, G.; Sahin, F. A Survey on Feature Selection Methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Hu, Y.; Shangguan, J.; Song, Q.; Xu, J.; Wang, H.; Xue, M.; Wang, L.; Zhang, Y. Identification of Key Biomarkers for STAD Using Filter Feature Selection Approaches. Sci. Rep. 2022, 12, 19854. [Google Scholar] [CrossRef] [PubMed]
- Sugai, Y.; Kadoya, N.; Tanaka, S.; Tanabe, S.; Umeda, M.; Yamamoto, T.; Takeda, K.; Dobashi, S.; Ohashi, H.; Takeda, K.; et al. Impact of Feature Selection Methods and Subgroup Factors on Prognostic Analysis with CT-Based Radiomics in Non-Small Cell Lung Cancer Patients. Radiat. Oncol. 2021, 16, 80. [Google Scholar] [CrossRef] [PubMed]
- Khanna, M.; Singh, L.K.; Shrivastava, K.; Singh, R. An Enhanced and Efficient Approach for Feature Selection for Chronic Human Disease Prediction: A Breast Cancer Study. Heliyon 2024, 10, e26799. [Google Scholar] [CrossRef] [PubMed]
- Taghizadeh, E.; Heydarheydari, S.; Saberi, A.; JafarpoorNesheli, S.; Rezaeijo, S.M. Breast Cancer Prediction with Transcriptome Profiling Using Feature Selection and Machine Learning Methods. BMC Bioinform. 2022, 23, 410. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, X.; Ru, X.; Sun, P.; Wang, J. Identification of Gene Signatures for COAD Using Feature Selection and Bayesian Network Approaches. Sci. Rep. 2022, 12, 8761. [Google Scholar] [CrossRef]
- Cai, Z.; Xu, D.; Zhang, Q.; Zhang, J.; Ngai, S.-M.; Shao, J. Classification of Lung Cancer Using Ensemble-Based Feature Selection and Machine Learning Methods. Mol. BioSystems 2015, 11, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, X.; Wang, J. Functional Proteomic Profiling Analysis in Four Major Types of Gastrointestinal Cancers. Biomolecules 2023, 13, 701. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.d.R.; Islam, M.M.M.; Kim, J.-M. Feature Selection Techniques for Increasing Reliability of Fault Diagnosis of Bearings. In Proceedings of the 2016 9th International Conference on Electrical and Computer Engineering (ICECE), Dhaka, Bangladesh, 20–22 December 2016; pp. 396–399. [Google Scholar]
- Saeys, Y.; Inza, I.; Larrañaga, P. A Review of Feature Selection Techniques in Bioinformatics. Bioinformatics 2007, 23, 2507–2517. [Google Scholar] [CrossRef] [PubMed]
- Fortino, V.; Kinaret, P.; Fyhrquist, N.; Alenius, H.; Greco, D. A Robust and Accurate Method for Feature Selection and Prioritization from Multi-Class OMICs Data. PLoS ONE 2014, 9, e107801. [Google Scholar] [CrossRef] [PubMed]
- Loughrey, J.; Cunningham, P. Overfitting in Wrapper-Based Feature Subset Selection: The Harder You Try the Worse It Gets. In Proceedings of the Research and Development in Intelligent Systems XXI, Cambridge, UK, 13–15 December 2004; Springer: London, UK, 2005; pp. 33–43. [Google Scholar]
- Abiodun, E.O.; Alabdulatif, A.; Abiodun, O.I.; Alawida, M.; Alabdulatif, A.; Alkhawaldeh, R.S. A Systematic Review of Emerging Feature Selection Optimization Methods for Optimal Text Classification: The Present State and Prospective Opportunities. Neural Comput. Applic 2021, 33, 15091–15118. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Rincon, A.; Mendoza-Maldonado, L.; Martinez-Archundia, M.; Schönhuth, A.; Kraneveld, A.D.; Garssen, J.; Tonda, A. Machine Learning-Based Ensemble Recursive Feature Selection of Circulating miRNAs for Cancer Tumor Classification. Cancers 2020, 12, 1785. [Google Scholar] [CrossRef] [PubMed]
- Remeseiro, B.; Bolon-Canedo, V. A Review of Feature Selection Methods in Medical Applications. Comput. Biol. Med. 2019, 112, 103375. [Google Scholar] [CrossRef] [PubMed]
- Pudjihartono, N.; Fadason, T.; Kempa-Liehr, A.W.; O’Sullivan, J.M. A Review of Feature Selection Methods for Machine Learning-Based Disease Risk Prediction. Front. Bioinform. 2022, 2, 927312. [Google Scholar] [CrossRef]
- Alhassan, A.M.; Wan Zainon, W.M.N. Review of Feature Selection, Dimensionality Reduction and Classification for Chronic Disease Diagnosis. IEEE Access 2021, 9, 87310–87317. [Google Scholar] [CrossRef]
- Jiang, Q.; Jin, M. Feature Selection for Breast Cancer Classification by Integrating Somatic Mutation and Gene Expression. Front. Genet. 2021, 12, 629946. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; An, N.; Chen, G.; Li, L.; Alterovitz, G. Accelerating Wrapper-Based Feature Selection with K-Nearest-Neighbor. Knowl.-Based Syst. 2015, 83, 81–91. [Google Scholar] [CrossRef]
- Hou, C.; Nie, F.; Yi, D.; Wu, Y. Feature Selection via Joint Embedding Learning and Sparse Regression. In Proceedings of the Twenty-Second International Joint Conference on Artificial Intelligence—Volume Volume Two, Barcelona, Spain, 16–22 July 2022; AAAI Press: Barcelona, Spain, 2011; pp. 1324–1329. [Google Scholar]
- Rostami, M.; Berahmand, K.; Nasiri, E.; Forouzandeh, S. Review of Swarm Intelligence-Based Feature Selection Methods. Eng. Appl. Artif. Intell. 2021, 100, 104210. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Sánchez-Maroño, N.; Alonso-Betanzos, A.; Benítez, J.M.; Herrera, F. A Review of Microarray Datasets and Applied Feature Selection Methods. Inf. Sci. 2014, 282, 111–135. [Google Scholar] [CrossRef]
- Zivkovic, M.; Stoean, C.; Chhabra, A.; Budimirovic, N.; Petrovic, A.; Bacanin, N. Novel Improved Salp Swarm Algorithm: An Application for Feature Selection. Sensors 2022, 22, 1711. [Google Scholar] [CrossRef]
- Liu, Y.; Mu, Y.; Chen, K.; Li, Y.; Guo, J. Daily Activity Feature Selection in Smart Homes Based on Pearson Correlation Coefficient. Neural Process Lett. 2020, 51, 1771–1787. [Google Scholar] [CrossRef]
- Liu, D.; Cho, S.-Y.; Sun, D.-M.; Qiu, Z.-D. A Spearman Correlation Coefficient Ranking for Matching-Score Fusion on Speaker Recognition. In Proceedings of the TENCON 2010—2010 IEEE Region 10 Conference, Fukuoka, Japan, 21–24 November 2010; pp. 736–741. [Google Scholar]
- Sulaiman, M.A.; Labadin, J. Feature Selection Based on Mutual Information. In Proceedings of the 2015 9th International Conference on IT in Asia (CITA), Sarawak, Malaysia, 4–5 August 2015; pp. 1–6. [Google Scholar]
- Lewis, D.D. Feature Selection and Feature Extraction for Text Categorization. In Proceedings of the Workshop on Speech and Natural Language, Harriman, NY, USA, 23–26 February 1992; pp. 212–217. [Google Scholar]
- Battiti, R. Using Mutual Information for Selecting Features in Supervised Neural Net Learning. IEEE Trans. Neural Netw. Learn. Syst. 1994, 5, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Long, F.; Ding, C. Feature Selection Based on Mutual Information: Criteria of Max-Dependency, Max-Relevance, and Min-Redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Fleuret, F. Fast Binary Feature Selection with Conditional Mutual Information. J. Mach. Learn. Res. 2004, 5, 1531–1555. [Google Scholar]
- Bennasar, M.; Hicks, Y.; Setchi, R. Feature Selection Using Joint Mutual Information Maximisation. Expert. Syst. Appl. 2015, 42, 8520–8532. [Google Scholar] [CrossRef]
- Lin, D.; Tang, X. Conditional Infomax Learning: An Integrated Framework for Feature Extraction and Fusion; Springer: Berlin/Heidelberg, Germany, 2006; pp. 68–82. [Google Scholar]
- Wang, J.; Wei, J.-M.; Yang, Z.; Wang, S.-Q. Feature Selection by Maximizing Independent Classification Information. IEEE Trans. Knowl. Data Eng. 2017, 29, 828–841. [Google Scholar] [CrossRef]
- Wang, L.; Jiang, S.; Jiang, S. A Feature Selection Method via Analysis of Relevance, Redundancy, and Interaction. Expert. Syst. Appl. 2021, 183, 115365. [Google Scholar] [CrossRef]
- Meyer, P.E.; Schretter, C.; Bontempi, G. Information-Theoretic Feature Selection in Microarray Data Using Variable Complementarity. IEEE J. Sel. Top. Signal Process. 2008, 2, 261–274. [Google Scholar] [CrossRef]
- Bommert, A.; Sun, X.; Bischl, B.; Rahnenführer, J.; Lang, M. Benchmark for Filter Methods for Feature Selection in High-Dimensional Classification Data. Comput. Stat. Data Anal. 2020, 143, 106839. [Google Scholar] [CrossRef]
- Ververidis, D.; Kotropoulos, C. Sequential Forward Feature Selection with Low Computational Cost. In Proceedings of the 2005 13th European Signal Processing Conference, Antalya, Turkey, 4–8 September 2005; pp. 1–4. [Google Scholar]
- Borboudakis, G.; Tsamardinos, I. Forward-Backward Selection with Early Dropping. J. Mach. Learn. Res. 2019, 20, 1–39. [Google Scholar]
- Haq, A.U.; Li, J.; Memon, M.H.; Hunain Memon, M.; Khan, J.; Marium, S.M. Heart Disease Prediction System Using Model of Machine Learning and Sequential Backward Selection Algorithm for Features Selection. In Proceedings of the 2019 IEEE 5th International Conference for Convergence in Technology (I2CT), Bombay, India, 29–31 March 2019; pp. 1–4. [Google Scholar]
- Chen, X.; Jeong, J.C. Enhanced Recursive Feature Elimination. In Proceedings of the Sixth International Conference on Machine Learning and Applications (ICMLA 2007), Cincinnati, OH, USA, 13–15 December 2007; pp. 429–435. [Google Scholar]
- Darst, B.F.; Malecki, K.C.; Engelman, C.D. Using Recursive Feature Elimination in Random Forest to Account for Correlated Variables in High Dimensional Data. BMC Genet. 2018, 19, 65. [Google Scholar] [CrossRef] [PubMed]
- Tang, E.K.; Suganthan, P.N.; Yao, X. Feature Selection for Microarray Data Using Least Squares SVM and Particle Swarm Optimization. In Proceedings of the 2005 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology, La Jolla, CA, USA, 15 November 2005; pp. 1–8. [Google Scholar]
- Shoorehdeli, M.A.; Teshnehlab, M.; Moghaddam, H.A. Feature Subset Selection for Face Detection Using Genetic Algorithms and Particle Swarm Optimization. In Proceedings of the 2006 IEEE International Conference on Networking, Sensing and Control, Ft. Lauderdale, FL, USA, 23–25 April 2006; pp. 686–690. [Google Scholar]
- Tan, F.; Fu, X.; Zhang, Y.; Bourgeois, A.G. A Genetic Algorithm-Based Method for Feature Subset Selection. Soft Comput. 2007, 12, 111–120. [Google Scholar] [CrossRef]
- Unler, A.; Murat, A.; Chinnam, R.B. mr2PSO: A Maximum Relevance Minimum Redundancy Feature Selection Method Based on Swarm Intelligence for Support Vector Machine Classification. Inf. Sci. 2011, 181, 4625–4641. [Google Scholar] [CrossRef]
- Xue, B.; Zhang, M.; Browne, W.N.; Yao, X. A Survey on Evolutionary Computation Approaches to Feature Selection. IEEE Trans. Evol. Computat. 2016, 20, 606–626. [Google Scholar] [CrossRef]
- Alba, E.; Garcia-Nieto, J.; Jourdan, L.; Talbi, E.-G. Gene Selection in Cancer Classification Using PSO/SVM and GA/SVM Hybrid Algorithms. In Proceedings of the 2007 IEEE Congress on Evolutionary Computation, Singapore, 25–28 September 2007; pp. 284–290. [Google Scholar]
- Huang, C.-L.; Dun, J.-F. A Distributed PSO–SVM Hybrid System with Feature Selection and Parameter Optimization. Appl. Soft Comput. 2008, 8, 1381–1391. [Google Scholar] [CrossRef]
- Jeong, Y.-S.; Shin, K.S.; Jeong, M.K. An Evolutionary Algorithm with the Partial Sequential Forward Floating Search Mutation for Large-Scale Feature Selection Problems. J. Oper. Res. Soc. 2015, 66, 529–538. [Google Scholar] [CrossRef]
- Cho, H.-W.; Kim, S.B.; Jeong, M.K.; Park, Y.; Ziegler, T.R.; Jones, D.P. Genetic Algorithm-Based Feature Selection in High-Resolution NMR Spectra. Expert. Syst. Appl. 2008, 35, 967–975. [Google Scholar] [CrossRef]
- Xue, B.; Zhang, M.; Browne, W.N. New Fitness Functions in Binary Particle Swarm Optimisation for Feature Selection. In Proceedings of the 2012 IEEE Congress on Evolutionary Computation, Brisbane, QLD, Australia, 10–15 June 2012; pp. 1–8. [Google Scholar]
- Xue, B.; Zhang, M.; Browne, W.N. Novel Initialisation and Updating Mechanisms in PSO for Feature Selection in Classification. In Proceedings of the Applications of Evolutionary Computation, Vienna, Austria, 3–5 April 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 428–438. [Google Scholar]
- da Silva, S.F.; Ribeiro, M.X.; Neto, J.D.E.B.; Traina, C., Jr.; Traina, A.J. Improving the Ranking Quality of Medical Image Retrieval Using a Genetic Feature Selection Method. Decis. Support. Syst. 2011, 51, 810–820. [Google Scholar] [CrossRef]
- Sousa, P.; Cortez, P.; Vaz, R.; Rocha, M.; Rio, M. Email Spam Detection: A Symbiotic Feature Selection Approach Fostered by Evolutionary Computation. Int. J. Inf. Technol. Decis. Mak. 2013, 12, 863–884. [Google Scholar] [CrossRef]
- Canuto, A.M.P.; Nascimento, D.S.C. A Genetic-Based Approach to Features Selection for Ensembles Using a Hybrid and Adaptive Fitness Function. In Proceedings of the 2012 International Joint Conference on Neural Networks (IJCNN), Brisbane, QLD, Australia, 10–15 June 2012; pp. 1–8. [Google Scholar]
- Yusta, S.C. Different Metaheuristic Strategies to Solve the Feature Selection Problem. Pattern Recognit. Lett. 2009, 30, 525–534. [Google Scholar] [CrossRef]
- Muthukrishnan, R.; Rohini, R. LASSO: A Feature Selection Technique in Predictive Modeling for Machine Learning. In Proceedings of the 2016 IEEE International Conference on Advances in Computer Applications (ICACA), Coimbatore, India, 24 October 2016; pp. 18–20. [Google Scholar]
- Yamada, M.; Jitkrittum, W.; Sigal, L.; Xing, E.P.; Sugiyama, M. High-Dimensional Feature Selection by Feature-Wise Kernelized Lasso. Neural Comput. 2014, 26, 185–207. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Sun, L.; Wang, W.; Chen, T.; Guo, M.; Zhang, P. A Feature Selection Method Combined with Ridge Regression and Recursive Feature Elimination in Quantitative Analysis of Laser Induced Breakdown Spectroscopy. Plasma Sci. Technol. 2020, 22, 074002. [Google Scholar] [CrossRef]
- Cawley, G.C. Causal & Non-Causal Feature Selection for Ridge Regression. In Proceedings of the Workshop on the Causation and Prediction Challenge at WCCI 2008, Hong Kong, China, 3–4 June 2018; pp. 107–128. [Google Scholar]
- Rashid, M.; Kamruzzaman, J.; Imam, T.; Wibowo, S.; Gordon, S. A Tree-Based Stacking Ensemble Technique with Feature Selection for Network Intrusion Detection. Appl. Intell. 2022, 52, 9768–9781. [Google Scholar] [CrossRef]
- Pham, N.T.; Foo, E.; Suriadi, S.; Jeffrey, H.; Lahza, H.F.M. Improving Performance of Intrusion Detection System Using Ensemble Methods and Feature Selection. In Proceedings of the Australasian Computer Science Week Multiconference, Brisband, Australia, 29 January–2 February 2018; Association for Computing Machinery: New York, NY, USA, 2018; pp. 1–6. [Google Scholar]
- Xu, Z.; Huang, G.; Weinberger, K.Q.; Zheng, A.X. Gradient Boosted Feature Selection. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; Association for Computing Machinery: New York, NY, USA, 2014; pp. 522–531. [Google Scholar]
- Chen, C.; Zhang, Q.; Yu, B.; Yu, Z.; Lawrence, P.J.; Ma, Q.; Zhang, Y. Improving Protein-Protein Interactions Prediction Accuracy Using XGBoost Feature Selection and Stacked Ensemble Classifier. Comput. Biol. Med. 2020, 123, 103899. [Google Scholar] [CrossRef]
- Cantor, E.; Guauque-Olarte, S.; León, R.; Chabert, S.; Salas, R. Knowledge-Slanted Random Forest Method for High-Dimensional Data and Small Sample Size with a Feature Selection Application for Gene Expression Data. BioData Min. 2024, 17, 34. [Google Scholar] [CrossRef]
- Prasetiyowati, M.I.; Maulidevi, N.U.; Surendro, K. Feature Selection to Increase the Random Forest Method Performance on High Dimensional Data. Int. J. Adv. Intell. Inform. 2020, 6, 303. [Google Scholar] [CrossRef]
- Fakhr, M.W.; Youssef, E.-N.S.; El-Mahallawy, M.S. L1-Regularized Least Squares Sparse Extreme Learning Machine for Classification. In Proceedings of the 2015 International Conference on Information and Communication Technology Research (ICTRC), Abu Dhabi, United Arab Emirates, 17–19 May 2015; pp. 222–225. [Google Scholar]
- Demir-Kavuk, O.; Kamada, M.; Akutsu, T.; Knapp, E.-W. Prediction Using Step-Wise L1, L2 Regularization and Feature Selection for Small Data Sets with Large Number of Features. BMC Bioinform. 2011, 12, 412. [Google Scholar] [CrossRef] [PubMed]
- Amini, F.; Hu, G. A Two-Layer Feature Selection Method Using Genetic Algorithm and Elastic Net. Expert. Syst. Appl. 2021, 166, 114072. [Google Scholar] [CrossRef]
- Jenul, A.; Schrunner, S.; Liland, K.H.; Indahl, U.G.; Futsæther, C.M.; Tomic, O. RENT—Repeated Elastic Net Technique for Feature Selection. IEEE Access 2021, 9, 152333–152346. [Google Scholar] [CrossRef]
- Rasmussen, M.A.; Bro, R. A Tutorial on the Lasso Approach to Sparse Modeling. Chemom. Intell. Lab. Syst. 2012, 119, 21–31. [Google Scholar] [CrossRef]
- Xie, Z.; Xu, Y. Sparse Group LASSO Based Uncertain Feature Selection. Int. J. Mach. Learn. Cyber. 2014, 5, 201–210. [Google Scholar] [CrossRef]
- Gui, J.; Sun, Z.; Ji, S.; Tao, D.; Tan, T. Feature Selection Based on Structured Sparsity: A Comprehensive Study. IEEE Trans. Neural Netw. Learn. Syst. 2017, 28, 1490–1507. [Google Scholar] [CrossRef]
- Zhao, L.; Hu, Q.; Wang, W. Heterogeneous Feature Selection With Multi-Modal Deep Neural Networks and Sparse Group LASSO. IEEE Trans. Multimed. 2015, 17, 1936–1948. [Google Scholar] [CrossRef]
- Cui, L.; Bai, L.; Wang, Y.; Yu, P.S.; Hancock, E.R. Fused Lasso for Feature Selection Using Structural Information. Pattern Recognit. 2021, 119, 108058. [Google Scholar] [CrossRef]
- Xin, B.; Kawahara, Y.; Wang, Y.; Hu, L.; Gao, W. Efficient Generalized Fused Lasso and Its Applications. ACM Trans. Intell. Syst. Technol. 2016, 7, 60. [Google Scholar] [CrossRef]
- Zou, H. The Adaptive Lasso and Its Oracle Properties. J. Am. Stat. Assoc. 2006, 101, 1418–1429. [Google Scholar] [CrossRef]
- Shortreed, S.M.; Ertefaie, A. Outcome-Adaptive Lasso: Variable Selection for Causal Inference. Biometrics 2017, 73, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.H.; Xue, B.; Zhang, M. A Survey on Swarm Intelligence Approaches to Feature Selection in Data Mining. Swarm Evol. Comput. 2020, 54, 100663. [Google Scholar] [CrossRef]
- Brezočnik, L.; Fister, I.; Podgorelec, V. Swarm Intelligence Algorithms for Feature Selection: A Review. Appl. Sci. 2018, 8, 1521. [Google Scholar] [CrossRef]
- Kashef, S.; Nezamabadi-pour, H. An Advanced ACO Algorithm for Feature Subset Selection. Neurocomputing 2015, 147, 271–279. [Google Scholar] [CrossRef]
- Nayar, N.; Gautam, S.; Singh, P.; Mehta, G. Ant Colony Optimization: A Review of Literature and Application in Feature Selection. In Proceedings of the Inventive Computation and Information Technologies, Coimbatore, India, 24–25 September 2020; Springer Nature: Singapore, 2021; pp. 285–297. [Google Scholar]
- Haznedar, B.; Arslan, M.T.; Kalinli, A. Optimizing ANFIS Using Simulated Annealing Algorithm for Classification of Microarray Gene Expression Cancer Data. Med. Biol. Eng. Comput. 2021, 59, 497–509. [Google Scholar] [CrossRef]
- Acharya, S.; Saha, S.; Thadisina, Y. Multiobjective Simulated Annealing-Based Clustering of Tissue Samples for Cancer Diagnosis. IEEE J. Biomed. Health Inform. 2016, 20, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Hancer, E.; Xue, B.; Zhang, M. Differential Evolution for Filter Feature Selection Based on Information Theory and Feature Ranking. Knowl.-Based Syst. 2018, 140, 103–119. [Google Scholar] [CrossRef]
- El Aboudi, N.; Benhlima, L. Review on Wrapper Feature Selection Approaches. In Proceedings of the 2016 International Conference on Engineering & MIS (ICEMIS), Agadir, Morocco, 22–24 September 2016; pp. 1–5. [Google Scholar]
- Maldonado, J.; Riff, M.C.; Neveu, B. A Review of Recent Approaches on Wrapper Feature Selection for Intrusion Detection. Expert. Syst. Appl. 2022, 198, 116822. [Google Scholar] [CrossRef]
- Bajer, D.; Dudjak, M.; Zorić, B. Wrapper-Based Feature Selection: How Important Is the Wrapped Classifier? In Proceedings of the 2020 International Conference on Smart Systems and Technologies (SST), Osijek, Croatia, 14–16 October 2020; pp. 97–105. [Google Scholar]
- Wang, S.; Tang, J.; Liu, H. Embedded Unsupervised Feature Selection. In Proceedings of the AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015; pp. 470–476. [Google Scholar] [CrossRef]
- Venkatesh, B.; Anuradha, J. A Review of Feature Selection and Its Methods. Cybern. Inf. Technol. 2019, 19, 3–26. [Google Scholar] [CrossRef]
- Ramos-González, J.; López-Sánchez, D.; Castellanos-Garzón, J.A.; de Paz, J.F.; Corchado, J.M. A CBR Framework with Gradient Boosting Based Feature Selection for Lung Cancer Subtype Classification. Comput. Biol. Med. 2017, 86, 98–106. [Google Scholar] [CrossRef]
- Chen, J.W.; Dhahbi, J. Lung Adenocarcinoma and Lung Squamous Cell Carcinoma Cancer Classification, Biomarker Identification, and Gene Expression Analysis Using Overlapping Feature Selection Methods. Sci. Rep. 2021, 11, 13323. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, X.; Ru, X.; Sun, P.; Wang, J. The Weight-Based Feature Selection (WBFS) Algorithm Classifies Lung Cancer Subtypes Using Proteomic Data. Entropy 2023, 25, 1003. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, X.; Ru, X.; Sun, P.; Wang, J. Using Feature Selection and Bayesian Network Identify Cancer Subtypes Based on Proteomic Data. J. Proteom. 2023, 280, 104895. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Qiu, Q.; Fu, J.; Cui, K.; Chen, X.; Xing, L.; Sun, X. Stage-Specific PET Radiomic Prediction Model for the Histological Subtype Classification of Non-Small-Cell Lung Cancer. Cancer Manag. Res. 2021, 13, 307–317. [Google Scholar] [CrossRef]
- Ma, M.; Liu, R.; Wen, C.; Xu, W.; Xu, Z.; Wang, S.; Wu, J.; Pan, D.; Zheng, B.; Qin, G.; et al. Predicting the Molecular Subtype of Breast Cancer and Identifying Interpretable Imaging Features Using Machine Learning Algorithms. Eur. Radiol. 2022, 32, 1652–1662. [Google Scholar] [CrossRef] [PubMed]
- Pozzoli, S.; Soliman, A.; Bahri, L.; Branca, R.M.; Girdzijauskas, S.; Brambilla, M. Domain Expertise–Agnostic Feature Selection for the Analysis of Breast Cancer Data*. Artif. Intell. Med. 2020, 108, 101928. [Google Scholar] [CrossRef] [PubMed]
- Cascianelli, S.; Galzerano, A.; Masseroli, M. Supervised Relevance-Redundancy Assessments for Feature Selection in Omics-Based Classification Scenarios. J. Biomed. Inf. 2023, 144, 104457. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Chen, W.; Wei, H.; Zhang, X.; Yuan, S.; Qiao, X.; Chen, Y.-W. Machine Learning for Histologic Subtype Classification of Non-Small Cell Lung Cancer: A Retrospective Multicenter Radiomics Study. Front. Oncol. 2020, 10, 608598. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, J.P.; Saha, I.; Sarkar, A.; Maulik, U. Machine Learning Integrated Ensemble of Feature Selection Methods Followed by Survival Analysis for Predicting Breast Cancer Subtype Specific miRNA Biomarkers. Comput. Biol. Med. 2021, 131, 104244. [Google Scholar] [CrossRef]
- MotieGhader, H.; Masoudi-Sobhanzadeh, Y.; Ashtiani, S.H.; Masoudi-Nejad, A. mRNA and microRNA Selection for Breast Cancer Molecular Subtype Stratification Using Meta-Heuristic Based Algorithms. Genomics 2020, 112, 3207–3217. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Hicks, C. Breast Cancer Type Classification Using Machine Learning. J. Pers. Med. 2021, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Feng, Y.; Wu, X.; Li, R.; Shi, J.; Chang, W.; Wang, J. ForestSubtype: A Cancer Subtype Identifying Approach Based on High-Dimensional Genomic Data and a Parallel Random Forest. BMC Bioinform. 2023, 24, 289. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yu, L.; Gao, L. Cancer Classification Based on Multiple Dimensions: SNV Patterns. Comput. Biol. Med. 2022, 151, 106270. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Mali, K. Breast Cancer Subtypes Classification with Hybrid Machine Learning Model. Methods Inf. Med. 2022, 61, 68–83. [Google Scholar] [CrossRef]
- Huang, M.; Ye, X.; Imakura, A.; Sakurai, T. Sequential Reinforcement Active Feature Learning for Gene Signature Identification in Renal Cell Carcinoma. J. Biomed. Inform. 2022, 128, 104049. [Google Scholar] [CrossRef]
- Maulik, U.; Mukhopadhyay, A.; Chakraborty, D. Gene-Expression-Based Cancer Subtypes Prediction Through Feature Selection and Transductive SVM. IEEE Trans. Biomed. Eng. 2013, 60, 1111–1117. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Bø, T.H.; Jonassen, I.; Myklebost, O.; Hovig, E. Tumor Classification and Marker Gene Prediction by Feature Selection and Fuzzy C-Means Clustering Using Microarray Data. BMC Bioinform. 2003, 4, 60. [Google Scholar] [CrossRef] [PubMed]
- Mendonca-Neto, R.; Li, Z.; Fenyö, D.; Silva, C.T.; Nakamura, F.G.; Nakamura, E.F. A Gene Selection Method Based on Outliers for Breast Cancer Subtype Classification. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 2547–2559. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Pascual, T.; De Angelis, C.; Gutierrez, C.; Llombart-Cussac, A.; Wang, T.; Cortés, J.; Rexer, B.; Paré, L.; Forero, A.; et al. HER2-Enriched Subtype and ERBB2 Expression in HER2-Positive Breast Cancer Treated with Dual HER2 Blockade. JNCI J. Natl. Cancer Inst. 2020, 112, 46–54. [Google Scholar] [CrossRef]
- Llombart-Cussac, A.; Cortés, J.; Paré, L.; Galván, P.; Bermejo, B.; Martínez, N.; Vidal, M.; Pernas, S.; López, R.; Muñoz, M.; et al. HER2-Enriched Subtype as a Predictor of Pathological Complete Response Following Trastuzumab and Lapatinib without Chemotherapy in Early-Stage HER2-Positive Breast Cancer (PAMELA): An Open-Label, Single-Group, Multicentre, Phase 2 Trial. Lancet Oncol. 2017, 18, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Potlitz, F.; Link, A.; Schulig, L. Advances in the Discovery of New Chemotypes through Ultra-Large Library Docking. Expert. Opin. Drug Discov. 2023, 18, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Bhinder, B.; Gilvary, C.; Madhukar, N.S.; Elemento, O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 2021, 11, 900–915. [Google Scholar] [CrossRef]
- Speiser, J.L. A Random Forest Method with Feature Selection for Developing Medical Prediction Models with Clustered and Longitudinal Data. J. Biomed. Inf. 2021, 117, 103763. [Google Scholar] [CrossRef]
- Karalis, V.D. The Integration of Artificial Intelligence into Clinical Practice. Appl. Biosci. 2024, 3, 14–44. [Google Scholar] [CrossRef]
- Yao, J.; Wang, S.; Zhu, X.; Huang, J. Imaging Biomarker Discovery for Lung Cancer Survival Prediction. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016, Athens, Greece, 17–21 October 2016; Springer International Publishing: Cham, Switzerland, 2016; pp. 649–657. [Google Scholar]
- Miller, H.A.; van Berkel, V.H.; Frieboes, H.B. Lung Cancer Survival Prediction and Biomarker Identification with an Ensemble Machine Learning Analysis of Tumor Core Biopsy Metabolomic Data. Metabolomics 2022, 18, 57. [Google Scholar] [CrossRef]
- Leite, A.F.; Vasconcelos, K.D.F.; Willems, H.; Jacobs, R. Radiomics and Machine Learning in Oral Healthcare. Proteom. Clin. Appl. 2020, 14, e1900040. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, T.; Liu, H. Recent Advances in Feature Selection and Its Applications. Knowl. Inf. Syst. 2017, 53, 551–577. [Google Scholar] [CrossRef]
- Goh, W.W.B.; Wong, L. Evaluating Feature-Selection Stability in next-Generation Proteomics. J. Bioinform. Comput. Biol. 2016. [Google Scholar] [CrossRef]
- Khaire, U.M.; Dhanalakshmi, R. Stability of Feature Selection Algorithm: A Review. J. King Saud. Univ.—Comput. Inf. Sci. 2022, 34, 1060–1073. [Google Scholar] [CrossRef]
- Sangeetha, S.K.B.; Mathivanan, S.K.; Karthikeyan, P.; Rajadurai, H.; Shivahare, B.D.; Mallik, S.; Qin, H. An Enhanced Multimodal Fusion Deep Learning Neural Network for Lung Cancer Classification. Syst. Soft Comput. 2024, 6, 200068. [Google Scholar] [CrossRef]
- Waqas, A.; Tripathi, A.; Ramachandran, R.P.; Stewart, P.A.; Rasool, G. Multimodal Data Integration for Oncology in the Era of Deep Neural Networks: A Review. Front. Artif. Intell. 2024, 7, 1408843. [Google Scholar] [CrossRef]
- Boehm, K.M.; Aherne, E.A.; Ellenson, L.; Nikolovski, I.; Alghamdi, M.; Vázquez-García, I.; Zamarin, D.; Long Roche, K.; Liu, Y.; Patel, D.; et al. Multimodal Data Integration Using Machine Learning Improves Risk Stratification of High-Grade Serous Ovarian Cancer. Nat. Cancer 2022, 3, 723–733. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).