Shotgun Proteomics Links Proteoglycan-4+ Extracellular Vesicles to Cognitive Protection in Amyotrophic Lateral Sclerosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Quantification of EVs by Nanoparticle Tracking Analysis (NTA)
2.3. Scanning Electron Microscopy (SEM)
2.4. Western Blot Analysis
2.5. Proteomics Analysis
2.6. Analysis of PRG-4 Expression in EVs of ALS Patients and HCs
2.7. Statistical Analysis
3. Results
3.1. Clinical Characteristics
3.2. The Size and Concentration of Plasmatic EVs Are Similar between ALS Patients and HCs
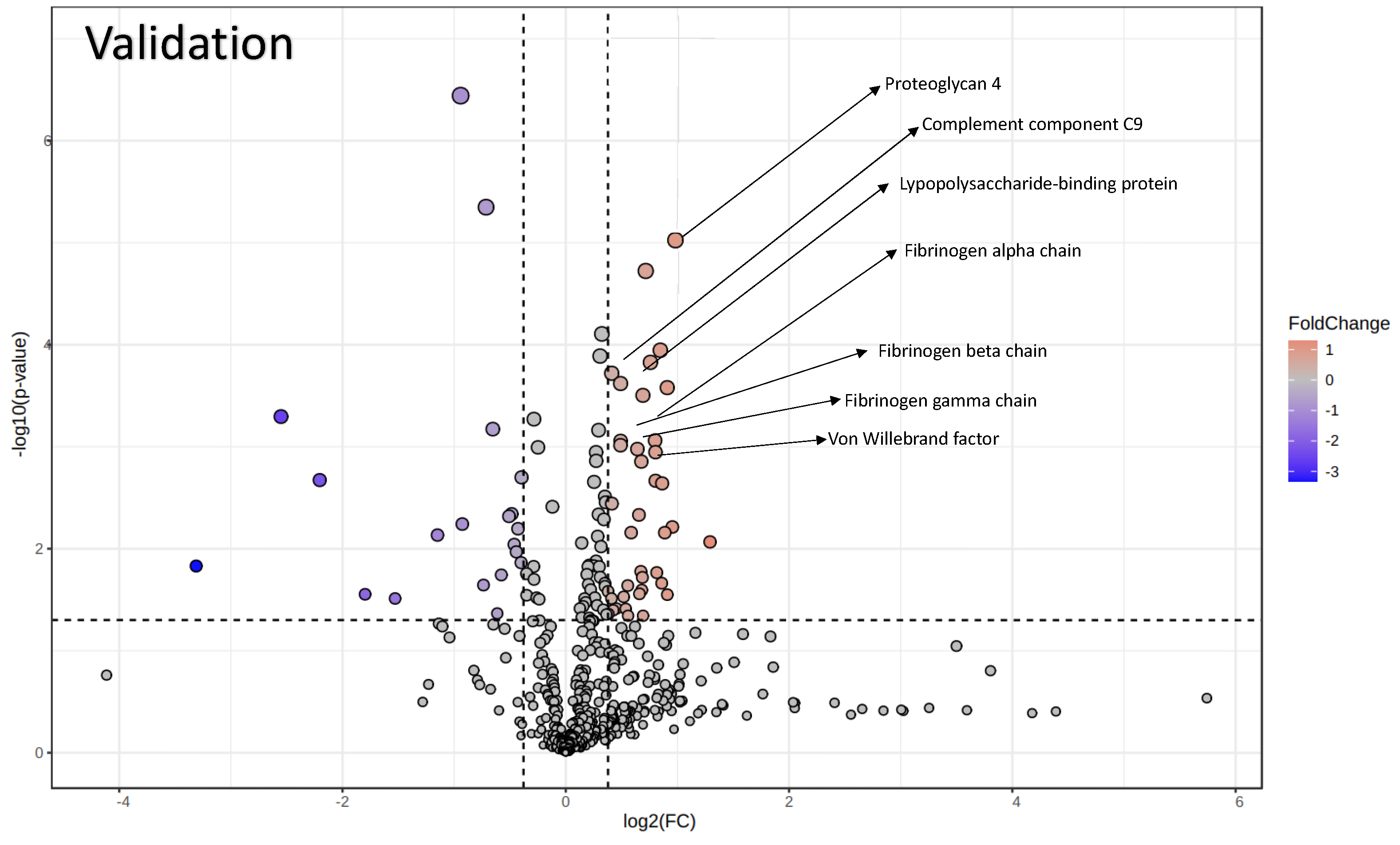
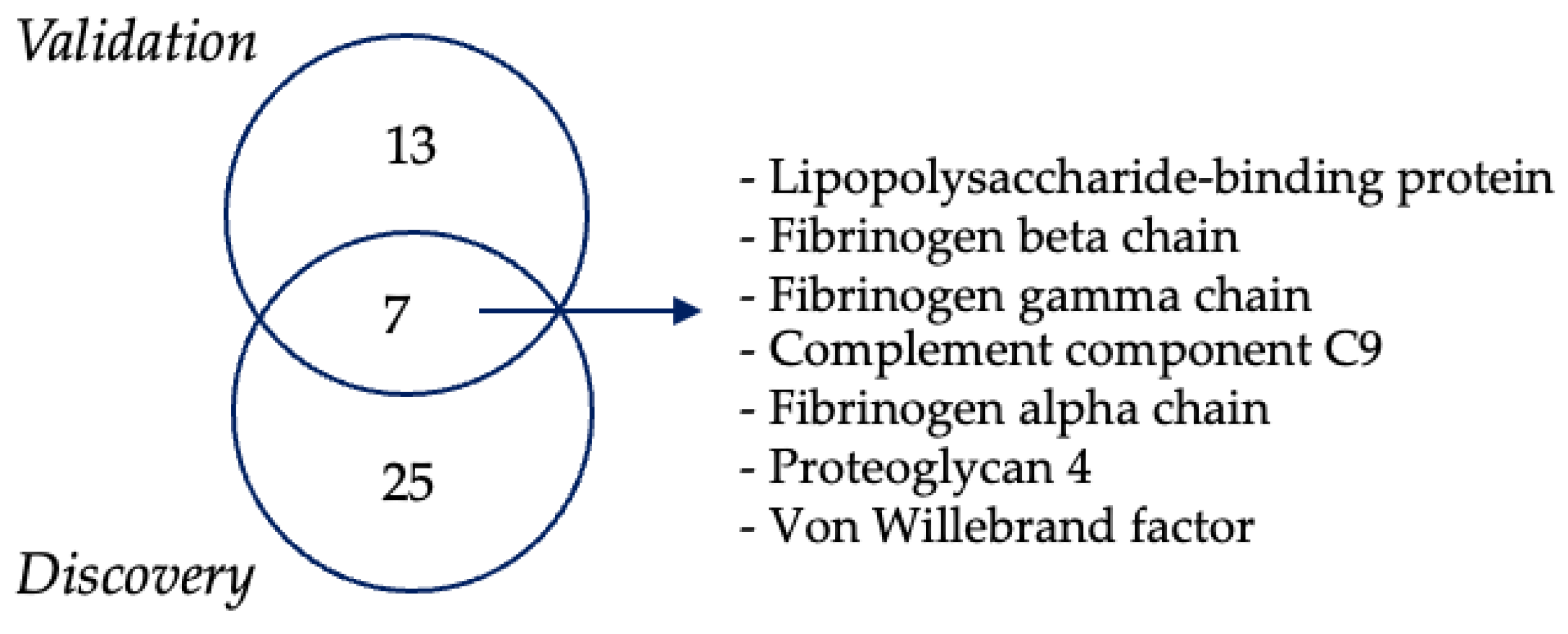
3.3. Proteomics Analysis of EVs
3.4. PRG-4-Enriched EVs Are Associated with Normal Cognitive Function in ALS Patients
3.5. ELISA Confirms Increased PRG-4 Expression Levels in EVs of ALS Patients Compared to HCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rojas, P.; Ramírez, A.I.; Fernández-Albarral, J.A.; López-Cuenca, I.; Salobrar-García, E.; Cadena, M.; Elvira-Hurtado, L.; Salazar, J.J.; de Hoz, R.; Ramírez, J.M. Amyotrophic Lateral Sclerosis: A Neurodegenerative Motor Neuron Disease With Ocular Involvement. Front. Neurosci. 2020, 14, 566858. [Google Scholar] [CrossRef]
- Feldman, E.L.; A Goutman, S.; Petri, S.; Mazzini, L.; Savelieff, M.G.; Shaw, P.J.; Sobue, G. Amyotrophic lateral sclerosis. Lancet 2022, 400, 1363–1380. [Google Scholar] [CrossRef]
- Zarei, S.; Carr, K.; Reiley, L.; Diaz, K.; Guerra, O.; Altamirano, P.; Pagani, W.; Lodin, D.; Orozco, G.; Chinea, A. A comprehensive review of amyotrophic lateral sclerosis. Surg. Neurol. Int. 2015, 6, 171. [Google Scholar] [CrossRef]
- Blasco, H.; Mavel, S.; Corcia, P.; Gordon, P.H. The glutamate hypothesis in ALS: Pathophysiology and drug development. Curr. Med. Chem. 2014, 21, 3551–3575. [Google Scholar] [CrossRef]
- Chen, J.; Bassot, A.; Giuliani, F.; Simmen, T. Amyotrophic Lateral Sclerosis (ALS): Stressed by Dysfunctional Mitochondria-Endoplasmic Reticulum Contacts (MERCs). Cells 2021, 10, 1789. [Google Scholar] [CrossRef]
- Ross, C.; Poirier, M. Protein aggregation and neurodegenerative disease. Nat. Med. 2004, 10, S10–S17. [Google Scholar] [CrossRef]
- Chua, J.P.; De Calbiac, H.; Kabashi, E.; Barmada, S.J. Autophagy and ALS: Mechanistic insights and therapeutic implications. Autophagy 2022, 18, 254–282. [Google Scholar] [CrossRef]
- Lee, S.; Kim, H.-J. Prion-like Mechanism in Amyotrophic Lateral Sclerosis: Are Protein Aggregates the Key? Exp. Neurobiol. 2015, 24, 1–7. [Google Scholar] [CrossRef]
- Komine, O.; Yamanaka, K. Neuroinflammation in motor neuron disease. Nagoya J. Med. Sci. 2015, 77, 537–549. [Google Scholar]
- Delic, V.; Kurien, C.; Cruz, J.; Zivkovic, S.; Barretta, J.; Thomson, A.; Hennessey, D.; Joseph, J.; Ehrhart, J.; Willing, A.E.; et al. Discrete mitochondrial aberrations in the spinal cord of sporadic ALS patients. J. Neurosci. Res. 2018, 96, 1353–1366. [Google Scholar] [CrossRef]
- Kuo, J.J.; Siddique, T.; Fu, R.; Heckman, C.J. Increased persistent Na+ current and its effect on excitability in motoneurones cultured from mutant SOD1 mice. J. Physiol. 2005, 563, 843–854. [Google Scholar] [CrossRef]
- Hammad, M.; Silva, A.; Glass, J.; Sladky, J.T.; Benatar, M. Clinical, electrophysiologic, and pathologic evidence for sensory abnormalities in ALS. Neurology 2007, 69, 2236–2242. [Google Scholar] [CrossRef]
- Pugdahl, K.; Fuglsang-Frederiksen, A.; de Carvalho, M.; Johnsen, B.; Fawcett, P.R.W.; Labarre-Vila, A.; Liguori, R.; A Nix, W.; Schofield, I.S. Generalised sensory system abnormalities in amyotrophic lateral sclerosis: A European multicentre study. J. Neurol. Neurosurg. Psychiatry 2007, 78, 746–749. [Google Scholar] [CrossRef]
- García, D.J.L.S.; Sleutjes, B.T.H.M.; van Schelven, L.J.; Goedee, H.S.; van den Berg, L.H. Diagnostic accuracy of nerve excitability and compound muscle action potential scan derived biomarkers in amyotrophic lateral sclerosis. Eur. J. Neurol. 2023, 30, 3068–3078. [Google Scholar] [CrossRef]
- Nishijima, H.; Tomiyama, M.; Suzuki, C.; Kon, T.; Funamizu, Y.; Ueno, T.; Haga, R.; Miki, Y.; Arai, A.; Kimura, T.; et al. Amyotrophic Lateral Sclerosis with Demyelinating Neuropathy. Intern. Med. 2012, 51, 1917–1921. [Google Scholar] [CrossRef]
- Pender, N.; Pinto-Grau, M.; Hardiman, O. Cognitive and behavioural impairment in amyotrophic lateral sclerosis. Curr. Opin. Neurol. 2020, 33, 649–654. [Google Scholar] [CrossRef]
- Baron, D.M.; Fenton, A.R.; Saez-Atienzar, S.; Giampetruzzi, A.; Sreeram, A.; Shankaracharya; Keagle, P.J.; Doocy, V.R.; Smith, N.J.; Danielson, E.W.; et al. ALS-associated KIF5A mutations abolish autoinhibition resulting in a toxic gain of function. Cell Rep. 2022, 39, 110598. [Google Scholar] [CrossRef]
- De Marchi, F.; Franjkic, T.; Schito, P.; Russo, T.; Nimac, J.; Chami, A.A.; Mele, A.; Vidatic, L.; Kriz, J.; Julien, J.-P.; et al. Emerging Trends in the Field of Inflammation and Proteinopathy in ALS/FTD Spectrum Disorder. Biomedicines 2023, 11, 1599. [Google Scholar] [CrossRef]
- Oh, S.; Jang, Y.; Na, C.H. Discovery of Biomarkers for Amyotrophic Lateral Sclerosis from Human Cerebrospinal Fluid Using Mass-Spectrometry-Based Proteomics. Biomedicines 2023, 11, 1250. [Google Scholar] [CrossRef]
- Witzel, S.; Mayer, K.; Oeckl, P. Biomarkers for amyotrophic lateral sclerosis. Curr. Opin. Neurol. 2022, 35, 699–704. [Google Scholar] [CrossRef]
- Sproviero, D.; La Salvia, S.; Giannini, M.; Crippa, V.; Gagliardi, S.; Bernuzzi, S.; Diamanti, L.; Ceroni, M.; Pansarasa, O.; Poletti, A.; et al. Pathological Proteins Are Transported by Extracellular Vesicles of Sporadic Amyotrophic Lateral Sclerosis Patients. Front. Neurosci. 2018, 12, 487. [Google Scholar] [CrossRef]
- Maione, F.; Cappellano, G.; Bellan, M.; Raineri, D. Chicken-or-egg question: Which came first, extracellular vesicles or autoimmune diseases? J. Leukoc. Biol. 2020, 108, 601–616. [Google Scholar] [CrossRef]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; You, S.; Spinelli, C.; Morley, S.; Zandian, M.; Aspuria, P.-J.; Cavallini, L.; Ciardiello, C.; Sobreiro, M.R.; Morello, M.; et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget 2015, 6, 11327–11341. [Google Scholar] [CrossRef]
- Vassileff, N.; Vella, L.J.; Rajapaksha, H.; Shambrook, M.; Kenari, A.N.; McLean, C.; Hill, A.F.; Cheng, L. Revealing the Proteome of Motor Cortex Derived Extracellular Vesicles Isolated from Amyotrophic Lateral Sclerosis Human Postmortem Tissues. Cells 2020, 9, 1709. [Google Scholar] [CrossRef]
- Abramowicz, A.; Widłak, P.; Pietrowska, M. Different Types of Cellular Stress Affect the Proteome Composition of Small Extracellular Vesicles: A Mini Review. Proteomes 2019, 7, 23. [Google Scholar] [CrossRef]
- Strong, M.J.; Grace, G.M.; Freedman, M.; Lomen-Hoerth, C.; Woolley, S.; Goldstein, L.H.; Murphy, J.; Shoesmith, C.; Rosenfeld, J.; Leigh, P.N.; et al. Consensus criteria for the diagnosis of frontotemporal cognitive and behavioural syndromes in amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 2009, 10, 131–146. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Manfredi, M.; Martinotti, S.; Gosetti, F.; Ranzato, E.; Marengo, E. The secretome signature of malignant mesothelioma cell lines. J. Proteom. 2016, 145, 3–10. [Google Scholar] [CrossRef]
- Sódar, B.W.; Kittel, Á.; Pálóczi, K.; Vukman, K.V.; Osteikoetxea, X.; Szabó-Taylor, K.; Németh, A.; Sperlágh, B.; Baranyai, T.; Giricz, Z.; et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci. Rep. 2016, 6, 24316. [Google Scholar] [CrossRef]
- Hallal, S.; Tűzesi, Á.; Grau, G.E.; Buckland, M.E.; Alexander, K.L. Understanding the extracellular vesicle surface for clinical molecular biology. J. Extracell. Vesicles 2022, 11, e12260. [Google Scholar] [CrossRef]
- Kooijmans, S.A.A.; Aleza, C.G.; Roffler, S.R.; van Solinge, W.W.; Vader, P.; Schiffelers, R.M. Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J. Extracell. Vesicles 2016, 5, 31053. [Google Scholar] [CrossRef]
- Al-Sharif, A.; Jamal, M.; Zhang, L.X.; Larson, K.; Schmidt, T.A.; Jay, G.D.; Elsaid, K.A. Lubricin/Proteoglycan 4 Binding to CD44 Receptor: A Mechanism of the Suppression of Proinflammatory Cytokine-Induced Synoviocyte Proliferation by Lubricin. Arthritis Rheumatol. 2015, 67, 1503–1513. [Google Scholar] [CrossRef]
- Brennan, K.; Iversen, K.F.; Blanco-Fernández, A.; Lund, T.; Plesner, T.; Gee, M.M.M. Extracellular Vesicles Isolated from Plasma of Multiple Myeloma Patients Treated with Daratumumab Express CD38, PD-L1, and the Complement Inhibitory Proteins CD55 and CD59. Cells 2022, 11, 21. [Google Scholar] [CrossRef]
- Tsuchiya, M.; Ohashi, Y.; Kodera, Y.; Satoh, M.; Matsui, T.; Fukushima, K.; Iwase, D.; Aikawa, J.; Mukai, M.; Inoue, G.; et al. CD39+CD55− Fb Subset Exhibits Myofibroblast-Like Phenotype and Is Associated with Pain in Osteoarthritis of the Knee. Biomedicines 2023, 11, 3047. [Google Scholar] [CrossRef]
- Lane, R.E.; Korbie, D.; Hill, M.M.; Trau, M. Extracellular vesicles as circulating cancer biomarkers: Opportunities and challenges. Clin. Transl. Med. 2018, 7, 14. [Google Scholar] [CrossRef]
- Raghav, A.; Singh, M.; Jeong, G.-B.; Giri, R.; Agarwal, S.; Kala, S.; Gautam, K.A. Extracellular vesicles in neurodegenerative diseases: A systematic review. Front. Mol. Neurosci. 2022, 15, 1061076. [Google Scholar] [CrossRef]
- Petersen, M.A.; Ryu, J.K.; Akassoglou, K. Fibrinogen in neurological diseases: Mechanisms, imaging and therapeutics. Nat. Rev. Neurosci. 2018, 19, 283–301. [Google Scholar] [CrossRef]
- Keizman, D.; Rogowski, O.; Berliner, S.; Ish-Shalom, M.; Maimon, N.; Nefussy, B.; Artamonov, I.; Drory, V.E. Low-grade systemic inflammation in patients with amyotrophic lateral sclerosis. Acta Neurol. Scand. 2009, 119, 383–389. [Google Scholar] [CrossRef]
- Pronto-Laborinho, A.C.; Lopes, C.S.; Conceição, V.A.; Gromicho, M.; Santos, N.C.; de Carvalho, M.; Carvalho, F.A. γ’ Fibrinogen as a Predictor of Survival in Amyotrophic Lateral Sclerosis. Front. Cardiovasc. Med. 2021, 8, 715842. [Google Scholar] [CrossRef]
- Kjældgaard, A.-L.; Pilely, K.; Olsen, K.S.; Lauritsen, A.; Pedersen, S.W.; Svenstrup, K.; Karlsborg, M.; Thagesen, H.; Blaabjerg, M.; Theódórsdóttir, Á.; et al. Complement Profiles in Patients with Amyotrophic Lateral Sclerosis: A Prospective Observational Cohort Study. J. Inflamm. Res. 2021, 14, 1043–1053. [Google Scholar] [CrossRef]
- Karasu, E.; Eisenhardt, S.U.; Harant, J.; Huber-Lang, M. Extracellular Vesicles: Packages Sent With Complement. Front. Immunol. 2018, 9, 721. [Google Scholar] [CrossRef]
- Sta, M.; Sylva-Steenland, R.M.R.; Casula, M.; de Jong, J.M.B.V.; Troost, D.; Aronica, E.; Baas, F. Innate and adaptive immunity in amyotrophic lateral sclerosis: Evidence of complement activation. Neurobiol. Dis. 2011, 42, 211–220. [Google Scholar] [CrossRef]
- Peyvandi, F.; Garagiola, I.; Baronciani, L. Role of von Willebrand factor in the haemostasis. Blood Transfus. 2011, 9, s3–s8. [Google Scholar] [CrossRef]
- Vu, L.; Garcia-Mansfield, K.; Pompeiano, A.; An, J.; David-Dirgo, V.; Sharma, R.; Venugopal, V.; Halait, H.; Marcucci, G.; Kuo, Y.; et al. Proteomics and mathematical modeling of longitudinal CSF differentiates fast versus slow ALS progression. Ann. Clin. Transl. Neurol. 2023, 10, 2025–2042. [Google Scholar] [CrossRef]
- Zhou, J.; Zeng, Q.; Liao, Q.; Niu, Q.; Gu, W.; Su, D.; Li, S.; Xiao, B.; Bi, F. Biomarkers in cerebrospinal fluid for amyotrophic lateral sclerosis phenotypes. Ann. Clin. Transl. Neurol. 2023, 10, 1467–1480. [Google Scholar] [CrossRef]
- Jay, G.D.; Waller, K.A. The biology of lubricin: Near frictionless joint motion. Matrix Biol. 2014, 39, 17–24. [Google Scholar] [CrossRef]
- Gámez-Valero, A.; Campdelacreu, J.; Vilas, D.; Ispierto, L.; Reñé, R.; Álvarez, R.; Armengol, M.P.; Borràs, F.E.; Beyer, K. Exploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer’s disease and dementia with Lewy bodies. Transl. Neurodegener. 2019, 8, 31. [Google Scholar] [CrossRef]
- Watson, L.S.; Hamlett, E.D.; Stone, T.D.; Sims-Robinson, C. Neuronally derived extracellular vesicles: An emerging tool for understanding Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 22. [Google Scholar] [CrossRef]
- Al Kaabi, A.; Traupe, T.; Stutz, M.; Buchs, N.; Heller, M. Cause or effect of arteriogenesis: Compositional alterations of microparticles from CAD patients undergoing external counterpulsation therapy. PLoS ONE 2012, 7, e46822. [Google Scholar] [CrossRef]
- Whitham, M.; Parker, B.L.; Friedrichsen, M.; Hingst, J.R.; Hjorth, M.; Hughes, W.E.; Egan, C.L.; Cron, L.; Watt, K.I.; Kuchel, R.P.; et al. Extracellular Vesicles Provide a Means for Tissue Crosstalk during Exercise. Cell Metab. 2018, 27, 237–251.e4. [Google Scholar] [CrossRef]
- Manek, R.; Moghieb, A.; Yang, Z.; Kumar, D.; Kobessiy, F.; Sarkis, G.A.; Raghavan, V.; Wang, K.K.W. Protein Biomarkers and Neuroproteomics Characterization of Microvesicles/Exosomes from Human Cerebrospinal Fluid Following Traumatic Brain Injury. Mol. Neurobiol. 2018, 55, 6112–6128. [Google Scholar] [CrossRef]
- György, B.; Szabó, T.G.; Turiák, L.; Wright, M.; Herczeg, P.; Lédeczi, Z.; Kittel, Á.; Polgár, A.; Tóth, K.; Dérfalvi, B.; et al. Improved flow cytometric assessment reveals distinct microvesicle (cell-derived microparticle) signatures in joint diseases. PLoS ONE 2012, 7, e49726. [Google Scholar] [CrossRef]
- Liu, X.; Chinello, C.; Musante, L.; Cazzaniga, M.; Tataruch, D.; Calzaferri, G.; Smith, A.J.; De Sio, G.; Magni, F.; Zou, H.; et al. Intraluminal proteome and peptidome of human urinary extracellular vesicles. Proteom. Clin. Appl. 2015, 9, 568–573. [Google Scholar] [CrossRef]
- Looze, C.; Yui, D.; Leung, L.; Ingham, M.; Kaler, M.; Yao, X.; Wu, W.W.; Shen, R.-F.; Daniels, M.P.; Levine, S.J. Proteomic profiling of human plasma exosomes identifies PPARgamma as an exosome-associated protein. Biochem. Biophys. Res. Commun. 2009, 378, 433–438. [Google Scholar] [CrossRef]
- Sarker, S.; Scholz-Romero, K.; Perez, A.; Illanes, S.E.; Mitchell, M.D.; Rice, G.E.; Salomon, C. Placenta-derived exosomes continuously increase in maternal circulation over the first trimester of pregnancy. J. Transl. Med. 2014, 12, 204. [Google Scholar] [CrossRef]
- Toda, H.; Diaz-Varela, M.; Segui-Barber, J.; Roobsoong, W.; Baro, B.; Garcia-Silva, S.; Galiano, A.; Gualdrón-López, M.; Almeida, A.C.G.; Brito, M.A.M.; et al. Plasma-derived extracellular vesicles from Plasmodium vivax patients signal spleen fibroblasts via NF-kB facilitating parasite cytoadherence. Nat. Commun. 2020, 11, 2761. [Google Scholar] [CrossRef]
- El-Shennawy, L.; Hoffmann, A.D.; Dashzeveg, N.K.; McAndrews, K.M.; Mehl, P.J.; Cornish, D.; Yu, Z.; Tokars, V.L.; Nicolaescu, V.; Tomatsidou, A.; et al. Circulating ACE2-expressing extracellular vesicles block broad strains of SARS-CoV-2. Nat. Commun. 2022, 13, 405. [Google Scholar] [CrossRef]
- Bereman, M.S.; Beri, J.; Enders, J.R.; Nash, T. Machine Learning Reveals Protein Signatures in CSF and Plasma Fluids of Clinical Value for ALS. Sci. Rep. 2018, 8, 16334. [Google Scholar] [CrossRef]
- Bennett, M.; Chin, A.; Lee, H.J.; Cestero, E.M.; Strazielle, N.; Ghersi-Egea, J.-F.; Threlkeld, S.W.; Schmidt, T.A.; Richendrfer, H.A.; Szmydynger-Chodobska, J.; et al. Proteoglycan 4 Reduces Neuroinflammation and Protects the Blood-Brain Barrier after Traumatic Brain Injury. J. Neurotrauma 2021, 38, 385–398. [Google Scholar] [CrossRef]
- Kakaroubas, N.; Brennan, S.; Keon, M.; Saksena, N.K. Pathomechanisms of Blood-Brain Barrier Disruption in ALS. Neurosci. J. 2019, 2019, 2537698. [Google Scholar] [CrossRef]
- Alkhalifa, A.E.; Al-Ghraiybah, N.F.; Odum, J.; Shunnarah, J.G.; Austin, N.; Kaddoumi, A. Blood–Brain Barrier Breakdown in Alzheimer’s Disease: Mechanisms and Targeted Strategies. Int. J. Mol. Sci. 2023, 24, 16288. [Google Scholar] [CrossRef]
- Barisano, G.; Montagne, A.; Kisler, K.; Schneider, J.A.; Wardlaw, J.M.; Zlokovic, B.V. Blood-brain barrier link to human cognitive impairment and Alzheimer’s Disease. Nat. Cardiovasc. Res. 2022, 1, 108–115. [Google Scholar] [CrossRef]




| ALS N = 61 | |
|---|---|
| N (%) | |
| Gender | |
| Male | 39 (63.93%) |
| Female | 22 (36.07%) |
| Form of the Disease | |
| Spinal | 41 (67.21%) |
| Bulbar | 20 (32.79%) |
| Phenotype | |
| Classic | 32 (52.46%) |
| Bulbar | 20 (32.79%) |
| PUMN | 6 (9.84%) |
| Respiratory | 3 (4.92%) |
| Cognitive Impairment | |
| Normal | 39 (63.93%) |
| ALS-ci | 20 (32.79%) |
| ALS-bi | 1 (1.64%) |
| FTD | 1 (1.64%) |
| Progression | |
| Slow | 34 (55.74%) |
| Fast | 27 (44.26%) |
| Age at blood collection, mean ± SD) | 62.88 ± 12.60 |
| Average monthly change in FVC, median (Q1–Q3) | −1.93 (−3.78; −1.00) |
| Average monthly change in BMI, median (Q1–Q3) | −0.02 (−0.16; 0.15) |
| Average monthly change in ALFRSR, median (Q1–Q3) | −0.60 (−1.31; −0.27) |
| Protein Name | Fold Change (FC) | p Value |
|---|---|---|
| P20742|PZP_HUMAN Pregnancy zone protein | 3.97 | 0.01 |
| Q8TDD5|MCLN3_HUMAN Mucolipin-3 | 2.00 | 0.02 |
| P18428|LBP_HUMAN Lipopolysaccharide-binding protein | 1.84 | 0.02 |
| Q92954|PRG4_HUMAN Proteoglycan 4 | 1.72 | 0.02 |
| P04275|VWF_HUMAN von Willebrand factor | 1.58 | 0.01 |
| P02679|FIBG_HUMAN Fibrinogen gamma chain | 1.45 | 0.01 |
| O00187|MASP2_HUMAN Mannan-binding lectin serine protease 2 | 1.44 | 0.02 |
| P22792|CPN2_HUMAN Carboxypeptidase N subunit 2 | 1.41 | 0.03 |
| P02675|FIBB_HUMAN Fibrinogen beta chain | 1.41 | 0.01 |
| P01023|A2MG_HUMAN Alpha-2-macroglobulin | 1.39 | 0.02 |
| P02751|FINC_HUMAN Fibronectin | 1.38 | 0.01 |
| P04003|C4BPA_HUMAN C4b-binding protein alpha chain | 1.37 | 0.01 |
| P02671|FIBA_HUMAN Fibrinogen alpha chain | 1.36 | 0.02 |
| P01024|CO3_HUMAN Complement C3 | 1.34 | 0.005 |
| P02748|CO9_HUMAN Complement component C9 | 1.33 | 0.02 |
| P01031|CO5_HUMAN Complement C5 | 1.31 | 0.02 |
| P07225|PROS_HUMAN Vitamin K-dependent protein S | 1.31 | 0.04 |
| P02747|C1QC_HUMAN Complement C1q subcomponent subunit C | 1.27 | 0.04 |
| P01009|A1AT_HUMAN Alpha-1-antitrypsin | 1.26 | 0.01 |
| P05156|CFAI_HUMAN Complement factor I | 1.23 | 0.01 |
| P05452|TETN_HUMAN Tetranectin | 0.79 | 0.01 |
| P04004|VTNC_HUMAN Vitronectin | 0.79 | 0.02 |
| P01344|IGF2_HUMAN Insulin-like growth factor II | 0.78 | 0.03 |
| P06396|GELS_HUMAN Gelsolin | 0.72 | 0.002 |
| P25311|ZA2G_HUMAN Zinc-alpha-2-glycoprotein | 0.70 | 0.01 |
| P05090|APOD_HUMAN Apolipoprotein D | 0.64 | 0.004 |
| P07477|TRY1_HUMAN Trypsin-1 | 0.39 | 0.01 |
| Q12805|FBLN3_HUMAN EGF-containing fibulin-like extracellular matrix protein 1 | 0.33 | 0.03 |
| P15924|DESP_HUMAN Desmoplakin | 0.29 | 0.02 |
| Q08188|TGM3_HUMAN Protein-glutamine gamma-glutamyltransferase E | 0.25 | 0.04 |
| P14923|PLAK_HUMAN Junction plakoglobin | 0.21 | 0.04 |
| P17931|LEG3_HUMAN Galectin-3 | 0.11 | 0.02 |
| Protein Name | Fold Change (FC) | p Value |
|---|---|---|
| Q92954|PRG4_HUMAN Proteoglycan 4 | 2.07 | 1.49 × 10−5 |
| O60306|AQR_HUMAN Intron-binding protein aquarius | 1.94 | 0.01 |
| P04275|VWF_HUMAN von Willebrand factor | 1.75 | 0.002 |
| P02671|FIBA_HUMAN Fibrinogen alpha chain | 1.75 | 0.001 |
| Q13103|SPP24_HUMAN Secreted phosphoprotein 24 | 1.74 | 0.009 |
| Q9BWP8|COL11_HUMAN Collectin-11 | 1.66 | 0.02 |
| Q9HDC9|APMAP_HUMAN Adipocyte plasma membrane-associated protein | 1.63 | 0.04 |
| P11169|GTR3_HUMAN Solute carrier family 2, facilitated glucose transporter member 3 | 1.60 | 0.02 |
| P61626|LYSC_HUMAN Lysozyme C | 1.58 | 0.043 |
| P02675|FIBB_HUMAN Fibrinogen beta chain | 1.58 | 0.005 |
| P18428|LBP_HUMAN Lipopolysaccharide-binding protein | 1.56 | 0.001 |
| P01833|PIGR_HUMAN Polymeric immunoglobulin receptor | 1.54 | 0.06 |
| P02679|FIBG_HUMAN Fibrinogen gamma chain | 1.50 | 0.007 |
| P01034|CYTC_HUMAN Cystatin-C | 1.47 | 0.02 |
| P02786|TFR1_HUMAN Transferrin receptor protein 1 | 1.45 | 0.04 |
| P02748|CO9_HUMAN Complement component C9 | 1.41 | 0.001 |
| Q6UXB8|PI16_HUMAN Peptidase inhibitor 16 | 1.34 | 0.04 |
| Q06033|ITIH3_HUMAN Inter-alpha-trypsin inhibitor heavy chain H3 | 1.28 | 0.02 |
| P02763|A1AG1_HUMAN Alpha-1-acid glycoprotein 1 | 1.26 | 0.04 |
| O00391|QSOX1_HUMAN Sulfhydryl oxidase 1 | 1.23 | 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vilardo, B.; De Marchi, F.; Raineri, D.; Manfredi, M.; De Giorgis, V.; Bebeti, A.; Scotti, L.; Kustrimovic, N.; Cappellano, G.; Mazzini, L.; et al. Shotgun Proteomics Links Proteoglycan-4+ Extracellular Vesicles to Cognitive Protection in Amyotrophic Lateral Sclerosis. Biomolecules 2024, 14, 727. https://doi.org/10.3390/biom14060727
Vilardo B, De Marchi F, Raineri D, Manfredi M, De Giorgis V, Bebeti A, Scotti L, Kustrimovic N, Cappellano G, Mazzini L, et al. Shotgun Proteomics Links Proteoglycan-4+ Extracellular Vesicles to Cognitive Protection in Amyotrophic Lateral Sclerosis. Biomolecules. 2024; 14(6):727. https://doi.org/10.3390/biom14060727
Chicago/Turabian StyleVilardo, Beatrice, Fabiola De Marchi, Davide Raineri, Marcello Manfredi, Veronica De Giorgis, Alen Bebeti, Lorenza Scotti, Natasa Kustrimovic, Giuseppe Cappellano, Letizia Mazzini, and et al. 2024. "Shotgun Proteomics Links Proteoglycan-4+ Extracellular Vesicles to Cognitive Protection in Amyotrophic Lateral Sclerosis" Biomolecules 14, no. 6: 727. https://doi.org/10.3390/biom14060727
APA StyleVilardo, B., De Marchi, F., Raineri, D., Manfredi, M., De Giorgis, V., Bebeti, A., Scotti, L., Kustrimovic, N., Cappellano, G., Mazzini, L., & Chiocchetti, A. (2024). Shotgun Proteomics Links Proteoglycan-4+ Extracellular Vesicles to Cognitive Protection in Amyotrophic Lateral Sclerosis. Biomolecules, 14(6), 727. https://doi.org/10.3390/biom14060727









