Pleiotropic Role of Rainbow Trout CXCRs in Response to Disease and Environment: Insights from Transcriptional Signatures and Structure Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Genome-Wide Identification and Sequence Analyses
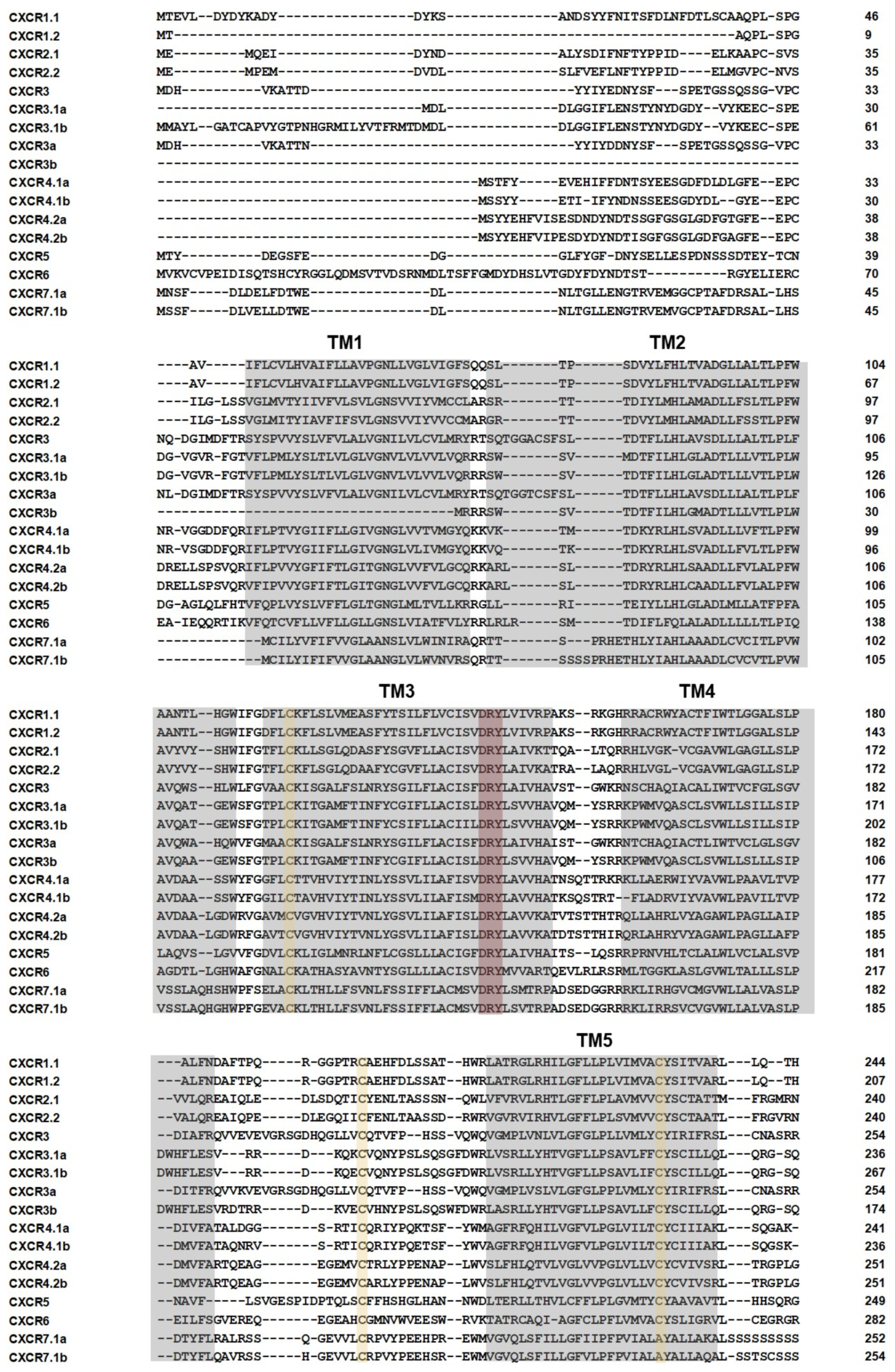
2.3. Gene Structure, Conserved Domains, and Motif Analysis of the CXCR
2.4. Expression Analysis Using Available RNA-Seq Datasets
- Brain, kidney, and spleen samples from rainbow trout challenged with Vibrio anguillarum (SRA ID: PRJNA667799 [40,41,42]). Brain, kidney, and spleen samples were collected from control, asymptomatic, and symptomatic rainbow trout after V. anguillarum challenge, and 27 libraries of RNA-Seq samples were used (3 phenotypes × 3 tissues × 3 replicates [40,42]).
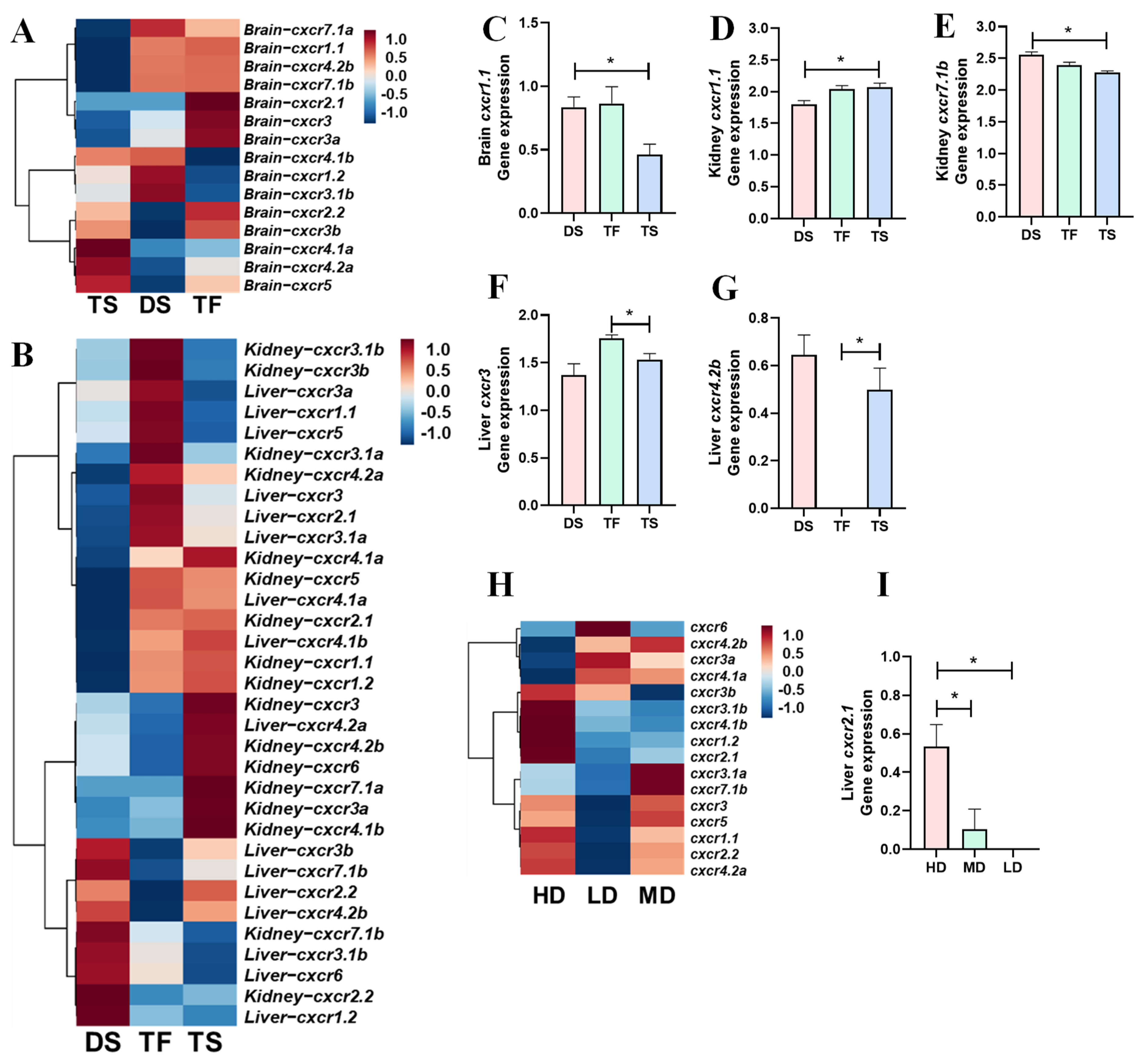
- Brain, kidney, and liver samples from rainbow trout with environmental salinity changes ([44]). Diploid and triploid trout were classified into diploid trout in freshwater (DF), diploid trout in saltwater (DS, at salinity of 15 parts-per-thousand (ppt)), triploid trout in freshwater (TF), and triploid trout in saltwater (TS, at salinity of 15 ppt). Brain, liver, and kidney samples were collected from DS, TS, and TF. Twenty-seven libraries of RNA-Seq samples were used (3 groups (TF, DF, DS) × 3 tissues × 3 replicates [44]).
- Liver samples from rainbow trout cultured in different stocking densities (unpublished data and count data are shown in Supplementary Materials). Rainbow trout were cultured in saltwater with initial densities at 9.15 kg/m3 (low density (LD)), 13.65 kg/m3 (moderate density (MD)), and 27.31 kg/m3 (high density (HD)) for 84 days. The final densities were 22.00 (LD), 32.05 (MD), and 52.24 (HD) kg/m3, respectively. Liver samples were collected from LD, MD, and HD on day 84.
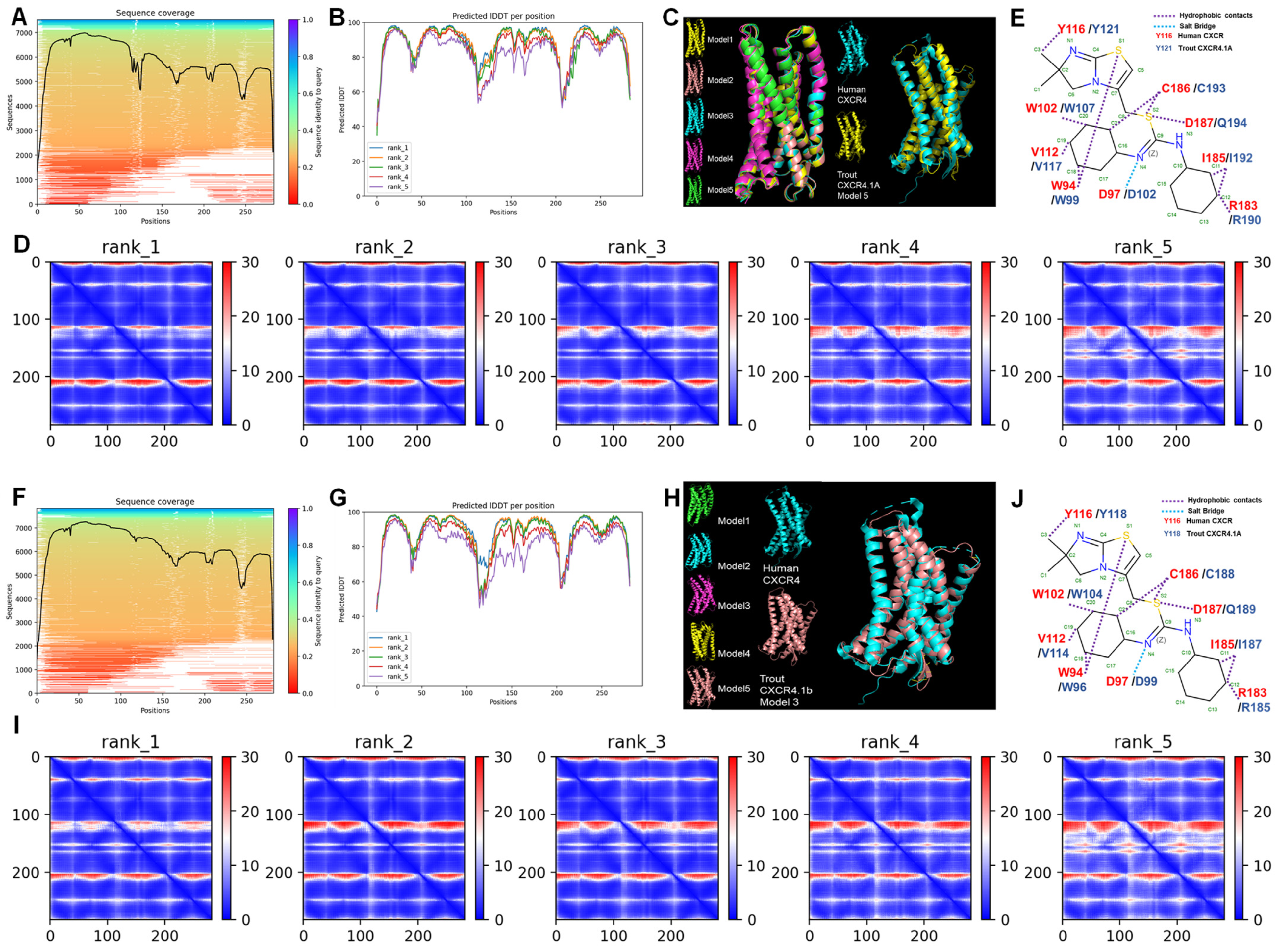
2.5. Structural Analysis of Trout CXCR4.1 Subtypes
2.6. Statistical Analysis
3. Results
3.1. Identification and Annotation of cxcr Genes in Rainbow Trout
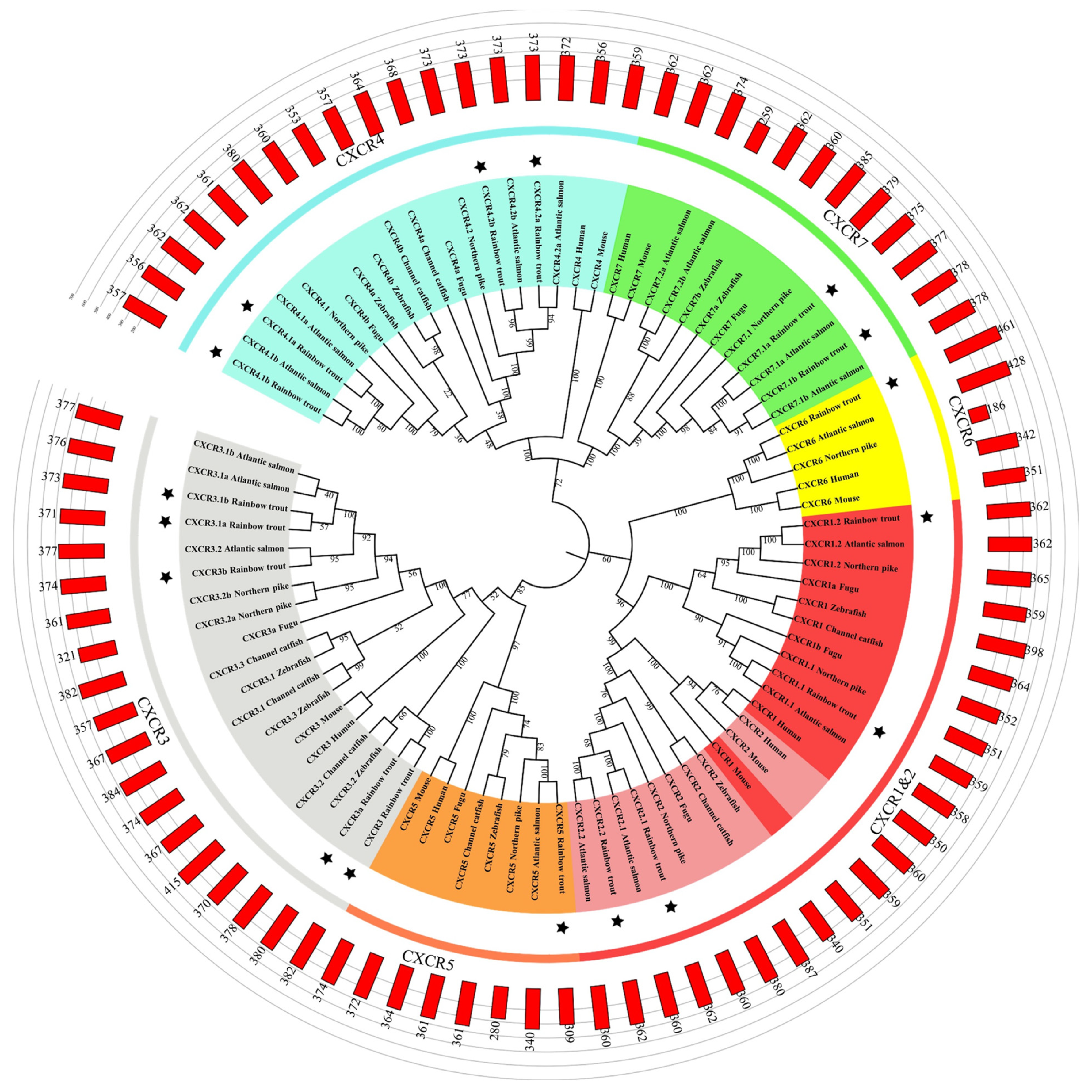
3.2. Phylogenetic Analysis and Gene Structure Analyses
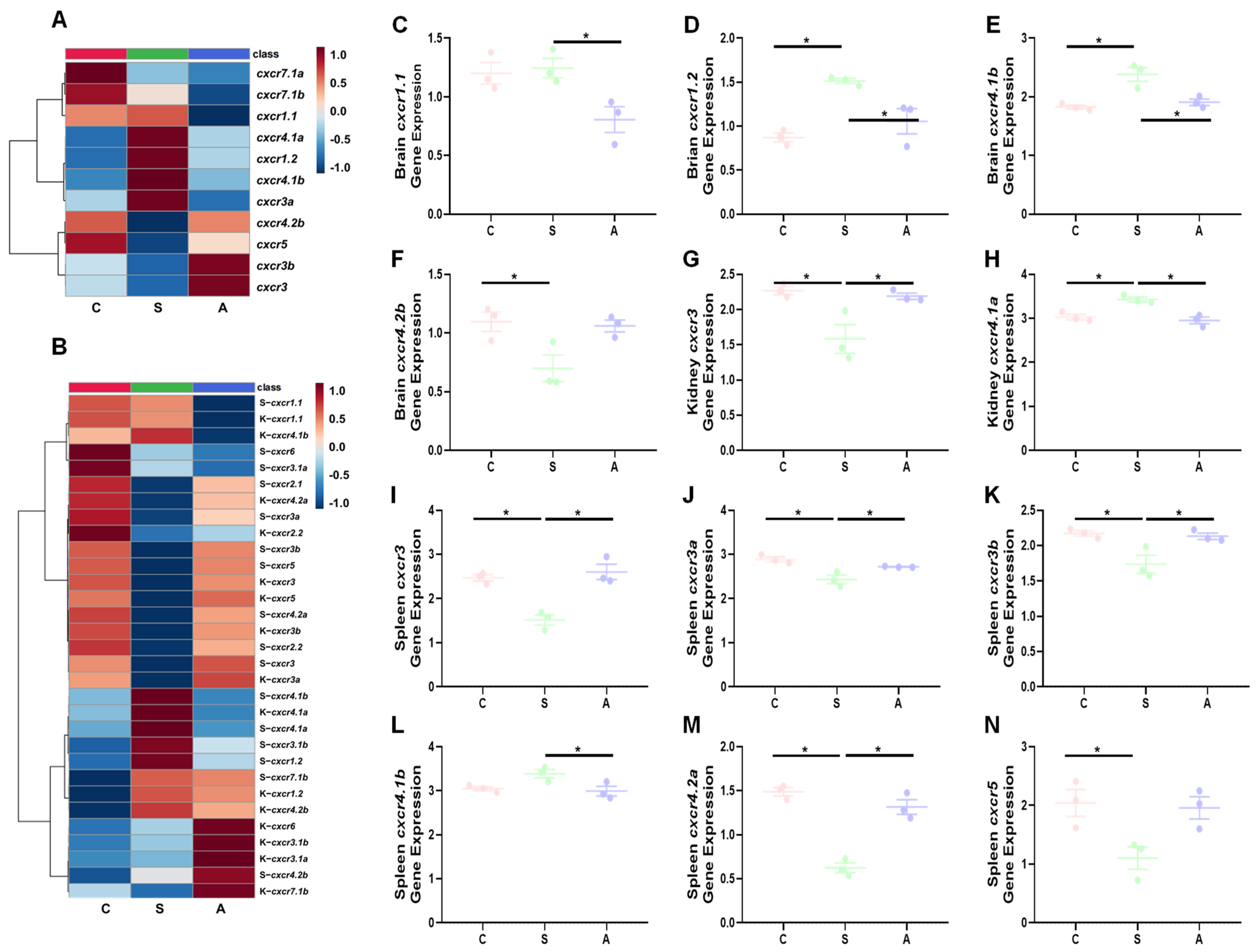
3.3. Transcriptional Profiles of cxcr in Trout after Bacterial Infection
3.3.1. V. anguillarum
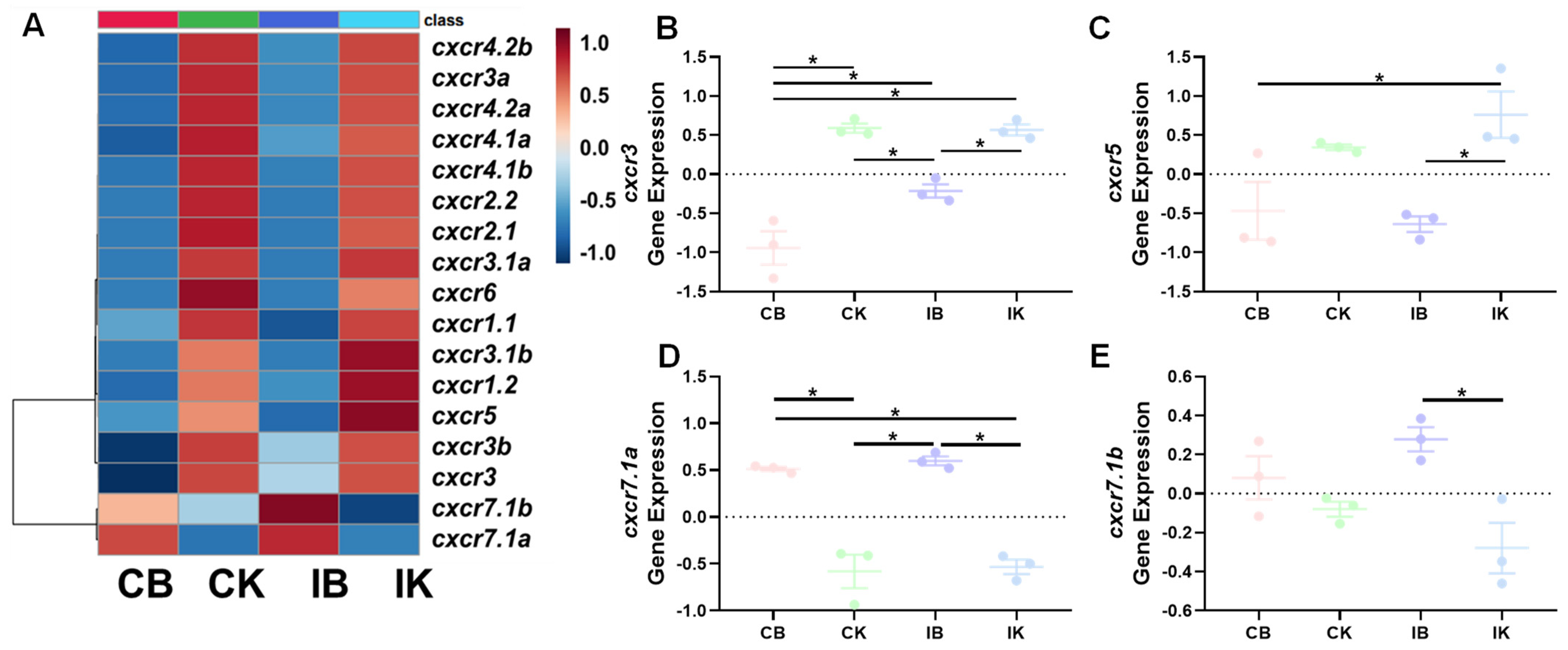
3.3.2. A. salmonicida
3.4. Transcriptional Profiles of cxcr in Trout in Response to Salinity Change and High Stocking Density
3.5. Structure Prediction of CXCR4.1a and CXCR4.1b
4. Discussion
4.1. Characterization of cxcr Genes
4.2. Physiological Functions of cxcr Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koelink, P.J.; Overbeek, S.A.; Braber, S.; de Kruijf, P.; Folkerts, G.; Smit, M.J.; Kraneveld, A.D. Targeting chemokine receptors in chronic inflammatory diseases: An extensive review. Pharmacol. Ther. 2012, 133, 1–18. [Google Scholar] [CrossRef]
- Allen, S.J.; Crown, S.E.; Handel, T.M. Chemokine: Receptor structure, interactions, and antagonism. Annu. Rev. Immunol. 2007, 25, 787–820. [Google Scholar] [CrossRef]
- Bacon, K.; Baggiolini, M.; Broxmeyer, H.; Horuk, R.; Lindley, I.; Mantovani, A.; Maysushima, K.; Murphy, P.; Nomiyama, H.; Oppenheim, J. Chemokine/chemokine receptor nomenclature. Cytokine 2003, 21, 48–49. [Google Scholar]
- Zlotnik, A.; Yoshie, O.; Nomiyama, H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006, 7, 243. [Google Scholar] [CrossRef]
- Zlotnik, A.; Burkhardt, A.M.; Homey, B. Homeostatic chemokine receptors and organ-specific metastasis. Nat. Rev. Immunol. 2011, 11, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Zlotnik, A.; Yoshie, O. Chemokines: A new classification system and their role in immunity. Immunity 2000, 12, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Yoshie, O.; Imai, T.; Nomiyama, H. Chemokines in immunity. In Advances in Immunology; Elsevier: Amsterdam, The Netherlands, 2001; pp. 57–110. [Google Scholar]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.-M.; Brunet, F.; Petit, J.-L.; Stange-Thomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozouf-Costaz, C.; Bernot, A.; et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef]
- Valdés, N.; Cortés, M.; Barraza, F.; Reyes-López, F.E.; Imarai, M. CXCL9-11 chemokines and CXCR3 receptor in teleost fish species. Fish Shellfish. Immunol. Rep. 2022, 3, 100068. [Google Scholar] [CrossRef]
- Dixon, B.; Shum, B.; Adams, E.J.; Magor, K.; Hedrick, R.P.; Muir, D.G.; Parham, P. CK-1, a putative chemokine of rainbow trout (Oncorhynchus mykiss). Immunol. Rev. 1998, 166, 341–348. [Google Scholar] [CrossRef]
- Peatman, E.; Liu, Z. Evolution of CC chemokines in teleost fish: A case study in gene duplication and implications for immune diversity. Immunogenetics 2007, 59, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chang, M.; Wu, S.; Nie, P. Characterization of C–C chemokine receptor subfamily in teleost fish. Mol. Immunol. 2009, 46, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Nomiyama, H.; Osada, N.; Yoshie, O. A family tree of vertebrate chemokine receptors for a unified nomenclature. Dev. Comp. Immunol. 2011, 35, 705–715. [Google Scholar] [CrossRef]
- Fu, Q.; Yang, Y.; Li, C.; Zeng, Q.; Zhou, T.; Li, N.; Liu, Y.; Liu, S.; Liu, Z. The CC and CXC chemokine receptors in channel catfish (Ictalurus punctatus) and their involvement in disease and hypoxia responses. Dev. Comp. Immunol. 2017, 77, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Leu, J.-H.; Tsai, C.-H.; Tsai, J.-M.; Yang, C.-H.; Hsueh, C.-Y.; Chou, H.-Y. Identification and expression analysis of 19 CC chemokine genes in orange-spotted grouper (Epinephelus coioides). Dev. Comp. Immunol. 2019, 97, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Thorgaard, G.H.; Bailey, G.S.; Williams, D.; Buhler, D.R.; Kaattari, S.L.; Ristow, S.S.; Hansen, J.D.; Winton, J.R.; Bartholomew, J.L.; Nagler, J.J.; et al. Status and opportunities for genomics research with rainbow trout. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2002, 133, 609–646. [Google Scholar] [CrossRef]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noël, B.; Bento, P.; Silva, C.D.; Labadie, K.; Alberti, A.; et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Valdés, N.; Gonzalez, A.; Garcia, V.; Tello, M. Analysis of the microbiome of rainbow trout (Oncorhynchus mykiss) exposed to the pathogen Flavobacterium psychrophilum 10094. Microbiol. Resour. Announc. 2020, 9, e01562-19. [Google Scholar] [CrossRef]
- Bowden, T.J. Modulation of the immune system of fish by their environment. Fish Shellfish. Immunol. 2008, 25, 373–383. [Google Scholar] [CrossRef]
- Makrinos, D.L.; Bowden, T.J. Natural environmental impacts on teleost immune function. Fish Shellfish. Immunol. 2016, 53, 50–57. [Google Scholar] [CrossRef]
- Macqueen, D.J.; Garcia de la serrana, D.; Johnston, I.A. Evolution of ancient functions in the vertebrate insulin-like growth factor system uncovered by study of duplicated salmonid fish genomes. Mol. Biol. Evol. 2013, 30, 1060–1076. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Thorgaard, G.H. Tetraploidy and the evolution of salmonid fishes. In Evolutionary Genetics of Fishes; Plenum Press: New York, NY, USA, 1984; pp. 1–53. [Google Scholar]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.R.; Kent, M.P.; Nome, T.; Hvidsten, T.R.; Leong, J.S.; Minkley, D.R.; Zimin, A.; et al. The Atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef]
- Macqueen, D.J.; Johnston, I.A. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc. R. Soc. B Biol. Sci. 2014, 281, 20132881. [Google Scholar] [CrossRef] [PubMed]
- Alzaid, A.; Martin, S.A.M.; Macqueen, D.J. The complete salmonid IGF-IR gene repertoire and its transcriptional response to disease. Sci. Rep. 2016, 6, 34806. [Google Scholar] [CrossRef] [PubMed]
- Alzaid, A.; Castro, R.; Wang, T.; Secombes, C.J.; Boudinot, P.; Macqueen, D.J.; Martin, S.A.M. Cross talk between growth and immunity: Coupling of the IGF axis to conserved cytokine pathways in rainbow trout. Endocrinology 2016, 157, 1942–1955. [Google Scholar] [CrossRef] [PubMed]
- Sequeida, A.; Castillo, A.; Cordero, N.; Wong, V.; Montero, R.; Vergara, C.; Valenzuela, B.; Vargas, D.; Valdés, N.; Morales, J.; et al. The Atlantic salmon interleukin 4/13 receptor family: Structure, tissue distribution and modulation of gene expression. Fish Shellfish. Immunol. 2020, 98, 773–787. [Google Scholar] [CrossRef]
- Conant, G.C.; Wolfe, K.H. Turning a hobby into a job: How duplicated genes find new functions. Nat. Rev. Genet. 2008, 9, 938–950. [Google Scholar] [CrossRef]
- Taylor, J.S.; Raes, J. Duplication and divergence: The evolution of new genes and old ideas. Annu. Rev. Genet. 2004, 38, 615–643. [Google Scholar] [CrossRef]
- Rebl, A.; Korytář, T.; Köbis, J.M.; Verleih, M.; Krasnov, A.; Jaros, J.; Kühn, C.; Köllner, B.; Goldammer, T. Transcriptome profiling reveals insight into distinct immune responses to Aeromonas salmonicida in gill of two rainbow trout strains. Mar. Biotechnol. 2014, 16, 333–348. [Google Scholar] [CrossRef]
- Frans, I.; Michiels, C.W.; Bossier, P.; Willems, K.A.; Lievens, B.; Rediers, H. Vibrio anguillarum as a fish pathogen: Virulence factors, diagnosis and prevention. J. Fish Dis. 2011, 34, 643–661. [Google Scholar] [CrossRef]
- Valdes, N.; Espinoza, C.; Sanhueza, L.; Gonzalez, A.; Corsini, G.; Tello, M. Draft genome sequence of the Chilean isolate Aeromonas salmonicida strain CBA100. FEMS Microbiol. Lett. 2015, 362, fnu062. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Bao, P.; Tang, B. Transcriptome analysis and discovery of genes involved in immune pathways in large yellow croaker (Larimichthys crocea) under high stocking density stress. Fish Shellfish. Immunol. 2017, 68, 332–340. [Google Scholar] [CrossRef]
- San, L.; Liu, B.; Liu, B.; Guo, H.; Guo, L.; Zhang, N.; Zhu, K.; Jiang, S.; Zhang, D. Transcriptome analysis of gills provides insights into translation changes under hypoxic stress and reoxygenation in golden pompano, Trachinotus ovatus (Linnaeus 1758). Front. Mar. Sci. 2021, 8, 763622. [Google Scholar] [CrossRef]
- de Fonseka, R.; Fjelldal, P.G.; Sambraus, F.; Nilsen, T.O.; Remø, S.C.; Stien, L.H.; Reinardy, H.C.; Madaro, A.; Hansen, T.J.; Fraser, T.W.K. Triploidy leads to a mismatch of smoltification biomarkers in the gill and differences in the optimal salinity for post-smolt growth in Atlantic salmon. Aquaculture 2022, 546, 737350. [Google Scholar] [CrossRef]
- Galbreath, P.F.; Thorgaard, G.H. Saltwater performance of all-female triploid Atlantic salmon. Aquaculture 1995, 138, 77–85. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37 (Suppl. 2), W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.-S.; Xin, Y.-R.; Zeng, C.; Zhao, H.-K.; Tian, Y.; Li, J.-F.; Wen, H.-S. GHRH-SST-GH-IGF Axis Regulates Crosstalk between Growth and Immunity in Rainbow Trout (Oncorhynchus mykiss) Infected with Vibrio anguillarum. Fish Shellfish. Immunol. 2020, 106, 887–897. [Google Scholar] [CrossRef]
- Zeng, C.; Hou, Z.-S.; Zhao, H.-K.; Xin, Y.-R.; Liu, M.-Q.; Yang, X.-D.; Wen, H.-S.; Li, J.-F. Identification and characterization of caspases genes in rainbow trout (Oncorhynchus mykiss) and their expression profiles after Aeromonas salmonicida and Vibrio anguillarum infection. Dev. Comp. Immunol. 2021, 118, 103987. [Google Scholar] [CrossRef]
- Hou, Z.-S.; Xin, Y.-R.; Yang, X.-D.; Zeng, C.; Zhao, H.-K.; Liu, M.-Q.; Zhang, M.-Z.; Daniel, J.G.; Li, J.-F.; Wen, H.-S. Transcriptional profiles of genes related to stress and immune response in Rainbow trout (Oncorhynchus mykiss) symptomatically or asymptomatically infected with Vibrio anguillarum. Front. Immunol. 2021, 12, 967. [Google Scholar] [CrossRef]
- Liu, M.; Yang, X.; Zeng, C.; Zhao, H.; Li, J.; Hou, Z.; Wen, H. Transcriptional Signatures of Immune, Neural, and Endocrine Functions in the Brain and Kidney of Rainbow Trout (Oncorhynchus mykiss) in Response to Aeromonas salmonicida Infection. Int. J. Mol. Sci. 2022, 23, 1340. [Google Scholar] [CrossRef] [PubMed]
- Xiang, K.; Yang, Q.; Liu, M.; Yang, X.; Li, J.; Hou, Z.; Wen, H. Crosstalk between Growth and Osmoregulation of GHRH-SST-GH-IGF Axis in Triploid Rainbow Trout (Oncorhynchus mykiss). Int. J. Mol. Sci. 2022, 23, 8691. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Yuan, S.; Chan, H.S.; Filipek, S.; Vogel, H. PyMOL and Inkscape bridge the data and the data visualization. Structure 2016, 24, 2041–2042. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Love, M.; Anders, S.; Huber, W. Beginner’s guide to using the DESeq2 package. Genome Biol. 2014, 15, 550. [Google Scholar]
- Johnson, K.A.; Krishnan, A. Robust normalization and transformation techniques for constructing gene coexpression networks from RNA-seq data. Genome Biol. 2022, 23, 1. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using metaboanalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Mouton, A.J.; Ma, Y.; Rivera Gonzalez, O.J.; Daseke, M.J., II; Flynn, E.R.; Freeman, T.C.; Garrett, M.R.; DeLeon-Pennell, K.Y.; Lindsey, M.L. Fibroblast polarization over the myocardial infarction time continuum shifts roles from inflammation to angiogenesis. Basic Res. Cardiol. 2019, 114, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Soufan, O.; Xia, J.; Tang, R.; Li, L.; Li, D. Transcriptome and physiological analysis reveal alterations in muscle metabolisms and immune responses of grass carp (Ctenopharyngodon idellus) cultured at different stocking densities. Aquaculture 2019, 503, 186–197. [Google Scholar] [CrossRef]
- Bird, S.; Tafalla, C. Teleost chemokines and their receptors. Biology 2015, 4, 756–784. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liu, F. Advances in chemokines of teleost fish species. Aquac. Fish. 2023, 9, 115–125. [Google Scholar] [CrossRef]
- Qi, Z.; Xu, Y.; Dong, B.; Pi, X.; Zhang, Q.; Wang, D.; Wang, Z. Molecular characterization, structural and expression analysis of twelve CXC chemokines and eight CXC chemokine receptors in largemouth bass (Micropterus salmoides). Dev. Comp. Immunol. 2023, 143, 104673. [Google Scholar] [CrossRef]
- Zhang, H.; Thorgaard, G.H.; Ristow, S.S. Molecular cloning and genomic structure of an interleukin-8 receptor-like gene from homozygous clones of rainbow trout (Oncorhynchus mykiss). Fish Shellfish. Immunol. 2002, 13, 251–258. [Google Scholar] [CrossRef]
- Xu, Q.; Li, R.; Monte, M.M.; Jiang, Y.; Nie, P.; Holland, J.W.; Secombes, C.J.; Wang, T. Sequence and expression analysis of rainbow trout CXCR2, CXCR3a and CXCR3b aids interpretation of lineage-specific conversion, loss and expansion of these receptors during vertebrate evolution. Dev. Comp. Immunol. 2014, 45, 201–213. [Google Scholar] [CrossRef]
- Daniels, G.D.; Zou, J.; Charlemagne, J.; Partula, S.; Cunningham, C.; Secombes, C.J. Cloning of two chemokine receptor homologs (CXC-R4 and CC-R7) in rainbow trout Oncorhynchus mykiss. J. Leukoc. Biol. 1999, 65, 684–690. [Google Scholar] [CrossRef]
- Grimholt, U.; Hauge, H.; Hauge, A.G.; Leong, J.; Koop, B.F. Chemokine receptors in Atlantic salmon. Dev. Comp. Immunol. 2015, 49, 79–95. [Google Scholar] [CrossRef]
- Alejo, A.; Tafalla, C. Chemokines in teleost fish species. Dev. Comp. Immunol. 2011, 35, 1215–1222. [Google Scholar] [CrossRef]
- Nomiyama, H.; Hieshima, K.; Osada, N.; Kato-Unoki, Y.; Otsuka-Ono, K.; Takegawa, S.; Izawa, T.; Yoshizawa, A.; Kikuchi, Y.; Tanase, S.; et al. Extensive expansion and diversification of the chemokine gene family in zebrafish: Identification of a novel chemokine subfamily CX. BMC Genom. 2008, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- DeVries, M.E.; Kelvin, A.A.; Xu, L.; Ran, L.; Robinson, J.; Kelvin, D.J. Defining the origins and evolution of the chemokine/chemokine receptor system. J. Immunol. 2006, 176, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Nomiyama, H.; Osada, N.; Yoshie, O. The evolution of mammalian chemokine genes. Cytokine Growth Factor Rev. 2010, 21, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu, Q.; Wang, T.; Collet, B.; Corripio-Miyar, Y.; Bird, S.; Xie, P.; Nie, P.; Secombes, C.J.; Zou, J. Phylogenetic analysis of vertebrate CXC chemokines reveals novel lineage specific groups in teleost fish. Dev. Comp. Immunol. 2013, 41, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Zlotnik, A.; Yoshie, O. The chemokine superfamily revisited. Immunity 2012, 36, 705–716. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, P.; Gao, C.; Cao, M.; Yang, N.; Li, X.; Li, C.; Fu, Q. CXC chemokines and their receptors in black rockfish (Sebastes schlegelii): Characterization, evolution analyses, and expression pattern after Aeromonas salmonicida infection. Int. J. Biol. Macromol. 2021, 186, 109–124. [Google Scholar] [CrossRef]
- Lefkowitz, R.J. Seven transmembrane receptors: Something old, something new. Acta Physiol. 2007, 190, 9–19. [Google Scholar] [CrossRef]
- Umasuthan, N.; Wan, Q.; Revathy, K.S.; Whang, I.; Noh, J.K.; Kim, S.; Park, M.-A.; Lee, J. Molecular aspects, genomic arrangement and immune responsive mRNA expression profiles of two CXC chemokine receptor homologs (CXCR1 and CXCR2) from rock bream, Oplegnathus fasciatus. Fish Shellfish. Immunol. 2014, 40, 304–318. [Google Scholar] [CrossRef]
- Liu, X.; Kang, L.; Liu, W.; Lou, B.; Wu, C.; Jiang, L. Molecular characterization and expression analysis of the large yellow croaker (Larimichthys crocea) chemokine receptors CXCR2, CXCR3, and CXCR4 after bacterial and poly I: C challenge. Fish Shellfish. Immunol. 2017, 70, 228–239. [Google Scholar] [CrossRef]
- Xu, T.; Zhu, Z.; Sun, Y.; Ren, L.; Wang, R. Characterization and expression of the CXCR1 and CXCR4 in miiuy croaker and evolutionary analysis shows the strong positive selection pressures imposed in mammal CXCR1. Dev. Comp. Immunol. 2014, 44, 133–144. [Google Scholar] [CrossRef]
- Fu, Q.; Yang, Y.; Li, C.; Zeng, Q.; Zhou, T.; Li, N.; Liu, Y.; Li, Y.; Wang, X.; Liu, S.; et al. The chemokinome superfamily: II. The 64 CC chemokines in channel catfish and their involvement in disease and hypoxia responses. Dev. Comp. Immunol. 2017, 73, 97–108. [Google Scholar] [CrossRef]
- Danik, M.; Puma, C.; Quirion, R.; Williams, S. Widely expressed transcripts for chemokine receptor CXCR1 in identified glutamatergic, γ-aminobutyric acidergic, and cholinergic neurons and astrocytes of the rat brain: A single-cell reverse transcription-multiplex polymerase chain reaction study. J. Neurosci. Res. 2003, 74, 286–295. [Google Scholar] [CrossRef]
- Puma, C.; Danik, M.; Quirion, R.; Ramon, F.; Williams, S. The chemokine interleukin-8 acutely reduces Ca2+ currents in identified cholinergic septal neurons expressing CXCR1 and CXCR2 receptor mRNAs. J. Neurochem. 2001, 78, 960–971. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Redmond, A.K.; Qi, Z.; Dooley, H.; Secombes, C.J. The CXC chemokine receptors of fish: Insights into CXCR evolution in the vertebrates. Gen. Comp. Endocrinol. 2015, 215, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Li, S.; Xu, Q.; Zhu, D.; Zhang, Q.; Luo, K.; Zhang, W. Molecular characterization and expression analysis of Asian swamp eel (Monopterus albus) CXC chemokine receptor (CXCR) 1a, CXCR1b, CXCR2, CXCR3a, CXCR3b, and CXCR4 after bacteria and poly I: C challenge. Fish Shellfish. Immunol. 2019, 84, 572–586. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, Y.; Cao, M.; Yang, N.; Hu, J.; Xue, T.; Li, C.; Fu, Q. The CC and CXC chemokine receptors in turbot (Scophthalmus maximus L.) and their response to Aeromonas salmonicida infection. Dev. Comp. Immunol. 2021, 123, 104155. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, S.H.; Van Der Meer, P.; Hesselgesser, J.; Jaffer, S.; Kolson, D.L.; Albright, A.V.; González-Scarano, F.; Lavi, E. CXCR3 expression in human central nervous system diseases. Neuropathol. Appl. Neurobiol. 2001, 27, 127–138. [Google Scholar] [CrossRef]
- Soto, H.; Wang, W.; Strieter, R.M.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Hedrick, J.; Zlotnik, A. The CC chemokine 6Ckine binds the CXC chemokine receptor CXCR3. Proc. Natl. Acad. Sci. USA 1998, 95, 8205–8210. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.X.; Sun, B.J.; Nie, P. The first non-mammalian CXCR3 in a teleost fish: Gene and expression in blood cells and central nervous system in the grass carp (Ctenopharyngodon idella). Mol. Immunol. 2007, 44, 1123–1134. [Google Scholar] [CrossRef]
- Aquilino, C.; Castro, R.; Fischer, U.; Tafalla, C. Transcriptomic responses in rainbow trout gills upon infection with viral hemorrhagic septicemia virus (VHSV). Dev. Comp. Immunol. 2014, 44, 12–20. [Google Scholar] [CrossRef]
- Wang, T.; Hanington, P.C.; Belosevic, M.; Secombes, C.J. Two macrophage colony-stimulating factor genes exist in fish that differ in gene organization and are differentially expressed. J. Immunol. 2008, 181, 3310–3322. [Google Scholar] [CrossRef] [PubMed]
- Ehlert, J.E.; Addison, C.A.; Burdick, M.D.; Kunkel, S.L.; Strieter, R.M. Identification and partial characterization of a variant of human CXCR3 generated by posttranscriptional exon skipping. J. Immunol. 2004, 173, 6234–6240. [Google Scholar] [CrossRef]
- Wu, Q.; Dhir, R.; Wells, A. Altered CXCR3 isoform expression regulates prostate cancer cell migration and invasion. Mol. Cancer 2012, 11, 1–16. [Google Scholar] [CrossRef]
- Müller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef]
- Teicher, B.A.; Fricker, S.P. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Cancer Res. 2010, 16, 2927–2931. [Google Scholar] [CrossRef]
- Oberlin, E.; Amara, A.; Bachelerie, F.; Bessia, C.; Virelizier, J.-L.; Arenzana-Seisdedos, F.; Schwartz, O.; Heard, J.-M.; Clark-Lewis, I.; Legler, D.F.; et al. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature 1996, 382, 833–835. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Broder, C.C.; Kennedy, P.E.; Berger, E.A. HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 1996, 272, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-Y.; Chen, Y.-M.; Hsu, H.-H.; Shiu, C.-T.; Kuo, H.-C.; Chen, T.-Y. Grouper (Epinephelus coioides) CXCR4 is expressed in response to pathogens infection and early stage of development. Dev. Comp. Immunol. 2012, 36, 112–120. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, X.L.; Chen, L.P.; Chen, N.; Li, Y.H.; Wang, W.M.; Wang, H.L. Sequence analysis and expression differentiation of chemokine receptor CXCR4b among three populations of Megalobrama amblycephala. Dev. Comp. Immunol. 2013, 40, 195–201. [Google Scholar] [CrossRef]
- Xu, Q.Q.; Chang, M.X.; Sun, R.H.; Xiao, F.S.; Nie, P. The first non-mammalian CXCR5 in a teleost fish: Molecular cloning and expression analysis in grass carp (Ctenopharyngodon idella). BMC Immunol. 2010, 11, 25. [Google Scholar] [CrossRef]
- Kizil, C.; Dudczig, S.; Kyritsis, N.; Machate, A.; Blaesche, J.; Kroehne, V.; Brand, M. The chemokine receptor cxcr5 regulates the regenerative neurogenesis response in the adult zebrafish brain. Neural Dev. 2012, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Chien, E.Y.T.; Mol, C.D.; Fenalti, G.; Liu, W.; Katritch, V.; Abagyan, R.; Brooun, A.; Wells, P.; Bi, F.C.; et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 2010, 330, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- Skerlj, R.T.; Bridger, G.J.; Kaller, A.; McEachern, E.J.; Crawford, J.B.; Zhou, Y.; Atsma, B.; Langille, J.; Nan, S.; Veale, D.; et al. Discovery of Novel Small Molecule Orally Bioavailable C− X− C Chemokine Receptor 4 Antagonists That Are Potent Inhibitors of T-Tropic (X4) HIV-1 Replication. J. Med. Chem. 2010, 53, 3376–3388. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene ID | Chromosome | Position (bp) | Protein Length (aa) | MW (kDa) | pI | Derived from | Chemokines [10,55,56,57] | Reference |
|---|---|---|---|---|---|---|---|---|---|
| cxcr1.1 | LOC100135914 | Chr18 | 3,898,505–3,899,881 | 359 | 39.98 | 9.2 | AF260964.1 | CXCL8 | [55,58] |
| cxcr1.2 | LOC110501285 | Chr22 | 5,736,707–5,742,123 | 362 | 40.1 | 8.31 | Newly Identified | CXCL8 | |
| cxcr2.1 | LOC110520605 | Chr3 | 78,906,276–78,908,301 | 362 | 40.3 | 8.94 | HG794530.1 | CXCL8 | [55,59] |
| cxcr2.2 | LOC110501383 | Chr22 | 11,509,532–11,512,348 | 362 | 40.07 | 8.78 | Newly Identified | CXCL8 | |
| cxcr3.1a | LOC110537629 | Chr2 | 7,072,125–7,076,959 | 371 | 41.7 | 6.08 | Newly Identified | CXCL9, 10, 11 | |
| cxcr3.1b | LOC110514317 | Chr2 | 102–3242 | 373 | 41.94 | 5.88 | Newly Identified | CXCL9, 10, 11 | |
| cxcr3b | LOC100136126 | Chr3 | 16,782,015–16,785,624 | 374 | 42.19 | 8.17 | AJ888881.1 | CXCL9, 10, 11 | [10,55,56,59] |
| cxcr3 | LOC110537622 | Chr2 | 7,038,779–7,047,349 | 382 | 42.37 | 9.1 | Newly Identified | CXCL9, 10, 11 | |
| cxcr3a | LOC100136649 | Chr3 | 16,764,415–16,768,463 | 380 | 42.27 | 9.24 | AJ888878.1 | CXCL9, 10, 11 | [10,55,56,59] |
| cxcr4.1a | LOC110520024 | Chr3 | 48,667,827–48,669,883 | 362 | 40.57 | 8.74 | AJ001039.1 | CXCL12 | [55,60] |
| cxcr4.1b | LOC110501543 | Chr22 | 18,785,356–18,787,371 | 357 | 39.99 | 8.86 | Newly Identified | CXCL12 | |
| cxcr4.2a | LOC110530627 | Chr8 | 70,897,183–70,922,718 | 373 | 40.82 | 8.58 | Newly Identified | CXCL12 | |
| cxcr4.2b | LOC110516585 | Chr28 | 1208–2756 | 373 | 41.08 | 8.54 | Newly Identified | CXCL12 | |
| cxcr5 | LOC110503290 | Chr24 | 38,512,905–38,516,651 | 309 | 33.92 | 5.96 | Newly Identified | CXCL13 | |
| cxcr6 | LOC110492888 | Chr16 | 21,961,814–21,965,417 | 461 | 50.85 | 8.68 | Newly Identified | ||
| cxcr7.1a | LOC110520437 | Chr3 | 70,094,498–70,106,981 | 375 | 42.01 | 6.94 | Newly Identified | CXCL11, 12 | |
| cxcr7.1b | LOC110501640 | Chr22 | 25,651,985–25,660,915 | 378 | 42.22 | 6.66 | Newly Identified | CXCL11, 12 |
| Name | Human | Mouse | Chicken | Frog | Zebrafish | Channel Catfish | Atlantic Salmon | Fugu | Northern Pike | Rainbow Trout |
|---|---|---|---|---|---|---|---|---|---|---|
| cxcr1 | 1 (~14%) | 1 (~14%) | 1 (~33%) | 0 | 1 (10%) | 1 (~11%) | 2 (~11%) | 2 (25%) | 2 (20%) | 2 (~11%) |
| cxcr2 | 1 (~14%) | 1 (~14%) | 0 | 0 | 1 (10%) | 1 (~11%) | 2 (~11%) | 1 (12.5%) | 1 (10%) | 2 (~11%) |
| cxcr3 | 1 (~14%) | 1 (~14%) | 0 | 1 (25%) | 3 (30%) | 3 (~33%) | 3 (~17%) | 1 (12.5%) | 2 (20%) | 5 (~29%) |
| cxcr4 | 1 (~14%) | 1 (~14%) | 1 (~33%) | 1 (25%) | 2 (20%) | 2 (~22%) | 4 (~23%) | 2 (25%) | 2 (20%) | 4 (~23%) |
| cxcr5 | 1 (~14%) | 1 (~14%) | 1 (~33%) | 1 (25%) | 1 (10%) | 1 (~11%) | 1 (~5%) | 1 (12.5%) | 1 (10%) | 1 (~5%) |
| cxcr6 | 1 (~14%) | 1 (~14%) | 0 | 0 | 0 | 0 | 1 (~5%) | 0 | 1 (10%) | 1 (~5%) |
| cxcr7 | 1 (~14%) | 1 (~14%) | 0 | 1 (25%) | 2 (20%) | 1 (~11%) | 4 (~23%) | 1 (12.5%) | 1 (10%) | 2 (~11%) |
| Total | 7 | 7 | 3 | 4 | 10 | 9 | 17 | 8 | 10 | 17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Z.-S.; Zhao, H.-K.; Perdiguero, P.; Liu, M.-Q.; Xiang, K.-W.; Zeng, C.; Li, Z.; Yang, X.-D.; Yang, Q.; Xin, Y.-R.; et al. Pleiotropic Role of Rainbow Trout CXCRs in Response to Disease and Environment: Insights from Transcriptional Signatures and Structure Analysis. Biomolecules 2024, 14, 337. https://doi.org/10.3390/biom14030337
Hou Z-S, Zhao H-K, Perdiguero P, Liu M-Q, Xiang K-W, Zeng C, Li Z, Yang X-D, Yang Q, Xin Y-R, et al. Pleiotropic Role of Rainbow Trout CXCRs in Response to Disease and Environment: Insights from Transcriptional Signatures and Structure Analysis. Biomolecules. 2024; 14(3):337. https://doi.org/10.3390/biom14030337
Chicago/Turabian StyleHou, Zhi-Shuai, Hong-Kui Zhao, Pedro Perdiguero, Meng-Qun Liu, Kai-Wen Xiang, Chu Zeng, Zhao Li, Xiao-Dong Yang, Qian Yang, Yuan-Ru Xin, and et al. 2024. "Pleiotropic Role of Rainbow Trout CXCRs in Response to Disease and Environment: Insights from Transcriptional Signatures and Structure Analysis" Biomolecules 14, no. 3: 337. https://doi.org/10.3390/biom14030337
APA StyleHou, Z.-S., Zhao, H.-K., Perdiguero, P., Liu, M.-Q., Xiang, K.-W., Zeng, C., Li, Z., Yang, X.-D., Yang, Q., Xin, Y.-R., Li, J.-F., Tafalla, C., & Wen, H.-S. (2024). Pleiotropic Role of Rainbow Trout CXCRs in Response to Disease and Environment: Insights from Transcriptional Signatures and Structure Analysis. Biomolecules, 14(3), 337. https://doi.org/10.3390/biom14030337






