Biocontrol Potential of Sodin 5, Type 1 Ribosome-Inactivating Protein from Salsola soda L. Seeds
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Protein Purification
2.3. Analytical Methods
2.4. Polynucleotide:Adenosine Glycosidase Activity
2.5. rRNA N-Glycosylase Assay
2.6. Endonuclease Activity on Supercoiled pUC18 DNA
2.7. Fungal Cultures
2.8. Antifungal Activity
2.9. Cytotoxic Activity
2.10. N-Terminal Amino Acid Sequencing
2.11. Mass Spectrometry
2.12. Statistical Analysis
3. Results and Discussion
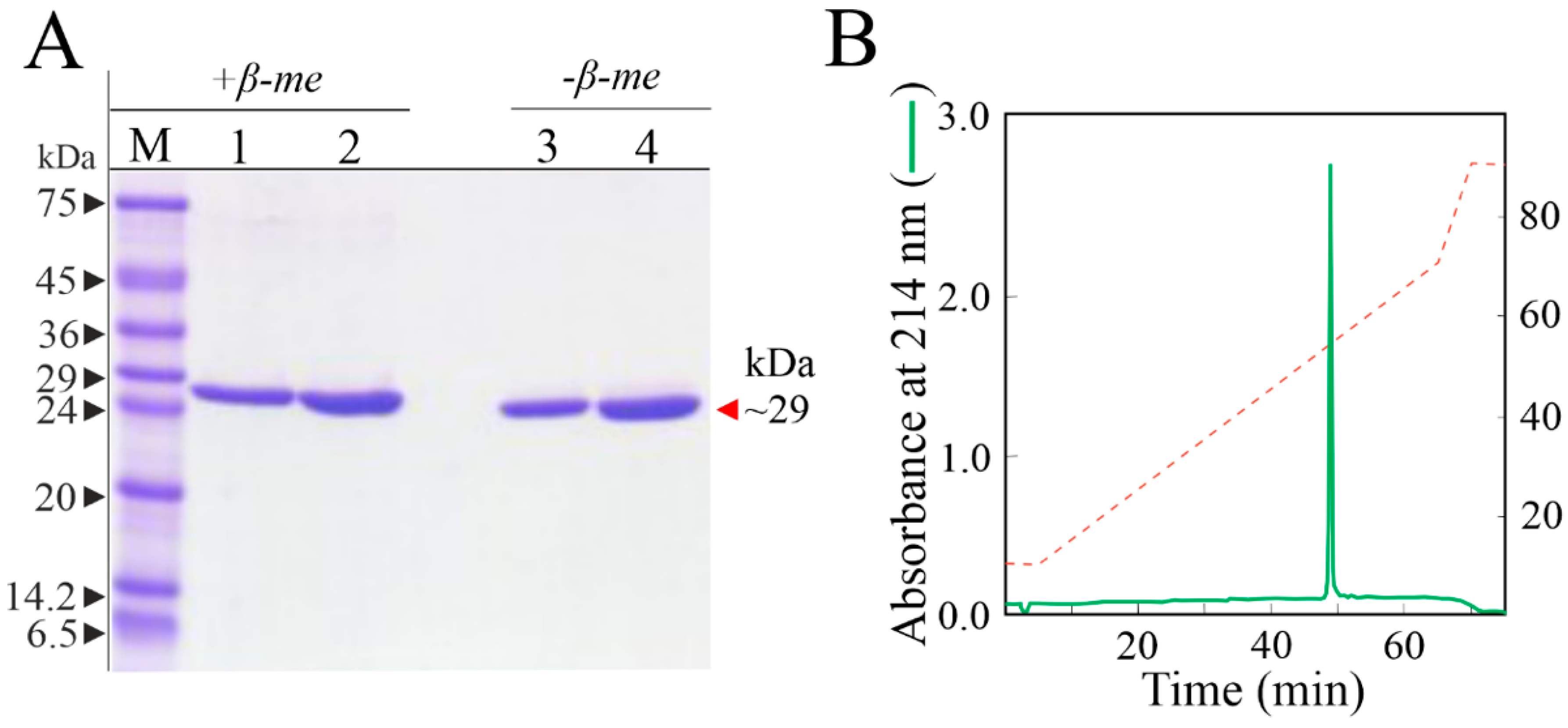
3.1. Purification of Sodin 5
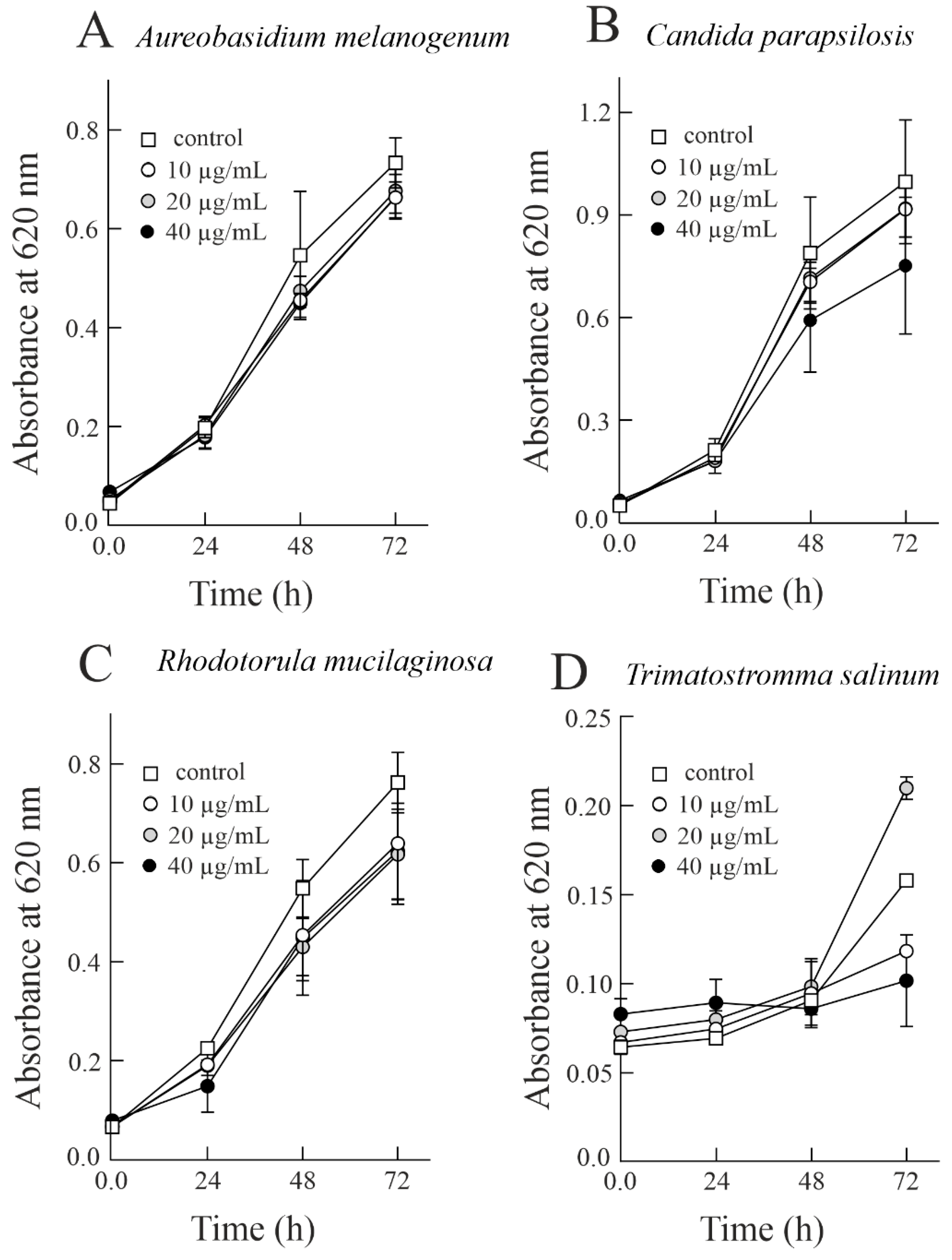
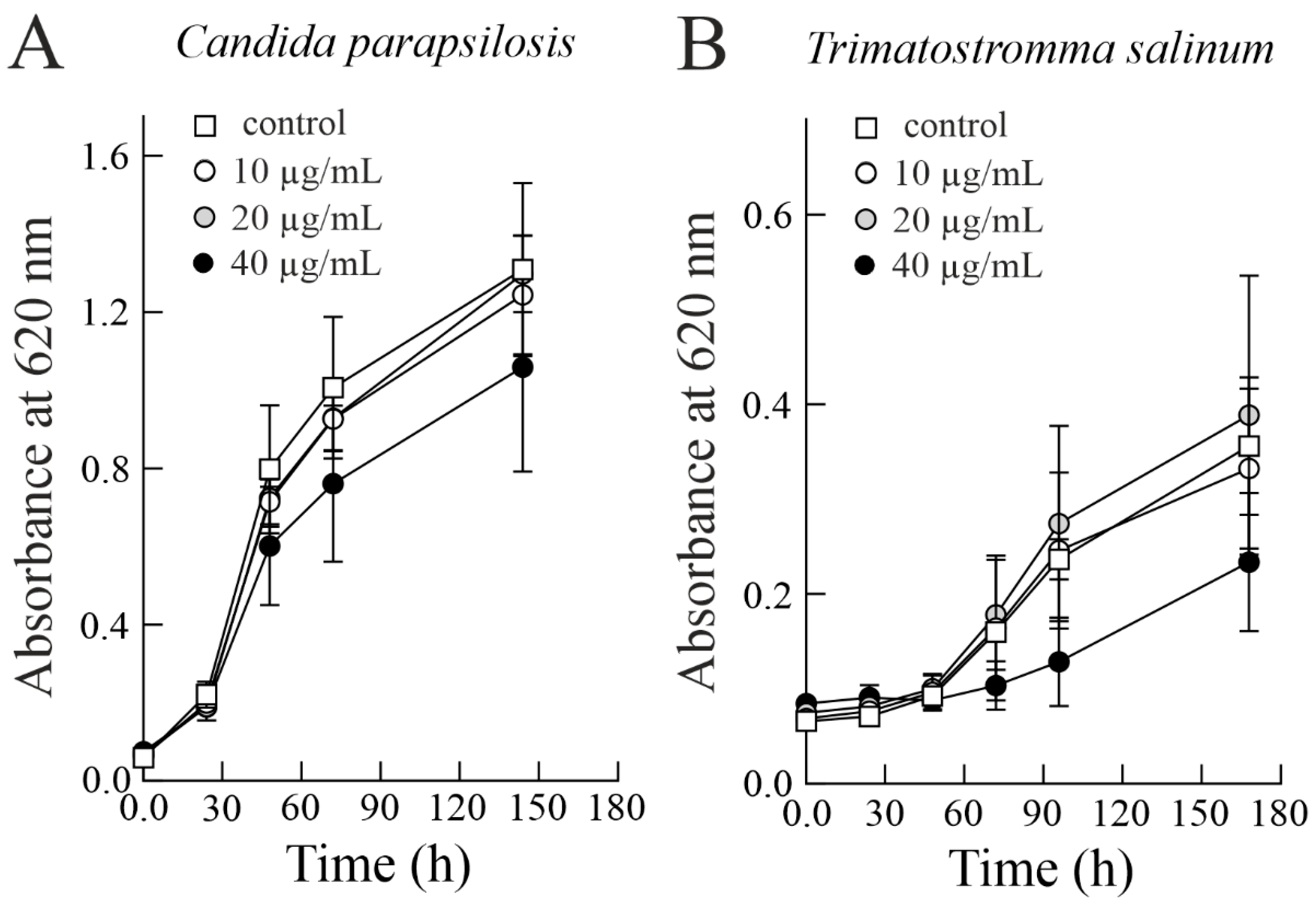
3.2. Effect of Sodin 5 on the Growth of Halophilic Fung
3.3. Cytotoxicity of Sodin 5 to Insect Sf9 Cell Line
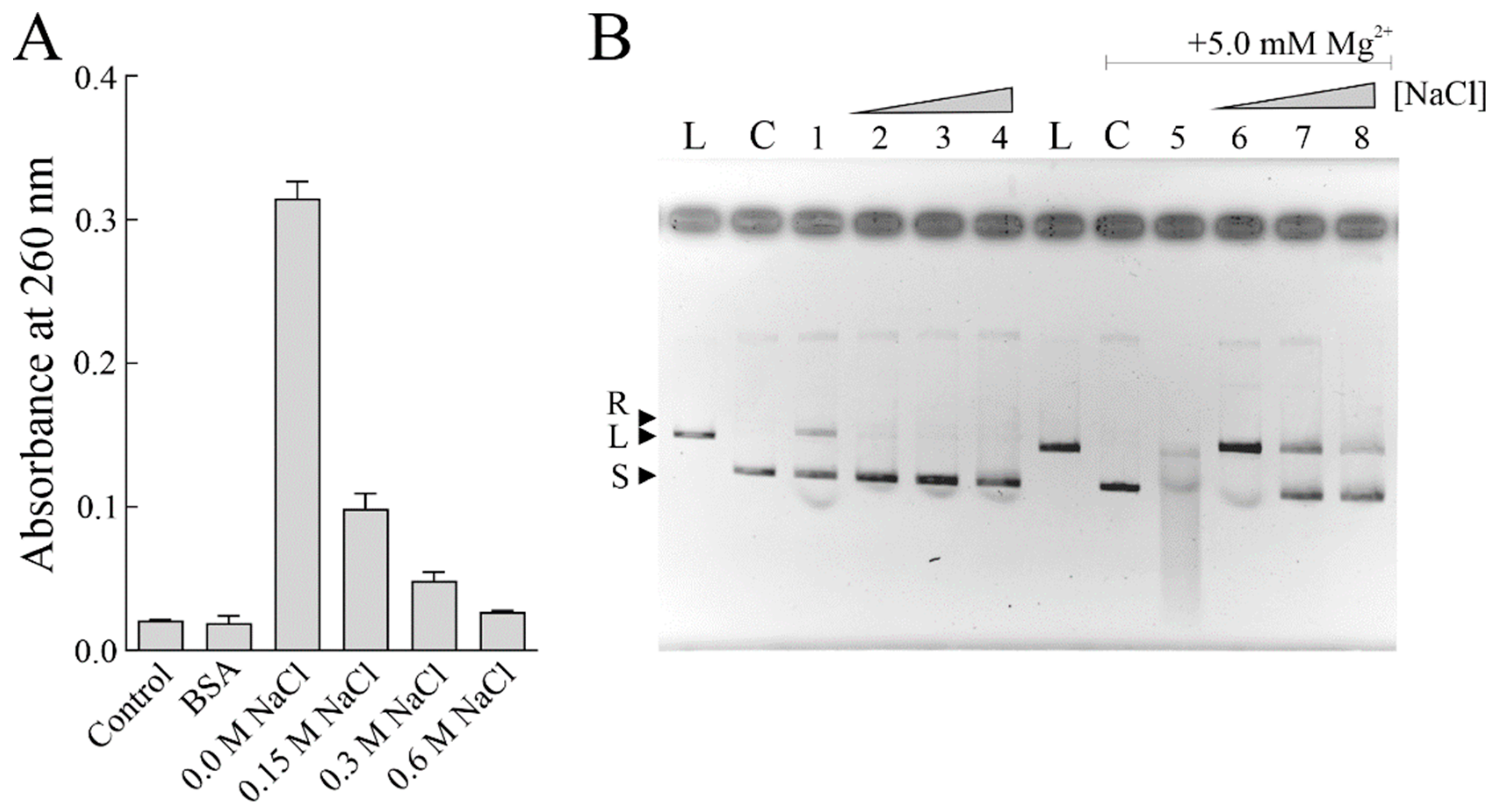
3.4. PNAG Activity of Sodin 5 in the Presence of NaCl
3.5. The Partial Amino Acid Sequence of Sodin 5
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barbieri, L.; Battelli, M.G.; Stirpe, F. Ribosome-inactivating proteins from plants. Biochim. Biophys. Acta 1993, 1154, 237–282. [Google Scholar] [CrossRef]
- Stirpe, F. Ribosome-inactivating proteins: From toxins to useful proteins. Toxicon 2013, 67, 12–16. [Google Scholar] [CrossRef]
- Girbés, T.; Ferreras, J.M.; Arias, F.J.; Stirpe, F. Description, distribution, activity and phylogenetic relationship of ribosome-inactivating proteins in plants, fungi and bacteria. Mini Rev. Med. Chem. 2004, 4, 461–476. [Google Scholar] [CrossRef]
- Landi, N.; Hussain, H.Z.F.; Pedone, P.V.; Ragucci, S.; Di Maro, A. Ribotoxic proteins, known as Inhibitors of protein synthesis, from mushrooms and other fungi according to Endo’s fragment detection. Toxins 2022, 14, 403. [Google Scholar] [CrossRef] [PubMed]
- Ferreras, J.M.; Barbieri, L.; Girbés, T.; Battelli, M.G.; Rojo, M.A.; Arias, F.J.; Rocher, M.A.; Soriano, F.; Mendéz, E.; Stirpe, F. Distribution and properties of major ribosome-inactivating proteins (28 S rRNA N-glycosidases) of the plant Saponaria officinalis L. (Caryophyllaceae). Biochim. Biophys. Acta 1993, 1216, 31–42. [Google Scholar] [CrossRef]
- Iglesias, R.; Russo, R.; Landi, N.; Valletta, M.; Chambery, A.; Di Maro, A.; Bolognesi, A.; Ferreras, J.M.; Citores, L. Structure and Biological Properties of Ribosome-Inactivating Proteins and Lectins from Elder (Sambucus nigra L.) Leaves. Toxins 2022, 14, 611. [Google Scholar] [CrossRef] [PubMed]
- Stirpe, F.; Barbieri, L.; Gorini, P.; Valbonesi, P.; Bolognesi, A.; Polito, L. Activities associated with the presence of ribosome-inactivating proteins increase in senescent and stressed leaves. FEBS Lett. 1996, 382, 309–312. [Google Scholar] [CrossRef]
- Polito, L.; Bortolotti, M.; Mercatelli, D.; Mancuso, R.; Baruzzi, G.; Faedi, W.; Bolognesi, A. Protein synthesis inhibition activity by strawberry tissue protein extracts during plant life cycle and under biotic and abiotic stresses. Int. J. Mol. Sci. 2013, 14, 15532–15545. [Google Scholar] [CrossRef]
- Endo, Y.; Tsurugi, K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J. Biol. Chem. 1987, 262, 8128–8130. [Google Scholar] [CrossRef] [PubMed]
- Grela, P.; Szajwaj, M.; Horbowicz-Drożdżal, P.; Tchórzewski, M. How Ricin Damages the Ribosome. Toxins 2019, 11, 241. [Google Scholar] [CrossRef]
- Barbieri, L.; Valbonesi, P.; Righi, F.; Zuccheri, G.; Monti, F.; Gorini, P.; Samorí, B.; Stirpe, F. Polynucleotide:Adenosine glycosidase is the sole activity of ribosome-inactivating proteins on DNA. J. Biochem. 2000, 128, 883–889. [Google Scholar] [CrossRef]
- Citores, L.; Iglesias, R.; Ferreras, J.M. Antiviral Activity of Ribosome-Inactivating Proteins. Toxins 2021, 13, 80. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.; Boston, R.S. RIBOSOME-INACTIVATING PROTEINS: A Plant Perspective. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 785–816. [Google Scholar] [CrossRef] [PubMed]
- Stirpe, F.; Gilabert-Oriol, R. Ribosome-Inactivating Proteins: An Overview. In Plant Toxins; Gopalakrishnakone, P., Carlini, C.R., Ligabue-Braun, R., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2015; pp. 1–29. [Google Scholar]
- Van Damme, E.J.M.; Hao, Q.; Chen, Y.; Barre, A.; Vandenbussche, F.; Desmyter, S.; Rougé, P.; Peumans, W.J. Ribosome-Inactivating Proteins: A Family of Plant Proteins That Do More Than Inactivate Ribosomes. Crit. Rev. Plant Sci. 2001, 20, 395–465. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, S.; Sharma, N.R.; Paul, K. Expanding role of ribosome-inactivating proteins: From toxins to therapeutics. IUBMB Life 2023, 75, 82–96. [Google Scholar] [CrossRef]
- Zhu, F.; Zhou, Y.K.; Ji, Z.L.; Chen, X.R. The Plant Ribosome-Inactivating Proteins Play Important Roles in Defense against Pathogens and Insect Pest Attacks. Front. Plant Sci. 2018, 9, 146. [Google Scholar] [CrossRef]
- Fabbrini, M.S.; Katayama, M.; Nakase, I.; Vago, R. Plant Ribosome-Inactivating Proteins: Progesses, Challenges and Biotechnological Applications (and a Few Digressions). Toxins 2017, 9, 314. [Google Scholar] [CrossRef]
- de Virgilio, M.; Lombardi, A.; Caliandro, R.; Fabbrini, M.S. Ribosome-inactivating proteins: From plant defense to tumor attack. Toxins 2010, 2, 2699–2737. [Google Scholar] [CrossRef]
- Mishra, V.; Mishra, R.; Shamra, R.S. Ribosome inactivating proteins—An unfathomed biomolecule for developing multi-stress tolerant transgenic plants. Int. J. Biol. Macromol. 2022, 210, 107–122. [Google Scholar] [CrossRef]
- Landi, N.; Ragucci, S.; Citores, L.; Clemente, A.; Hussain, H.Z.F.; Iglesias, R.; Ferreras, J.M.; Di Maro, A. Isolation, Characterization and Biological Action of Type-1 Ribosome-Inactivating Proteins from Tissues of Salsola soda L. Toxins 2022, 14, 566. [Google Scholar] [CrossRef]
- Centofanti, T.; Bañuelos, G. Evaluation of the halophyte Salsola soda as an alternative crop for saline soils high in selenium and boron. J. Environ. Manag. 2015, 157, 96–102. [Google Scholar] [CrossRef]
- Hammer, K.; Pignone, D.; Cifarelli, S.; Perrino, P. Barilla (Salsola soda, Chenopodiaceae). Econ. Bot. 1990, 44, 410–412. [Google Scholar] [CrossRef]
- Atzori, G.; Guidi Nissim, W.; Mancuso, S.; Palm, E. Intercropping Salt-Sensitive Lactuca sativa L. and Salt-Tolerant Salsola soda L. in a Saline Hydroponic Medium: An Agronomic and Physiological Assessment. Plants 2022, 11, 2924. [Google Scholar] [CrossRef]
- Graifenberg, A.; Botrini, L.; Giustiniani, L.; Filippi, F.; Curadi, M. Tomato Growing in Saline Conditions with Biodesalinating Plants: Salsola soda L., and Portulaca oleracea L.; International Society for Horticultural Science (ISHS): Leuven, Belgium, 2003; pp. 301–305. [Google Scholar]
- Colla, G.; Rouphael, Y.; Fallovo, C.; Cardarelli, M.; Graifenberg, A. Use of Salsola soda as a companion plant to improve greenhouse pepper (Capsicum annuum) performance under saline conditions. N. Z. J. Crop Hortic. Sci. 2006, 34, 283–290. [Google Scholar] [CrossRef]
- Gunde-Cimermana, N.; Zalarb, P.; de Hoogc, S.; Plemenitasd, A. Hypersaline waters in salterns—Natural ecological niches for halophilic black yeasts. FEMS Microbiol. Ecol. 2000, 32, 235–240. [Google Scholar]
- Gunde-cimerman, N.; Zalar, P. Extremely Halotolerant and Halophilic Fungi Inhabit Brine in Solar Salterns Around the Globe. Food Technol. Biotechnol. 2014, 52, 170–179. [Google Scholar]
- Abu-Seidah, A. Effect of salt stress on amino acids, organic acids and ultra-structure of Aspergillus flavus and Penicillium roquefortii. Int. J. Agric. Biol. 2007, 9, 419–425. [Google Scholar]
- Mitchison-Field, L.M.Y.; Vargas-Muñiz, J.M.; Stormo, B.M.; Vogt, E.J.D.; Van Dierdonck, S.; Pelletier, J.F.; Ehrlich, C.; Lew, D.J.; Field, C.M.; Gladfelter, A.S. Unconventional Cell Division Cycles from Marine-Derived Yeasts. Curr. Biol. 2019, 29, 3439–3456.e5. [Google Scholar] [CrossRef] [PubMed]
- Śliżewska, W.; Struszczyk-Świta, K.; Marchut-Mikołajczyk, O. Metabolic Potential of Halophilic Filamentous Fungi-Current Perspective. Int. J. Mol. Sci. 2022, 23, 4189. [Google Scholar] [CrossRef] [PubMed]
- Razghandi, M.; Mohammadi, A.; Ghorbani, M.; Mirzaee, M.R. New fungal pathogens and endophytes associated with Salsola. J. Plant Prot. Res. 2020, 60, 362–368. [Google Scholar]
- Lombardi, T.; Bertacchi, A.; Pistelli, L.; Pardossi, A.; Pecchia, S.; Toffanin, A.; Sanmartin, C. Biological and Agronomic Traits of the Main Halophytes Widespread in the Mediterranean Region as Potential New Vegetable Crops. Horticulturae 2022, 8, 195. [Google Scholar] [CrossRef]
- Caicedo-Bejarano, L.D.; Osorio-Vanegas, L.S.; Ramírez-Castrillón, M.; Castillo, J.E.; Martínez-Garay, C.A.; Chávez-Vivas, M. Water Quality, Heavy Metals, and Antifungal Susceptibility to Fluconazole of Yeasts from Water Systems. Int. J. Environ. Res. Public Health 2023, 20, 3428. [Google Scholar] [CrossRef]
- Novak Babič, M.; Gunde-Cimerman, N.; Breskvar, M.; Džeroski, S.; Brandão, J. Occurrence, Diversity and Anti-Fungal Resistance of Fungi in Sand of an Urban Beach in Slovenia-Environmental Monitoring with Possible Health Risk Implications. J. Fungi 2022, 8, 860. [Google Scholar] [CrossRef]
- Cogliati, M.; Arikan-Akdagli, S.; Barac, A.; Bostanaru, A.C.; Brito, S.; Çerikçioğlu, N.; Efstratiou, M.A.; Ergin, Ç.; Esposto, M.C.; Frenkel, M.; et al. Environmental and bioclimatic factors influencing yeasts and molds distribution along European shores. Sci. Total Environ. 2023, 859, 160132. [Google Scholar] [CrossRef]
- Di Maro, A.; Chambery, A.; Daniele, A.; Casoria, P.; Parente, A. Isolation and characterization of heterotepalins, type 1 ribosome-inactivating proteins from Phytolacca heterotepala leaves. Phytochemistry 2007, 68, 767–776. [Google Scholar] [CrossRef]
- Di Maro, A.; Valbonesi, P.; Bolognesi, A.; Stirpe, F.; De Luca, P.; Siniscalco Gigliano, G.; Gaudio, L.; Delli Bovi, P.; Ferranti, P.; Malorni, A.; et al. Isolation and characterization of four type-1 ribosome-inactivating proteins, with polynucleotide:adenosine glycosidase activity, from leaves of Phytolacca dioica L. Planta 1999, 208, 125–131. [Google Scholar] [CrossRef]
- Aceto, S.; Di Maro, A.; Conforto, B.; Siniscalco, G.G.; Parente, A.; Delli Bovi, P.; Gaudio, L. Nicking activity on pBR322 DNA of ribosome inactivating proteins from Phytolacca dioica L. leaves. Biol. Chem. 2005, 386, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Landi, N.; Grundner, M.; Ragucci, S.; Pavšič, M.; Mravinec, M.; Pedone, P.V.; Sepčić, K.; Di Maro, A. Characterization and cytotoxic activity of ribotoxin-like proteins from the edible mushroom Pleurotus eryngii. Food Chem. 2022, 396, 133655. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Jain, K.; Desai, C.; Tiwari, O.; Madamwar, D. Chapter 18—Microbial Community Dynamics of Extremophiles/Extreme Environment. In Microbial Diversity in the Genomic Era; Das, S., Dash, H.R., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 323–332. [Google Scholar]
- Yike, I. Fungal proteases and their pathophysiological effects. Mycopathologia 2011, 171, 299–323. [Google Scholar] [CrossRef] [PubMed]
- Trigiano, R.N.; Poplawski, L.E. Extracellular Enzymes Produced by Fungi and Bacteria. In Plant Pathology Concepts and Laboratory Exercises; Ownley, B.H., Trigiano, R.N., Eds.; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Anthonies, S.; Vargas-Muñiz, J.M. Hortaea werneckii isolates exhibit different pathogenic potential in the invertebrate infection model Galleria mellonella. Front. Fungal Biol. 2022, 3, 941691. [Google Scholar] [CrossRef] [PubMed]
- Zalar, P.; Zupančič, J.; Gostinčar, C.; Zajc, J.; de Hoog, G.S.; De Leo, F.; Azua-Bustos, A.; Gunde-Cimerman, N. The extremely halotolerant black yeast Hortaea werneckii—A model for intraspecific hybridization in clonal fungi. IMA Fungus 2019, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.; Kim, H.; Choi, H.S. Fungi in salterns. J. Microbiol. 2019, 57, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Kogej, T.; Gorbushina, A.A.; Gunde-Cimerman, N. Hypersaline conditions induce changes in cell-wall melanization and colony structure in a halophilic and a xerophilic black yeast species of the genus Trimmatostroma. Mycol. Res. 2006, 110 Pt 6, 713–724. [Google Scholar] [CrossRef]
- Abbas, H.K.; Wilkinson, J.R.; Zablotowicz, R.M.; Accinelli, C.; Abel, C.A.; Bruns, H.A.; Weaver, M.A. Ecology of Aspergillus flavus, regulation of aflatoxin production, and management strategies to reduce aflatoxin contamination of corn. Toxin Rev. 2009, 28, 142–153. [Google Scholar] [CrossRef]
- Paulussen, C.; Hallsworth, J.E.; Álvarez-Pérez, S.; Nierman, W.C.; Hamill, P.G.; Blain, D.; Rediers, H.; Lievens, B. Ecology of aspergillosis: Insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb. Biotechnol. 2017, 10, 296–322. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Ramakrishna, S.; Vikineswary, S.; Das, D.; Bahkali, A.H.; Guo, S.Y.; Pang, K.L. How Do Fungi Survive in the Sea and Respond to Climate Change? J. Fungi 2022, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Zuluaga-Montero, A.; Ramírez-Camejo, L.; Rauscher, J.; Bayman, P. Marine isolates of Aspergillus flavus: Denizens of the deep or lost at sea? Fungal Ecol. 2010, 3, 386–391. [Google Scholar] [CrossRef]
- Natarajan, S.; Balachandar, D.; Senthil, N.; Velazhahan, R.; Paranidharan, V. Volatiles of antagonistic soil yeasts inhibit growth and aflatoxin production of Aspergillus flavus. Microbiol. Res. 2022, 263, 127150. [Google Scholar] [CrossRef]
- Černoša, A.; Sun, X.; Gostinčar, C.; Fang, C.; Gunde-Cimerman, N.; Song, Z. Virulence Traits and Population Genomics of the Black Yeast Aureobasidium melanogenum. J. Fungi 2021, 7, 665. [Google Scholar] [CrossRef]
- Suthar, M.; Dufossé, L.; Singh, S.K. The Enigmatic World of Fungal Melanin: A Comprehensive Review. J. Fungi 2023, 9, 891. [Google Scholar] [CrossRef]
- Babič, M.N.; Zupančič, J.; Gunde-Cimerman, N.; Zalar, P. Yeast in Anthropogenic and Polluted Environments. In Yeasts in Natural Ecosystems: Diversity; Buzzini, P., Lachance, M.-A., Yurkov, A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 145–169. [Google Scholar]
- Kulesza, K.; Biedunkiewicz, A.; Nowacka, K.; Dynowska, M.; Urbaniak, M.; Stępień, Ł. Dishwashers as an Extreme Environment of Potentially Pathogenic Yeast Species. Pathogens 2021, 10, 446. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.M.; Proenca, A.M.; Moreira-Silva, E.; Dos Santos, F.M.; Marconatto, L.; de Castro, A.M.; Medina-Silva, R. Biochemical features and early adhesion of marine Candida parapsilosis strains on high-density polyethylene. J. Appl. Microbiol. 2022, 132, 1954–1966. [Google Scholar] [CrossRef] [PubMed]
- Bessadok, B.; Jaouadi, B.; Brück, T.; Santulli, A.; Messina, C.M.; Sadok, S. Molecular Identification and Biochemical Characterization of Novel Marine Yeast Strains with Potential Application in Industrial Biotechnology. Fermentation 2022, 8, 538. [Google Scholar] [CrossRef]
- Sen, D.; Paul, K.; Saha, C.; Mukherjee, G.; Nag, M.; Ghosh, S.; Das, A.; Seal, A.; Tripathy, S. A unique life-strategy of an endophytic yeast Rhodotorula mucilaginosa JGTA-S1-a comparative genomics viewpoint. DNA Res. 2019, 26, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Cortes, A.; Garcia-Vásquez, J.A.; Aranguren, Y.; Ramirez-Castrillon, M. Pigment Production Improvement in Rhodotorula mucilaginosa AJB01 Using Design of Experiments. Microorganisms 2021, 9, 387. [Google Scholar] [CrossRef]
- Noguchi, H.; Matsumoto, T.; Kimura, U.; Hiruma, M.; Kano, R.; Yaguchi, T.; Fukushima, S.; Ihn, H. Fungal melanonychia caused by Candida parapsilosis successfully treated with oral fosravuconazole. J. Dermatol. 2019, 46, 911–913. [Google Scholar] [CrossRef]
- Zupančič, J.; Novak Babič, M.; Gunde-Cimerman, N. Chapter 5—High Incidence of an Emerging Opportunistic Pathogen Candida parapsilosis in Water-Related Domestic Environments. In Fungal Infection; de Loreto, É.S., Tondolo, J.S.M., Eds.; IntechOpen: Rijeka, Croatia, 2018; p. 81313. [Google Scholar]
- Morrow, C.A.; Fraser, J.A. Sexual reproduction and dimorphism in the pathogenic basidiomycetes. FEMS Yeast Res. 2009, 9, 161–177. [Google Scholar] [CrossRef]
- Bertholdo-Vargas, L.R.; Martins, J.N.; Bordin, D.; Salvador, M.; Schafer, A.E.; Barros, N.M.; Barbieri, L.; Stirpe, F.; Carlini, C.R. Type 1 ribosome-inactivating proteins—Entomotoxic, oxidative and genotoxic action on Anticarsia gemmatalis (Hübner) and Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae). J. Insect Physiol. 2009, 55, 51–58. [Google Scholar] [CrossRef]
- Chan, W.Y.; Huang, H.; Tam, S.C. Receptor-mediated endocytosis of trichosanthin in choriocarcinoma cells. Toxicology 2003, 186, 191–203. [Google Scholar] [CrossRef]
- Musidlak, O.; Nawrot, R.; Goździcka-Józefiak, A. Which Plant Proteins Are Involved in Antiviral Defense? Review on In Vivo and In Vitro Activities of Selected Plant Proteins against Viruses. Int. J. Mol. Sci. 2017, 18, 2300. [Google Scholar] [CrossRef]
- Tan, Z.J.; Chen, S.J. Nucleic acid helix stability: Effects of salt concentration, cation valence and size, and chain length. Biophys. J. 2006, 90, 1175–1190. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.J.; Zhang, C.; Zhou, E.; Dong, H.L.; Tan, Z.J.; Zhang, X.H.; Dai, L. Universality in RNA and DNA deformations induced by salt, temperature change, stretching force, and protein binding. Proc. Natl. Acad. Sci. USA 2023, 120, e2218425120. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, J.; Habuka, N.; Furuno, M.; Miyano, M.; Takanami, Y.; Koiwai, A. DNA sequence of Mirabilis antiviral protein (MAP), a ribosome-inactivating protein with an antiviral property, from Mirabilis jalapa L. and its expression in Escherichia coli. J. Biol. Chem. 1991, 266, 8426–8430. [Google Scholar] [CrossRef] [PubMed]
- Di Maro, A.; Ferranti, P.; Mastronicola, M.; Polito, L.; Bolognesi, A.; Stirpe, F.; Malorni, A.; Parente, A. Reliable sequence determination of ribosome- inactivating proteins by combining electrospray mass spectrometry and Edman degradation. J. Mass Spectrom. 2001, 36, 38–46. [Google Scholar] [CrossRef]
- Di Maro, A.; Citores, L.; Russo, R.; Iglesias, R.; Ferreras, J.M. Sequence comparison and phylogenetic analysis by the Maximum Likelihood method of ribosome-inactivating proteins from angiosperms. Plant Mol. Biol. 2014, 85, 575–588. [Google Scholar] [CrossRef]
- Monzingo, A.F.; Collins, E.J.; Ernst, S.R.; Irvin, J.D.; Robertus, J.D. The 2.5 A structure of pokeweed antiviral protein. J. Mol. Biol. 1993, 233, 705–715. [Google Scholar] [CrossRef]


| Fungal Strain | EXF-No 1 | Growth Inhibition (%) | ||
|---|---|---|---|---|
| 10 µg/mL | 20 µg/mL | 40 µg/mL | ||
| Aspergillus flavus | EXF-2368 | −14.12 | −8.48 | 12.16 |
| Aureobasidium melanogenum | EXF-9906 | 9.29 | 7.52 | 9.07 |
| Candida parapsilosis | EXF-517 | 7.98 | 7.82 | 24.38 |
| Hortaea werneckii | EXF-225 | −4.82 | 3.29 | 4.24 |
| Rhodotorula mucilaginosa | EXF-10514 | 16.17 | 19.06 | 18.23 |
| Trimatostromma salinum | EXF-295 | −2.71 | −11.48 | 35.28 |
| Peptide | Peptide Sequence | Number of Identified +2H Spectra | Number of Identified +3H Spectra | Calculated +1H Peptide Mass (Da) | Originating Protein [Species] | Accession Number | Molecular Mass (Da) |
|---|---|---|---|---|---|---|---|
| Trypsin cleavage | |||||||
| T-1 | NDLYVVAFADK | 2 | 0 | 1254.64 | antiviral ribosome-inactivating protein [Atriplex canescens] | AEY75258.1 | 30849.80 |
| T-2 | LSFPLGFDNLK | 8 | 0 | 1250.68 | antiviral ribosome-inactivating protein [Atriplex canescens] | AEY75258.1 | 30849.80 |
| T-1 | ILSLENNWGAISK | 1 | 0 | 1444.78 | antiviral ribosome-inactivating protein [Atriplex canescens] | AEY75258.1 | 30849.80 |
| T-4 | NDMGLLK | 44 | 0 | 790.41 | antiviral ribosome-inactivating protein [Atriplex canescens] | AEY75258.1 | 30849.80 |
| T-5 | FFLIAIQMVAEAAR | 23 | 21 | 1595.86 | ribosome-inactivating protein [Spinacia oleracea] | BAB83507.1, BAH56516.1 | 35769.30 |
| Chymotrypsin cleavage | |||||||
| C-1 | LIAIQMVAEAARF | 5 | 0 | 1448.79 | antiviral ribosome-inactivating protein [Atriplex canescens] ribosome-inactivating protein [Spinacia oleracea] | AEY75258.1 BAB83507.1, BAH56516.1 | 30849.80 35769.30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novak Babič, M.; Ragucci, S.; Leonardi, A.; Pavšič, M.; Landi, N.; Križaj, I.; Gunde-Cimerman, N.; Sepčić, K.; Di Maro, A. Biocontrol Potential of Sodin 5, Type 1 Ribosome-Inactivating Protein from Salsola soda L. Seeds. Biomolecules 2024, 14, 336. https://doi.org/10.3390/biom14030336
Novak Babič M, Ragucci S, Leonardi A, Pavšič M, Landi N, Križaj I, Gunde-Cimerman N, Sepčić K, Di Maro A. Biocontrol Potential of Sodin 5, Type 1 Ribosome-Inactivating Protein from Salsola soda L. Seeds. Biomolecules. 2024; 14(3):336. https://doi.org/10.3390/biom14030336
Chicago/Turabian StyleNovak Babič, Monika, Sara Ragucci, Adrijana Leonardi, Miha Pavšič, Nicola Landi, Igor Križaj, Nina Gunde-Cimerman, Kristina Sepčić, and Antimo Di Maro. 2024. "Biocontrol Potential of Sodin 5, Type 1 Ribosome-Inactivating Protein from Salsola soda L. Seeds" Biomolecules 14, no. 3: 336. https://doi.org/10.3390/biom14030336
APA StyleNovak Babič, M., Ragucci, S., Leonardi, A., Pavšič, M., Landi, N., Križaj, I., Gunde-Cimerman, N., Sepčić, K., & Di Maro, A. (2024). Biocontrol Potential of Sodin 5, Type 1 Ribosome-Inactivating Protein from Salsola soda L. Seeds. Biomolecules, 14(3), 336. https://doi.org/10.3390/biom14030336











