Extracellular Vesicle-Mediated Modulation of Stem-like Phenotype in Breast Cancer Cells under Fluid Shear Stress
Abstract
1. Introduction
2. Materials & Methods
2.1. Cell Culture
2.2. Fluid Shear Stress
2.3. EV Isolation
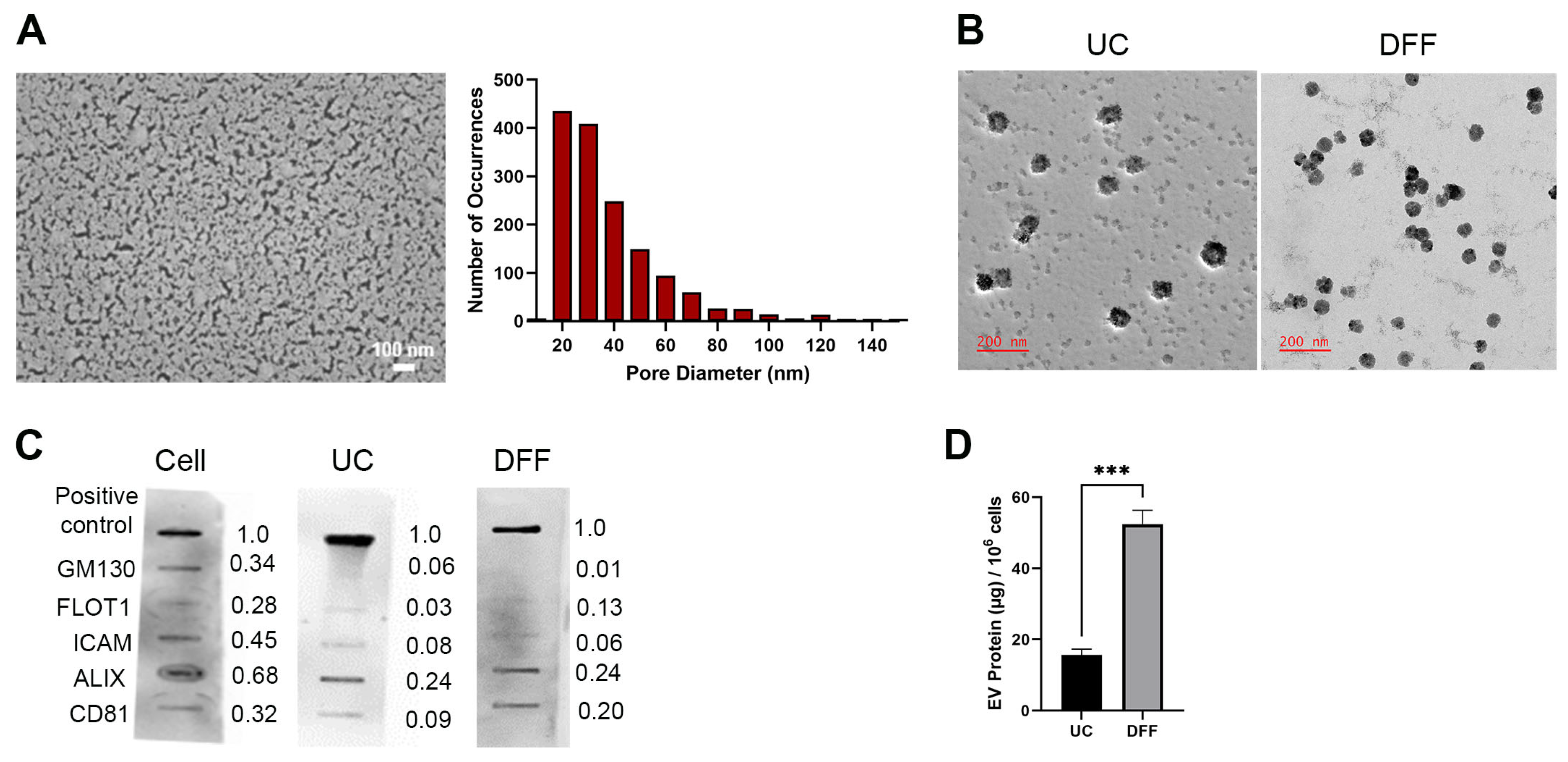
2.4. Scanning Electron Microscopy (SEM)
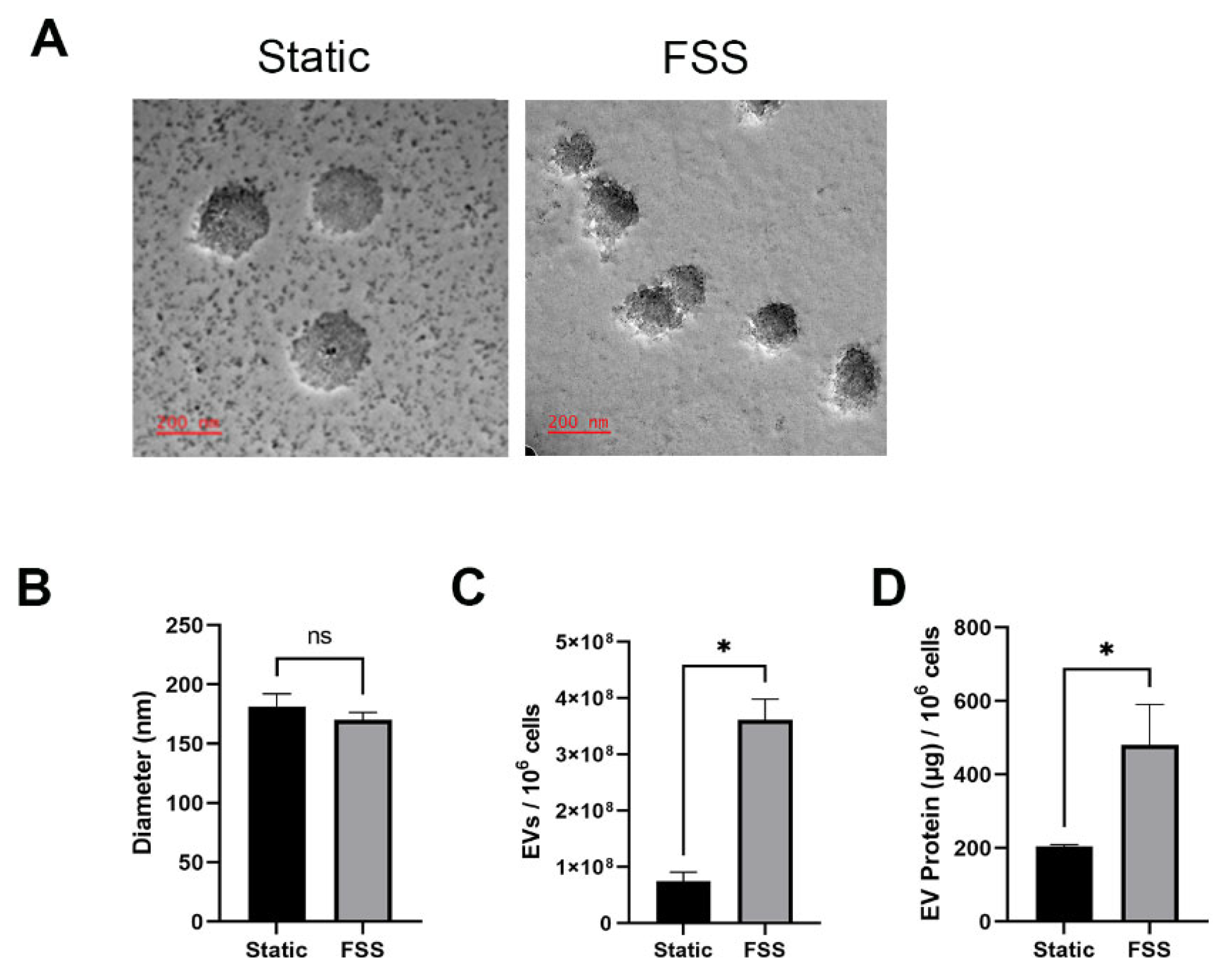
2.5. EV Verification and Quantification
2.6. Nanoparticle Tracking Analysis (NTA)
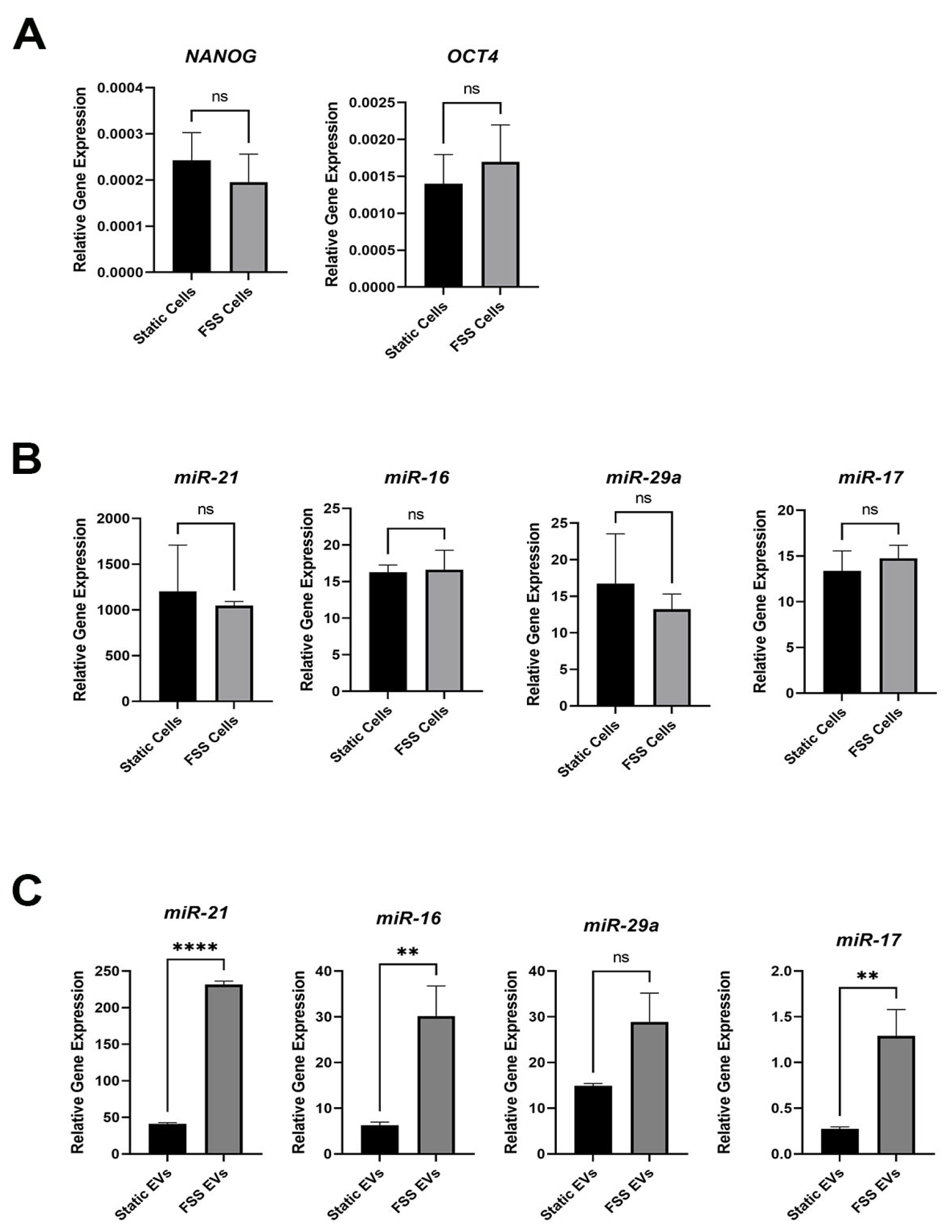
2.7. qRT-PCR
2.8. Fluorescent Imaging
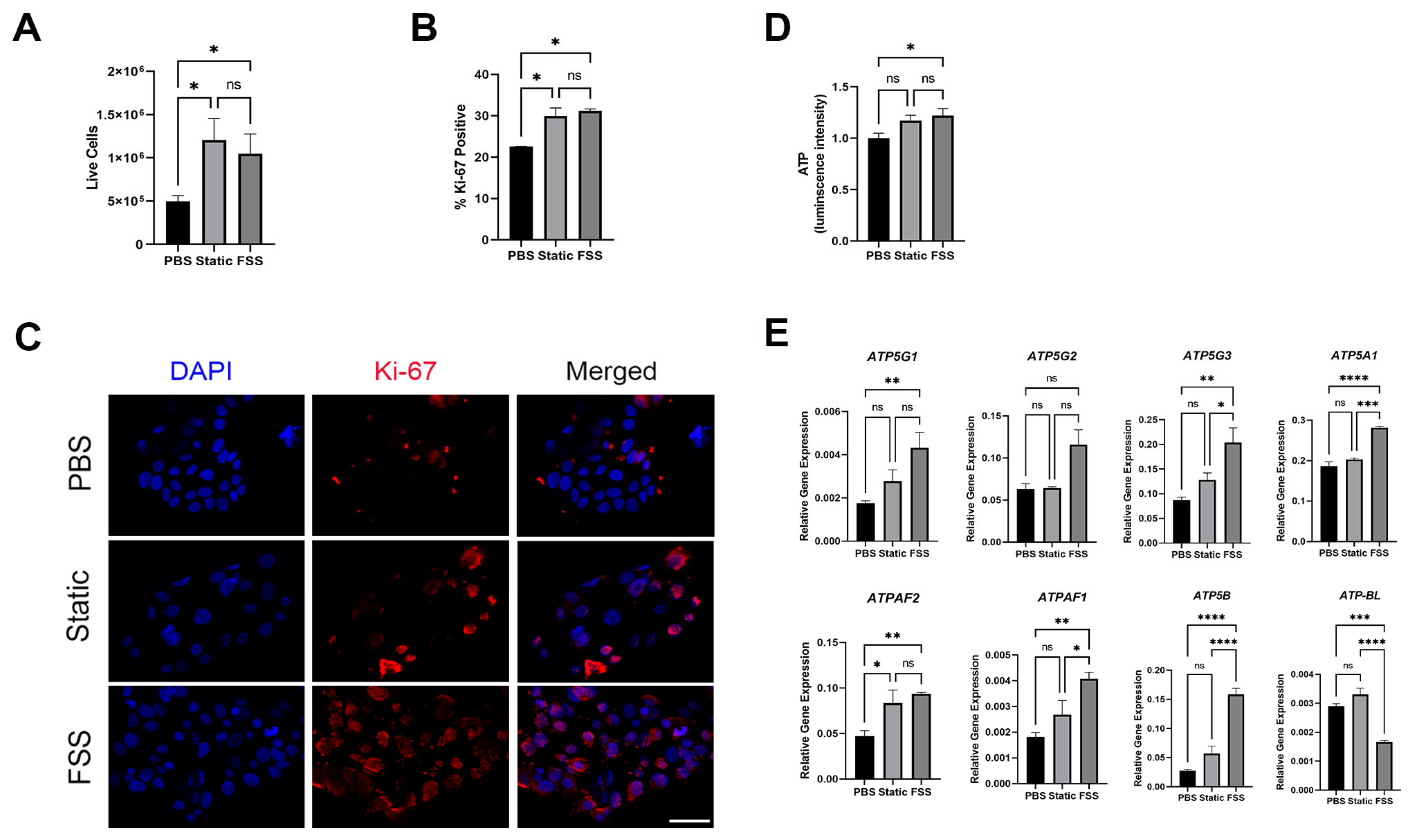
2.9. Flow Cytometry
2.10. Measurement of ATP Production
2.11. Statistical Analyses
3. Results
3.1. Direct Flow Filtration Produces Higher EV Recovery Than Differential Ultracentrifugation
3.2. Fluid Shear Stress Increases the Quantity of EVs Produced
3.3. Fluid Shear Stress Increases Stemness-Related miRNA in EVs
3.4. MCF-7 Cells Receiving EVs from MDA-MB-231 Showed Increased Stem-Like Signature
3.5. MCF-7 Cells Receiving EVs from MDA-MB-231 Showed Increased Proliferation and ATP Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Fidler, I.J.; Yano, S.; Zhang, R.-D.; Fujimaki, T.; Bucana, C.D. The seed and soil hypothesis: Vascularisation and brain metastases. Lancet Oncol. 2002, 3, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, D.; Konstantopoulos, K.; Searson, P.C. The physics of cancer: The role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer 2011, 11, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Muller, V.; Riethdorf, S.; Rack, B.; Janni, W.; Fasching, P.A.; Solomayer, E.; Aktas, B.; Kasimir-Bauer, S.; Pantel, K.; Fehm, T.; et al. Prognostic impact of circulating tumor cells assessed with the CellSearch System and AdnaTest Breast in metastatic breast cancer patients: The DETECT study. Breast Cancer Res. 2012, 14, R118. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, Y.; He, B.; Pan, Y.; Zhou, D.; Xiong, M.; Song, Y. An Enzyme-Loaded Metal-Organic Framework-Assisted Microfluidic Platform Enables Single-Cell Metabolite Analysis. Angew. Chem. Int. Ed. Engl. 2023, 62, e202302000. [Google Scholar] [CrossRef] [PubMed]
- Aktas, B.; Tewes, M.; Fehm, T.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009, 11, R46. [Google Scholar] [CrossRef] [PubMed]
- Kasimir-Bauer, S.; Hoffmann, O.; Wallwiener, D.; Kimmig, R.; Fehm, T. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res. 2012, 14, R15. [Google Scholar] [CrossRef] [PubMed]
- Theodoropoulos, P.A.; Polioudaki, H.; Agelaki, S.; Kallergi, G.; Saridaki, Z.; Mavroudis, D.; Georgoulias, V. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett. 2010, 288, 99–106. [Google Scholar] [CrossRef]
- Yang, M.H.; Imrali, A.; Heeschen, C. Circulating cancer stem cells: The importance to select. Chin. J. Cancer Res. 2015, 27, 437–449. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Clarke, M.F.; Hass, A.T. Cancer stem cells. In Reviews in Cell Biology and Molecular Medicine; Meyers, R.A., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006. [Google Scholar] [CrossRef]
- Najminejad, H.; Kalantar, S.M.; Abdollahpour-Alitappeh, M.; Karimi, M.H.; Seifalian, A.M.; Gholipourmalekabadi, M.; Sheikhha, M.H. Emerging roles of exosomal miRNAs in breast cancer drug resistance. IUBMB Life 2019, 71, 1672–1684. [Google Scholar] [CrossRef]
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A common pathway for a specialized function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef]
- Sharma, A. Role of stem cell derived exosomes in tumor biology. Int. J. Cancer 2018, 142, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Sung, B.H.; Ketova, T.; Hoshino, D.; Zijlstra, A.; Weaver, A.M. Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 2015, 6, 7164. [Google Scholar] [CrossRef]
- Antonyak, M.A.; Li, B.; Boroughs, L.K.; Johnson, J.L.; Druso, J.E.; Bryant, K.L.; Holowka, D.A.; Cerione, R.A. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc. Natl. Acad. Sci. USA 2011, 108, 4852–4857. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Fidler, I.J.; Poste, G. The “seed and soil” hypothesis revisited. Lancet Oncol. 2008, 9, 808. [Google Scholar] [CrossRef] [PubMed]
- Dos Anjos Pultz, B.; Andrés Cordero da Luz, F.; Socorro Faria, S.; Peixoto Ferreira de Souza, L.; Cristina Brígido Tavares, P.; Alonso Goulart, V.; Fontes, W.; Ricardo Goulart, L.; José Barbosa Silva, M. The multifaceted role of extracellular vesicles in metastasis: Priming the soil for seeding. Int. J. Cancer 2017, 140, 2397–2407. [Google Scholar] [CrossRef]
- Yang, J.; Pan, B.; Zeng, F.; He, B.; Gao, Y.; Liu, X.; Song, Y. Magnetic Colloid Antibodies Accelerate Small Extracellular Vesicles Isolation for Point-of-Care Diagnostics. Nano Lett. 2021, 21, 2001–2009. [Google Scholar] [CrossRef]
- De Jong, O.G.; Verhaar, M.C.; Chen, Y.; Vader, P.; Gremmels, H.; Posthuma, G.; Schiffelers, R.M.; Gucek, M.; van Balkom, B.W. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell. Vesicles 2012, 1, 18396. [Google Scholar] [CrossRef] [PubMed]
- Domenis, R.; Cifu, A.; Curcio, F. The Influence of a Stressful Microenvironment on Tumor Exosomes: A Focus on the DNA Cargo. Int. J. Mol. Sci. 2020, 21, 8728. [Google Scholar] [CrossRef]
- Hedlund, M.; Nagaeva, O.; Kargl, D.; Baranov, V.; Mincheva-Nilsson, L. Thermal- and oxidative stress causes enhanced release of NKG2D ligand-bearing immunosuppressive exosomes in leukemia/lymphoma T and B cells. PLoS ONE 2011, 6, e16899. [Google Scholar] [CrossRef]
- Li, J.; Lee, Y.; Johansson, H.J.; Mager, I.; Vader, P.; Nordin, J.Z.; Wiklander, O.P.; Lehtio, J.; Wood, M.J.; Andaloussi, S.E. Serum-free culture alters the quantity and protein composition of neuroblastoma-derived extracellular vesicles. J. Extracell. Vesicles 2015, 4, 26883. [Google Scholar] [CrossRef] [PubMed]
- Kucharzewska, P.; Belting, M. Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress. J. Extracell. Vesicles 2013, 2, 20304. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Eades, G.; Yao, Y.; Zhang, Y.; Zhou, Q. Characterization of a stem-like subpopulation in basal-like ductal carcinoma in situ (DCIS) lesions. J. Biol. Chem. 2014, 289, 1303–1312. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Di, K.; Fan, B.; Wu, J.; Gu, X.; Sun, Y.; Khan, A.; Li, P.; Li, Z. MicroRNAs in extracellular vesicles: Sorting mechanisms, diagnostic value, isolation, and detection technology. Front. Bioeng. Biotechnol. 2022, 10, 948959. [Google Scholar] [CrossRef]
- Humphries, B.; Wang, Z.; Yang, C. MicroRNA Regulation of Breast Cancer Stemness. Int. J. Mol. Sci. 2021, 22, 3756. [Google Scholar] [CrossRef] [PubMed]
- Joyce, D.P.; Kerin, M.J.; Dwyer, R.M. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int. J. Cancer 2016, 139, 1443–1448. [Google Scholar] [CrossRef]
- Wang, J.Y.; Zhang, Q.; Wang, D.D.; Yan, W.; Sha, H.H.; Zhao, J.H.; Yang, S.J.; Zhang, H.D.; Hou, J.C.; Xu, H.Z.; et al. MiR-29a: A potential therapeutic target and promising biomarker in tumors. Biosci. Rep. 2018, 38, BSR20171265. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, Y.; Nomura, S.; Miyake, T.; Kagawa, H.; Kitada, C.; Taniguchi, H.; Komiyama, Y.; Fujimura, Y.; Ikeda, Y.; Fukuhara, S. High Shear Stress Can Initiate Both Platelet Aggregation and Shedding of Procoagulant Containing Microparticles. Blood 1996, 88, 3456–3464. [Google Scholar] [CrossRef]
- Furukawa, H.; Kato, Y.; Inoue, Y.; Kato, T.; Tada, Y.; Hashimoto, S. Correlation of Power Consumption for Several Kinds of Mixing Impellers. Int. J. Chem. Eng. 2012, 2012, 106496. [Google Scholar] [CrossRef]
- Weinman, S.T.; Bass, M.; Pandit, S.; Herzberg, M.; Freger, V.; Husson, S.M. A switchable zwitterionic membrane surface chemistry for biofouling control. J. Membr. Sci. 2018, 548, 490–501. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Masselin, I.; Durand-Bourlier, L.; Laine, J.-M.; Sizaret, P.-Y.; Chasseray, X.; Lemordant, D. Membrane characterization using microscopic image analysis. J. Membr. Sci. 2001, 186, 85–96. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X. Study of membrane morphology by microscopic image analysis and membrane structure parameter model. J. Membr. Sci. 2006, 283, 109–115. [Google Scholar] [CrossRef]
- Danner, J.T.; Papier, L.G.; Callahan, M.L.; Jarrell, B.J.; Weinman, S.T. 2-Methylpyrazine: A Greener Solvent for Nonsolvent Induced Phase Separation (NIPS) Membrane Fabrication. Ind. Eng. Chem. Res. 2024, 63, 10735–10747. [Google Scholar] [CrossRef]
- Ferreira, T.; Rasband, W. ImageJ User Guide. Available online: https://imagej.net/ij/docs/guide/146-30.html#toc-Subsection-30.2 (accessed on 13 May 2024).
- Liang, Y.; Ridzon, D.; Wong, L.; Chen, C. Characterization of microRNA expression profiles in normal human tissues. BMC Genom. 2007, 8, 166. [Google Scholar] [CrossRef]
- Park, H.A.; Licznerski, P.; Mnatsakanyan, N.; Niu, Y.; Sacchetti, S.; Wu, J.; Polster, B.M.; Alavian, K.N.; Jonas, E.A. Inhibition of Bcl-xL prevents pro-death actions of DeltaN-Bcl-xL at the mitochondrial inner membrane during glutamate excitotoxicity. Cell Death Differ. 2017, 24, 1963–1974. [Google Scholar] [CrossRef] [PubMed]
- Park, H.A.; Brown, S.R.; Jansen, J.; Dunn, T.; Scott, M.; Mnatsakanyan, N.; Jonas, E.A.; Kim, Y. Fluid shear stress enhances proliferation of breast cancer cells via downregulation of the c-subunit of the F(1)F(O) ATP synthase. Biochem. Biophys. Res. Commun. 2022, 632, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.R.; Bates, J.C.; Avera, A.D.; Kim, Y. Relationship between Stemness, Reactive Oxygen Species, and Epithelial-to-Mesenchymal Transition in Model Circulating Tumor Cells. Cells Tissues Organs 2022, 211, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Nordin, J.Z.; Lee, Y.; Vader, P.; Mager, I.; Johansson, H.J.; Heusermann, W.; Wiklander, O.P.; Hallbrink, M.; Seow, Y.; Bultema, J.J.; et al. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine 2015, 11, 879–883. [Google Scholar] [CrossRef] [PubMed]
- McNamara, R.P.; Caro-Vegas, C.P.; Costantini, L.M.; Landis, J.T.; Griffith, J.D.; Damania, B.A.; Dittmer, D.P. Large-scale, cross-flow based isolation of highly pure and endocytosis-competent extracellular vesicles. J. Extracell. Vesicles 2018, 7, 1541396. [Google Scholar] [CrossRef] [PubMed]
- Mansur, S.; Habib, S.; Hawkins, M.; Brown, S.R.; Weinman, S.T.; Bao, Y. Preparation of Nanoparticle-Loaded Extracellular Vesicles Using Direct Flow Filtration. Pharmaceutics 2023, 15, 1551. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Deng, W.; Klinke, D.J., 2nd. Exosomes: Improved methods to characterize their morphology, RNA content, and surface protein biomarkers. Analyst 2015, 140, 6631–6642. [Google Scholar] [CrossRef]
- Triantafillu, U.L.; Park, S.; Klaassen, N.L.; Raddatz, A.D.; Kim, Y. Fluid shear stress induces cancer stem cell-like phenotype in MCF7 breast cancer cell line without inducing epithelial to mesenchymal transition. Int. J. Oncol. 2017, 50, 993–1001. [Google Scholar] [CrossRef] [PubMed]
- Park, H.A.; Brown, S.R.; Kim, Y. Cellular Mechanisms of Circulating Tumor Cells During Breast Cancer Metastasis. Int. J. Mol. Sci. 2020, 21, 5040. [Google Scholar] [CrossRef]
- Feng, Y.H.; Tsao, C.J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef]
- Yang, Y.; Chaerkady, R.; Beer, M.A.; Mendell, J.T.; Pandey, A. Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics 2009, 9, 1374–1384. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, W.; Li, J.; Song, E.; Liang, H.; Rong, W.; Jiang, X.; Xu, N.; Wang, W.; Qu, S.; et al. A Novel Pathway of Functional microRNA Uptake and Mitochondria Delivery. Adv. Sci. 2023, 10, e2300452. [Google Scholar] [CrossRef] [PubMed]
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Luongo, F.; Colonna, F.; Calapa, F.; Vitale, S.; Fiori, M.E.; de Maria, R. PTEN Tumor-Suppressor: The Dam of Stemness in Cancer. Cancers 2019, 11, 1076. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yang, H.S. The role of Pdcd4 in tumour suppression and protein translation. Biol. Cell 2018, 110, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Alavian, K.N.; Beutner, G.; Lazrove, E.; Sacchetti, S.; Park, H.A.; Licznerski, P.; Li, H.; Nabili, P.; Hockensmith, K.; Graham, M.; et al. An uncoupling channel within the c-subunit ring of the F1FO ATP synthase is the mitochondrial permeability transition pore. Proc. Natl. Acad. Sci. USA 2014, 111, 10580–10585. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, X.; Shan, T.; Ma, J.; Lin, W.; Li, W.; Kang, Y. MiR-21-mediated Metabolic Alteration of Cancer-associated Fibroblasts and Its Effect on Pancreatic Cancer Cell Behavior. Int. J. Biol. Sci. 2018, 14, 100–110. [Google Scholar] [CrossRef]
- Mnatsakanyan, N.; Park, H.A.; Wu, J.; He, X.; Llaguno, M.C.; Latta, M.; Miranda, P.; Murtishi, B.; Graham, M.; Weber, J.; et al. Mitochondrial ATP synthase c-subunit leak channel triggers cell death upon loss of its F(1) subcomplex. Cell Death Differ. 2022, 29, 1874–1887. [Google Scholar] [CrossRef]

| Primer | Sequence |
|---|---|
| ACTB forward | GCC CTG GAC TTC GAG CAA GAG A |
| ACTB reverse | ATG GTG ATG GAC CTG GCC GTC A |
| ATP5A1 forward | TGG AGC CCA GCA AGA TTA CA |
| ATP5A1 reverse | TGA TAG TGC CCA ACA AGG CT |
| ATP5B forward | CGC AAA CAT CTC CTT CGC CA |
| ATP5B reverse | AGT CCC TCA TCA AAC TGG ACG |
| ATP5G1 forward | GTG AGT CTG TCA CCT TGA GCC |
| ATP5G1 reverse | CTG CAC TCC TAC TAC CCT GCA A |
| ATP5G2 forward | GTC AAG AGC ACC TCA CAG C |
| ATP5G2 reverse | TCT GTC AGT ATC TCC GGT CGT |
| ATP5G3 forward | TTA ATG GGG CCC AGA ATG GTG |
| ATP5G3 reverse | CCA GCC ACT CCT ACT GTT GC |
| ATPAF1 forward | TAT GTG CTC TGC CAA GAA GGG |
| ATPAF1 reverse | AAG TGG AGT TCA GTA CCT GTC C |
| ATPAF2 forward | TTG AAG AAA CTG GGC GTG TC |
| ATPAF2 reverse | CTG CTT GAA CAT TCC TCA GCC |
| ATP-BL forward | ACC TCA CAT CTG ACC CTG GA |
| ATP-BL reverse | AAG CTT CCC TTC TTG GCC TC |
| NANOG forward | AAT ACC TCA GCC TCC AGC AGA TG |
| NANOG reverse | TGC GTC ACA CCA TTG CTA TTC TTC |
| OCT4 forward | GAG AAC CGA GTG AGA GGC AAC C |
| OCT4 reverse | CAT AGT CGC TGC TTG ATC GCT TG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brown, S.R.; Radcliffe, M.E.; Danner, J.T.; Andújar Cruz, W.J.; Lackey, K.H.; Park, H.-A.; Weinman, S.T.; Kim, Y. Extracellular Vesicle-Mediated Modulation of Stem-like Phenotype in Breast Cancer Cells under Fluid Shear Stress. Biomolecules 2024, 14, 757. https://doi.org/10.3390/biom14070757
Brown SR, Radcliffe ME, Danner JT, Andújar Cruz WJ, Lackey KH, Park H-A, Weinman ST, Kim Y. Extracellular Vesicle-Mediated Modulation of Stem-like Phenotype in Breast Cancer Cells under Fluid Shear Stress. Biomolecules. 2024; 14(7):757. https://doi.org/10.3390/biom14070757
Chicago/Turabian StyleBrown, Spenser R., Margaret E. Radcliffe, Joseph T. Danner, Wilmer J. Andújar Cruz, Kimberly H. Lackey, Han-A Park, Steven T. Weinman, and Yonghyun Kim. 2024. "Extracellular Vesicle-Mediated Modulation of Stem-like Phenotype in Breast Cancer Cells under Fluid Shear Stress" Biomolecules 14, no. 7: 757. https://doi.org/10.3390/biom14070757
APA StyleBrown, S. R., Radcliffe, M. E., Danner, J. T., Andújar Cruz, W. J., Lackey, K. H., Park, H.-A., Weinman, S. T., & Kim, Y. (2024). Extracellular Vesicle-Mediated Modulation of Stem-like Phenotype in Breast Cancer Cells under Fluid Shear Stress. Biomolecules, 14(7), 757. https://doi.org/10.3390/biom14070757









