Proteomic and Metabolomic Analysis of the Quercus ilex–Phytophthora cinnamomi Pathosystem Reveals a Population-Specific Response, Independent of Co-Occurrence of Drought
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Experimental Design
2.3. Morphological and Physiological Measurements
2.4. Protein Extraction and GeLC-MSMS Analysis
2.5. Protein Identification and Quantification
2.6. Metabolite Extraction and LC-MS Analysis
2.7. Metabolite Identification and Data Processing
2.8. Statistical Analysis
2.9. Integrative Analysis of Proteomics and Metabolomics Data
3. Results and Discussion
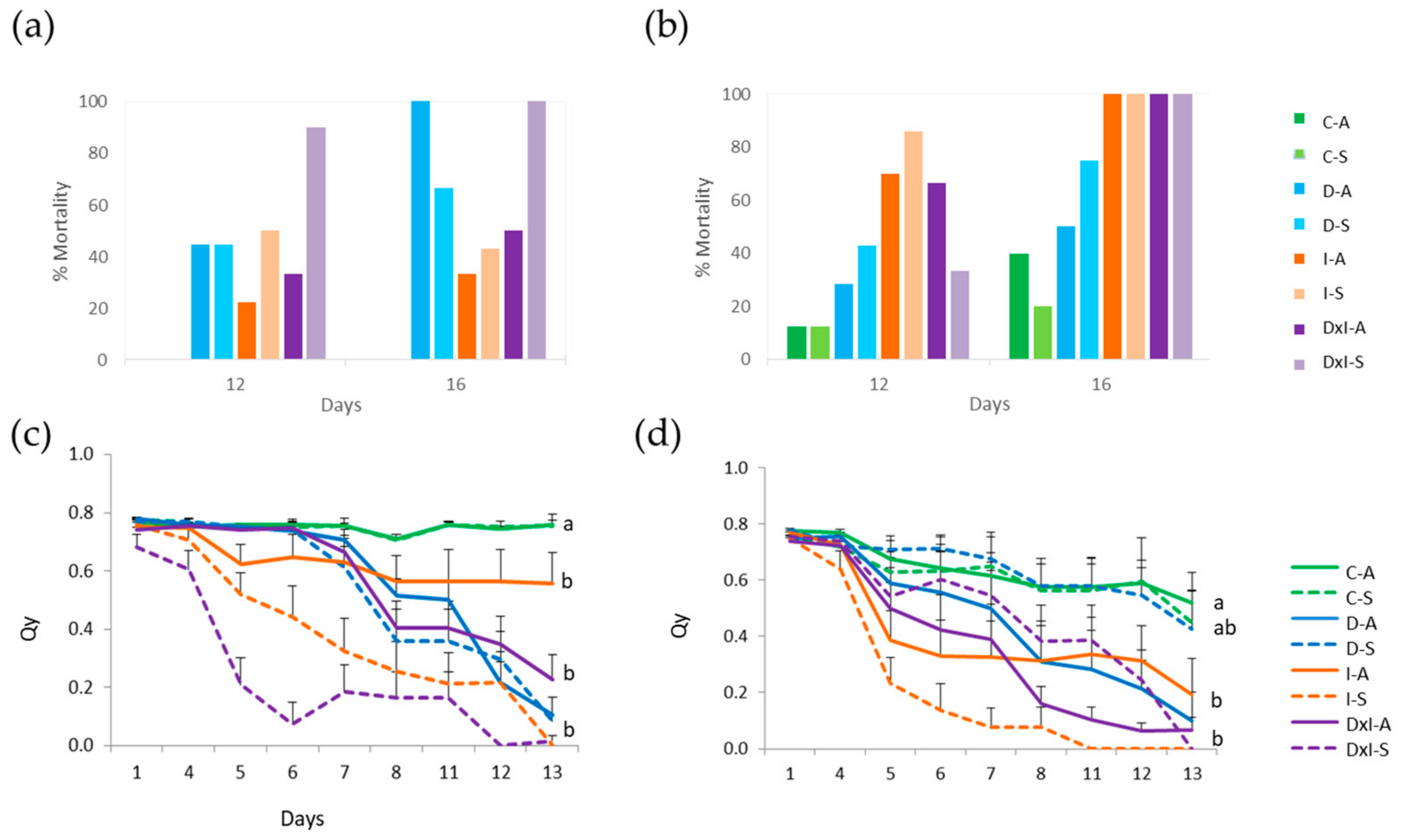
3.1. Seedling Damage Symptoms, Mortality, and Leaf Fluorescence
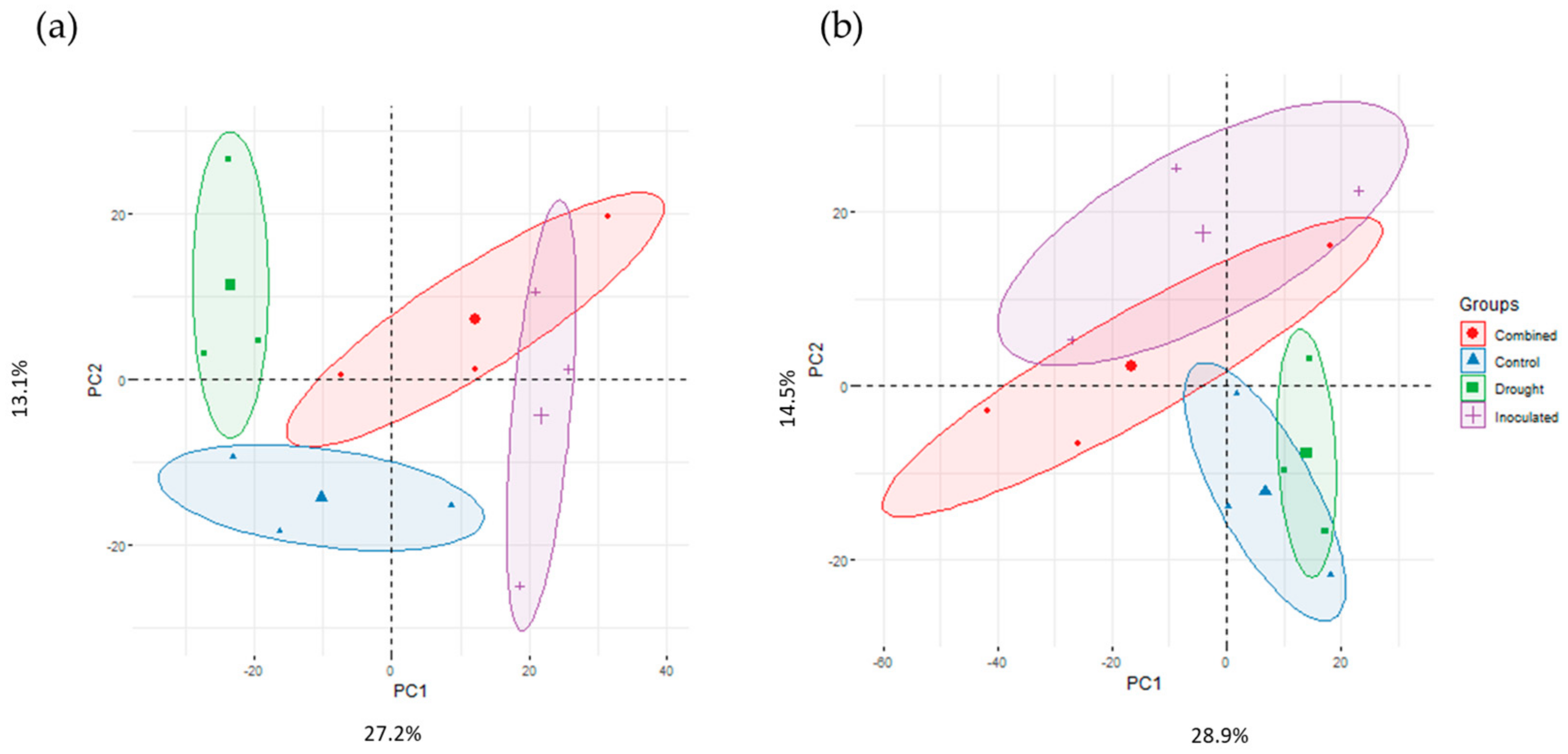
3.2. Proteomic Analysis
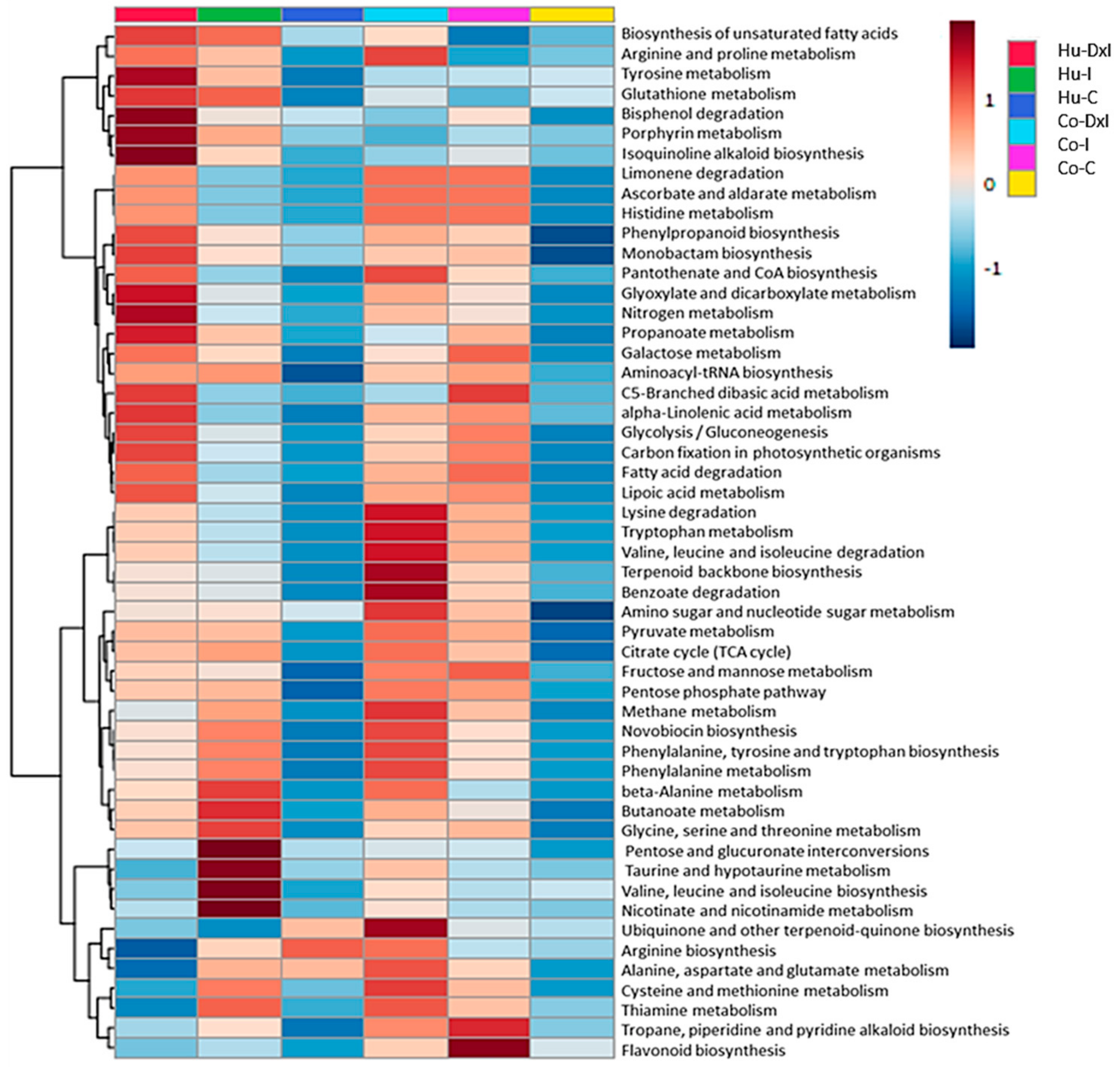
3.3. Metabolomic Analysis
3.4. Integrated Proteome–Metabolome Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allen, C.D.; Breshears, D.D.; McDowell, N.G. On Underestimation of Global Vulnerability to Tree Mortality and Forest Die-off from Hotter Drought in the Anthropocene. Ecosphere 2015, 6, 1–55. [Google Scholar] [CrossRef]
- Hansen, E.M. Phytophthora Species Emerging as Pathogens of Forest Trees. Curr. For. Rep. 2015, 1, 16–24. [Google Scholar] [CrossRef]
- Intergovernmental Panel on Climate Change (IPCC). Climate Change 2013: The Physical Science Basis. In Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Intergovernmental Panel on Climate Change, Ed.; Cambridge University Press: Cambridge, UK, 2013; Volume 1535, ISBN 9781107057999. [Google Scholar]
- Doblas-Miranda, E.; Alonso, R.; Arnan, X.; Bermejo, V.; Brotons, L.; de las Heras, J.; Estiarte, M.; Hódar, J.A.; Llorens, P.; Lloret, F.; et al. A Review of the Combination among Global Change Factors in Forests, Shrublands and Pastures of the Mediterranean Region: Beyond Drought Effects. Glob. Planet. Change 2017, 148, 42–54. [Google Scholar] [CrossRef]
- Burgess, T.I.; Scott, J.K.; Mcdougall, K.L.; Stukely, M.J.C.; Crane, C.; Dunstan, W.A.; Brigg, F.; Andjic, V.; White, D.; Rudman, T.; et al. Current and Projected Global Distribution of Phytophthora cinnamomi, One of the World’s Worst Plant Pathogens. Glob. Change Biol. 2017, 23, 1661–1674. [Google Scholar] [CrossRef]
- Homet, P.; Matías, L.; Godoy, O.; Gómez-Aparicio, L. Evidence for Antagonistic Effects of Climate Change and Exotic Pathogens on Regeneration of Mediterranean Forests. J. Ecol. 2023, 112, 174–188. [Google Scholar] [CrossRef]
- Kamoun, S.; Furzer, O.; Jones, J.D.G.; Judelson, H.S.; Ali, G.S.; Dalio, R.J.D.; Roy, S.G.; Schena, L.; Zambounis, A.; Panabières, F.; et al. The Top 10 Oomycete Pathogens in Molecular Plant Pathology. Mol. Plant Pathol. 2015, 16, 413–434. [Google Scholar] [CrossRef] [PubMed]
- Jung, T.; Pérez-Sierra, A.; Durán, A.; Jung, M.H.; Balci, Y.; Scanu, B. Canker and Decline Diseases Caused by Soil- and Airborne Phytophthora Species in Forests and Woodlands. Persoonia 2018, 40, 182–220. [Google Scholar] [CrossRef] [PubMed]
- Corcobado, T.; Miranda-Torres, J.J.; Martín-García, J.; Jung, T.; Solla, A. Early Survival of Quercus ilex Subspecies from Different Populations after Infections and Co-infections by Multiple Phytophthora Species. Plant Pathol. 2017, 66, 792–804. [Google Scholar] [CrossRef]
- Bezuidenhout, C.M.; Denman, S.; Kirk, S.A.; Botha, W.J.; Mostert, L.; McLeod, A. Phytophthora taxa associated with cultivated Agathosma, with emphasis on the P. citricola complex and P. capensis sp. nov. Persoonia 2010, 25, 32–49. [Google Scholar] [CrossRef]
- Marañón, T.; Ibáñez, B.; Anaya-Romero, M.; Muñoz-Rojas, M.; Pérez-Ramos, I.M. Oak trees and woodlands providing ecosystem services in Southern Spain. In Trees beyond the Wood. An Exploration of Concepts of Woods, Forests and Trees; Rotherham, I.D., Handley, C., Agnoletti, M., Samojlik, T., Eds.; Wildtrack Publishing: Sheffield, UK, 2012; pp. 369–378. [Google Scholar]
- Ruiz-Gómez, F.J.; Pérez-de-Luque, A.; Navarro-Cerrillo, R.M. The Involvement of Phytophthora Root Rot and Drought Stress in Holm Oak Decline: From Ecophysiology to Microbiome Influence. Curr. For. Rep. 2019, 5, 251–266. [Google Scholar] [CrossRef]
- Ducrey, M. Quelle Sylviculture et Quel Avenir Pour Les Taillis de Chêne Vert (Quercus ilex L.) de La Région Méditerranéenne Française. Rev. For. Francaise 1992, 44, 12–34. [Google Scholar] [CrossRef]
- De Rigo, D.; Caudullo, G. Quercus ilex in Europe: Distribution, Habitat, Usage and Threats. In European Atlas of Forest Tree Species; Publications Office of the European Union: Luxembourg, 2016; pp. 152–153. [Google Scholar]
- Ortego, J.; Bonal, R.; Munoz, A. Genetic Consequences of Habitat Fragmentation in Long-Lived Tree Species: The Case of the Mediterranean Holm Oak (Quercus ilex, L.). J. Hered. 2010, 101, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Guzmán, B.; Rodríguez López, C.M.; Forrest, A.; Cano, E.; Vargas, P. Protected Areas of Spain Preserve the Neutral Genetic Diversity of Quercus ilex L. Irrespective of Glacial Refugia. Tree Genet. Genomes 2015, 11, 124. [Google Scholar] [CrossRef]
- Fernández i Marti, A.; Romero-Rodríguez, C.; Navarro-Cerrillo, R.; Abril, N.; Jorrín-Novo, J.; Dodd, R. Population Genetic Diversity of Quercus ilex Subsp. ballota (Desf.) Samp. Reveals Divergence in Recent and Evolutionary Migration Rates in the Spanish Dehesas. Forests 2018, 9, 337. [Google Scholar] [CrossRef]
- Brasier, C.M. Oak Tree Mortality in Iberia. Nature 1992, 360, 539. [Google Scholar] [CrossRef]
- Corral-Ribera, M.; Fidalgo, C.H.; Peco, B.V. Environmental Variables in the Distribution of La Seca Disease in the Holm Oak (Quercus ilex Subsp. ballota). In Espacio Tiempo y Forma. Serie VI, Geografía; Universidad Nacional de Educación a Distancia: Madrid, Spain, 2018; p. 107. [Google Scholar] [CrossRef][Green Version]
- Surová, D.; Ravera, F.; Guiomar, N.; Martínez Sastre, R.; Pinto-Correia, T. Contributions of Iberian Silvo-Pastoral Landscapes to the Well-Being of Contemporary Society. Rangel. Ecol. Manag. 2018, 71, 560–570. [Google Scholar] [CrossRef]
- Corcobado, T.; Cubera, E.; Moreno, G.; Solla, A. Quercus ilex Forests Are Influenced by Annual Variations in Water Table, Soil Water Deficit and Fine Root Loss Caused by Phytophthora cinnamomi. Agric. For. Meteorol. 2013, 169, 92–99. [Google Scholar] [CrossRef]
- Corcobado, T.; Moreno, G.; Azul, A.M.; Solla, A. Seasonal Variations of Ectomycorrhizal Communities in Declining Quercus ilex Forests: Interactions with Topography, Tree Health Status and Phytophthora cinnamomi Infections. Forestry 2015, 88, 257–266. [Google Scholar] [CrossRef]
- Fernández-Habas, J.; Fernández-Rebollo, P.; Casado, M.R.; García Moreno, A.M.; Abellanas, B. Spatio-Temporal Analysis of Oak Decline Process in Open Woodlands: A Case Study in SW Spain. J. Environ. Manag. 2019, 248, 109308. [Google Scholar] [CrossRef]
- Sánchez-Cuesta, R.; Ruiz-Gómez, F.J.; Duque-Lazo, J.; González-Moreno, P.; Navarro-Cerrillo, R.M. The Environmental Drivers Influencing Spatio-Temporal Dynamics of Oak Defoliation and Mortality in Dehesas of Southern Spain. For. Ecol. Manag. 2021, 485, 118946. [Google Scholar] [CrossRef]
- Malcolm, J.R.; Liu, C.; Neilson, R.P.; Hansen, L.; Hannah, L. Global Warming and Extinctions of Endemic Species from Biodiversity Hotspots. Conserv. Biol. 2006, 20, 538–548. [Google Scholar] [CrossRef]
- Alderotti, F.; Verdiani, E. God Save the Queen! How and Why the Dominant Evergreen Species of the Mediterranean Basin Is Declining? AoB Plants 2023, 15, plad051. [Google Scholar] [CrossRef]
- Duque-Lazo, J.; Van-Gils, H.; Groen, T.A.; Navarro-Cerrillo, R.M. Transferability of Species Distribution Models: The Case of Phytophthora cinnamomi in Southwest Spain and Southwest Australia. Ecoll. Modell. 2016, 320, 62–70. [Google Scholar] [CrossRef]
- Duque-Lazo, J.; Navarro-Cerrillo, R.M.; Ruíz-Gómez, F.J. Assessment of the Future Stability of Cork Oak (Quercus suber L.) Afforestation under Climate Change Scenarios in Southwest Spain. For. Ecol. Manag. 2018, 409, 444–456. [Google Scholar] [CrossRef]
- Sghaier-Hammami, B.; Valero-Galvàn, J.; Romero-Rodríguez, M.C.; Navarro-Cerrillo, R.M.; Abdelly, C.; Jorrín-Novo, J. Physiological and Proteomics Analyses of Holm Oak (Quercus ilex Subsp. Ballota [Desf.] Samp.) Responses to Phytophthora cinnamomi. Plant Physiol. Biochem. 2013, 71, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Corcobado, T.; Cubera, E.; Juárez, E.; Moreno, G.; Solla, A. Drought Events Determine Performance of Quercus ilex Seedlings and Increase Their Susceptibility to Phytophthora cinnamomi. Agric. For. Meteorol. 2014, 192–193, 1–8. [Google Scholar] [CrossRef]
- Hardham, A.R.; Blackman, L.M. Phytophthora cinnamomi . Mol. Plant Pathol. 2018, 19, 260–285. [Google Scholar] [CrossRef] [PubMed]
- Saiz-Fernández, I.; Đorđević, B.; Kerchev, P.; Černý, M.; Jung, T.; Berka, M.; Fu, C.-H.; Horta Jung, M.; Brzobohatý, B. Differences in the Proteomic and Metabolomic Response of Quercus suber and Quercus variabilis During the Early Stages of Phytophthora cinnamomi Infection. Front. Microbiol. 2022, 13, 894533. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Gómez, F.; Pérez-de-Luque, A.; Sánchez-Cuesta, R.; Quero, J.; Navarro Cerrillo, R. Differences in the Response to Acute Drought and Phytophthora cinnamomi Rands Infection in Quercus ilex L. Seedlings. Forests 2018, 9, 634. [Google Scholar] [CrossRef]
- San-Eufrasio, B.; Castillejo, M.Á.; Labella-Ortega, M.; Ruiz-Gómez, F.J.; Navarro-Cerrillo, R.M.; Tienda-Parrilla, M.; Jorrín-Novo, J.V.; Rey, M.-D. Effect and Response of Quercus Ilex Subsp. Ballota [Desf.] Samp. Seedlings from Three Contrasting Andalusian Populations to Individual and Combined Phytophthora cinnamomi and Drought Stresses. Front. Plant Sci. 2021, 12, 722802. [Google Scholar] [CrossRef] [PubMed]
- Simova-Stoilova, L.P.; Romero-Rodríguez, M.C.; Sánchez-Lucas, R.; Navarro-Cerrillo, R.M.; Medina-Aunon, J.A.; Jorrín-Novo, J.V. 2-DE Proteomics Analysis of Drought Treated Seedlings of Quercus ilex Supports a Root Active Strategy for Metabolic Adaptation in Response to Water Shortage. Front. Plant Sci. 2015, 6, 627. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil. Circ.—Calif. Agric. Exp. Stn. 1950, 347, 32. [Google Scholar]
- Ruiz Gómez, F.J.; Navarro-Cerrillo, R.M.; Pérez-de-Luque, A.; Oβwald, W.; Vannini, A.; Morales-Rodríguez, C. Assessment of Functional and Structural Changes of Soil Fungal and Oomycete Communities in Holm Oak Declined Dehesas through Metabarcoding Analysis. Sci. Rep. 2019, 9, 5315. [Google Scholar] [CrossRef] [PubMed]
- San-Eufrasio, B.; Sánchez-Lucas, R.; López-Hidalgo, C.; Guerrero-Sánchez, V.M.; Castillejo, M.Á.; Maldonado-Alconada, A.M.; Jorrín-Novo, J.V.; Rey, M.-D. Responses and Differences in Tolerance to Water Shortage under Climatic Dryness Conditions in Seedlings from Quercus Spp. and Andalusian Q. ilex Populations. Forests 2020, 11, 707. [Google Scholar] [CrossRef]
- Wang, W.; Vignani, R.; Scali, M.; Cresti, M. A Universal and Rapid Protocol for Protein Extraction from Recalcitrant Plant Tissues for Proteomic Analysis. Electrophoresis 2006, 27, 2782–2786. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Castillejo, M.Á.; Bani, M.; Rubiales, D. Understanding Pea Resistance Mechanisms in Response to Fusarium oxysporum through Proteomic Analysis. Phytochemistry 2015, 115, 44–58. [Google Scholar] [CrossRef]
- Castillejo, M.Á.; Fondevilla-Aparicio, S.; Fuentes-Almagro, C.; Rubiales, D. Quantitative Analysis of Target Peptides Related to Resistance Against Ascochyta Blight (Peyronellaea pinodes) in Pea. Proteome Res. 2020, 19, 1000–1012. [Google Scholar] [CrossRef]
- Guerrero-Sanchez, V.M.; Maldonado-Alconada, A.M.; Amil-Ruiz, F.; Jorrin-Novo, J.V. Holm Oak (Quercus ilex) Transcriptome. De Novo Sequencing and Assembly Analysis. Front. Mol. Biosci. 2017, 4, 70. [Google Scholar] [CrossRef]
- Guerrero-Sanchez, V.M.; Maldonado-Alconada, A.M.; Amil-Ruiz, F.; Verardi, A.; Jorrín-Novo, J.V.; Rey, M.-D. Ion Torrent and Lllumina, Two Complementary RNA-Seq Platforms for Constructing the Holm Oak (Quercus ilex) Transcriptome. PLoS ONE 2019, 14, e0210356. [Google Scholar] [CrossRef]
- Al Shweiki, M.R.; Mönchgesang, S.; Majovsky, P.; Thieme, D.; Trutschel, D.; Hoehenwarter, W. Assessment of Label-Free Quantification in Discovery Proteomics and Impact of Technological Factors and Natural Variability of Protein Abundance. J. Proteome Res. 2017, 16, 1410–1424. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A Fast and Simple Web Server for Genome Scale Functional Annotation of Plant Sequence Data. Plant Cell Environ. 2014, 37, 1250–1258. [Google Scholar] [CrossRef]
- KEGG: Kyoto Encyclopedia of Genes and Genomes Database. Available online: https://www.genome.jp/kegg/ (accessed on 15 November 2023).
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated Chemical Classification with a Comprehensive, Computable Taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef]
- MetabolomicsWorkbench. Available online: https://www.metabolomicsworkbench.org (accessed on 12 December 2023).
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 10 October 2023).
- Yan, L. Ggvenn: Draw Venn Diagram by “Ggplot2”. R Package, Version 19; R Project: Vienna, Austria, 2021.
- Argelaguet, R.; Velten, B.; Arnol, D.; Dietrich, S.; Zenz, T.; Marioni, J.C.; Buettner, F.; Huber, W.; Stegle, O. Multi-Omics Factor Analysis—A Framework for Unsupervised Integration of Multi-omics Data Sets. Mol. Syst. Biol. 2018, 14, e8124. [Google Scholar] [CrossRef] [PubMed]
- Vivas, M.; Hernández, J.; Corcobado, T.; Cubera, E.; Solla, A. Transgenerational Induction of Resistance to Phytophthora cinnamomi in Holm Oak. Forests 2021, 12, 100. [Google Scholar] [CrossRef]
- Jeffers, S.N. Comparison of Two Media Selective for Phytophthora and Pythium Species. Plant Dis. 1986, 70, 1038. [Google Scholar] [CrossRef]
- San-Eufrasio, B.; Bigatton, E.D.; Guerrero-Sánchez, V.M.; Chaturvedi, P.; Jorrín-Novo, J.V.; Rey, M.-D.; Castillejo, M.Á. Proteomics Data Analysis for the Identification of Proteins and Derived Proteotypic Peptides of Potential Use as Putative Drought Tolerance Markers for Quercus ilex. Int. J. Mol. Sci. 2021, 22, 3191. [Google Scholar] [CrossRef] [PubMed]
- Amaral, J.; Ribeyre, Z.; Vigneaud, J.; Sow, M.D.; Fichot, R.; Messier, C.; Pinto, G.; Nolet, P.; Maury, S. Advances and Promises of Epigenetics for Forest Trees. Forests 2020, 11, 976. [Google Scholar] [CrossRef]
- Hofmeister, B.T.; Denkena, J.; Colomé-Tatché, M.; Shahryary, Y.; Hazarika, R.; Grimwood, J.; Mamidi, S.; Jenkins, J.; Grabowski, P.P.; Sreedasyam, A.; et al. A Genome Assembly and the Somatic Genetic and Epigenetic Mutation Rate in a Wild Long-Lived Perennial Populus trichocarpa. Genome Biol. 2020, 21, 259. [Google Scholar] [CrossRef]
- Bolhàr-Nordenkampf, H.R.; Öquist, G. Chlorophyll Fluorescence as a Tool in Photosynthesis Research. In Photosynthesis and Production in a Changing Environment; Springer: Dordrecht, The Netherlands, 1993; pp. 193–206. [Google Scholar]
- Peguero-Pina, J.J.; Sancho-Knapik, D.; Morales, F.; Flexas, J.; Gil-Pelegrín, E. Differential Photosynthetic Performance and Photoprotection Mechanisms of Three Mediterranean Evergreen Oaks under Severe Drought Stress. Funct. Plant Biol. 2009, 36, 453. [Google Scholar] [CrossRef] [PubMed]
- Sancho-Knapik, D.; Mendoza-Herrer, Ó.; Gil-Pelegrín, E.; Peguero-Pina, J. Chl Fluorescence Parameters and Leaf Reflectance Indices Allow Monitoring Changes in the Physiological Status of Quercus ilex L. under Progressive Water Deficit. Forests 2018, 9, 400. [Google Scholar] [CrossRef]
- Valero-Galván, J.; González-Fernández, R.; Navarro-Cerrillo, R.M.; Gil-Pelegrín, E.; Jorrín-Novo, J.V. Physiological and Proteomic Analyses of Drought Stress Response in Holm Oak Provenances. J. Proteome Res. 2013, 12, 5110–5123. [Google Scholar] [CrossRef]
- Castillejo, M.A.; Pascual, J.; Jorrín-Novo, J.V.; Balbuena, T.S. Proteomics Research in Forest Trees: A 2012–2022 Update. Front. Plant Sci. 2023, 14, 1130665. [Google Scholar] [CrossRef]
- Jeandet, P.; Formela-Luboińska, M.; Labudda, M.; Morkunas, I. The Role of Sugars in Plant Responses to Stress and Their Regulatory Function during Development. Int. J. Mol. Sci. 2022, 23, 5161. [Google Scholar] [CrossRef]
- Hartmann, H.; Trumbore, S. Understanding the Roles of Nonstructural Carbohydrates in Forest Trees—From What We Can Measure to What We Want to Know. New Phytol. 2016, 211, 386–403. [Google Scholar] [CrossRef]
- Niu, L.; Liu, L.; Wang, W. Digging for Stress-Responsive Cell Wall Proteins for Developing Stress-Resistant Maize. Front. Plant Sci. 2020, 11, 576385. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, L.; Asiegbu, F.O.; Yang, C.; Han, S.; Yang, S.; Zeng, Q.; Liu, Y. Molecular Identification and Antifungal Properties of Four Thaumatin-like Proteins in Spruce (Picea likiangensis). Forests 2021, 12, 1268. [Google Scholar] [CrossRef]
- Roosens, N.H.; Bitar, F.A.; Loenders, K.; Angenon, G.; Jacobs, M. Overexpression of Ornithine-δ-Aminotransferase Increases Proline Biosynthesis and Confers Osmotolerance in Transgenic Plants. Mol. Breed. 2002, 9, 73–80. [Google Scholar] [CrossRef]
- Anwar, A.; She, M.; Wang, K.; Riaz, B.; Ye, X. Biological Roles of Ornithine Aminotransferase (OAT) in Plant Stress Tolerance: Present Progress and Future Perspectives. Int. J. Mol. Sci. 2018, 19, 3681. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Luo, F.; Zou, R.; Liu, J.; Yan, Y. Integrated Physiological and Chloroplast Proteome Analysis of Wheat Seedling Leaves under Salt and Osmotic Stresses. J. Proteom. 2021, 234, 104097. [Google Scholar] [CrossRef]
- Guerrero-Sánchez, V.M.; López-Hidalgo, C.; Rey, M.-D.; Castillejo, M.Á.; Jorrín-Novo, J.V.; Escandón, M. Multiomic Data Integration in the Analysis of Drought-Responsive Mechanisms in Quercus ilex Seedlings. Plants 2022, 11, 3067. [Google Scholar] [CrossRef] [PubMed]
- Park, C.-J.; Seo, Y.-S. Heat Shock Proteins: A Review of the Molecular Chaperones for Plant Immunity. Plant Pathol. J. 2015, 31, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Sánchez, V.M.; Castillejo, M.Á.; López-Hidalgo, C.; Alconada, A.M.M.; Jorrín-Novo, J.V.; Rey, M.-D. Changes in the Transcript and Protein Profiles of Quercus ilex Seedlings in Response to Drought Stress. J. Proteom. 2021, 243, 104263. [Google Scholar] [CrossRef]
- Kong, F.; Deng, Y.; Zhou, B.; Wang, G.; Wang, Y.; Meng, Q. A Chloroplast-Targeted DnaJ Protein Contributes to Maintenance of Photosystem II under Chilling Stress. J. Exp. Bot. 2014, 65, 143–158. [Google Scholar] [CrossRef]
- Richardson, L.G.L.; Singhal, R.; Schnell, D.J. The Integration of Chloroplast Protein Targeting with Plant Developmental and Stress Responses. BMC Biol. 2017, 15, 118. [Google Scholar] [CrossRef]
- Richardson, L.G.L.; Schnell, D.J. Origins, Function, and Regulation of the TOC–TIC General Protein Import Machinery of Plastids. J Exp. Bot. 2020, 71, 1226–1238. [Google Scholar] [CrossRef]
- Seidel, T. The Plant V-ATPase. Front. Plant Sci. 2022, 13, 931777. [Google Scholar] [CrossRef]
- Wei, H.; Movahedi, A.; Liu, G.; Li, Y.; Liu, S.; Yu, C.; Chen, Y.; Zhong, F.; Zhang, J. Comprehensive Analysis of Carotenoid Cleavage Dioxygenases Gene Family and Its Expression in Response to Abiotic Stress in Poplar. Int. J. Mol. Sci. 2022, 23, 1418. [Google Scholar] [CrossRef]
- Shang, C.; Ye, T.; Zhou, Q.; Chen, P.; Li, X.; Li, W.; Chen, S.; Hu, Z.; Zhang, W. Genome-Wide Identification and Bioinformatics Analyses of Host Defense Peptides Snakin/GASA in Mangrove Plants. Genes 2023, 14, 923. [Google Scholar] [CrossRef]
- Malnoë, A.; Schultink, A.; Shahrasbi, S.; Rumeau, D.; Havaux, M.; Niyogi, K.K. The Plastid Lipocalin LCNP Is Required for Sustained Photoprotective Energy Dissipation in Arabidopsis. Plant Cell 2018, 30, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Henri, P.; Rumeau, D. Ectopic Expression of Human Apolipoprotein D in Arabidopsis Plants Lacking Chloroplastic Lipocalin Partially Rescues Sensitivity to Drought and Oxidative Stress. Plant Physiol. Biochem. 2021, 158, 265–274. [Google Scholar] [CrossRef]
- Garai, S.; Bhowal, B.; Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. What Signals the Glyoxalase Pathway in Plants? Physiol. Mol. Biol. Plants 2021, 27, 2407–2420. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Glutathione in Plants: Biosynthesis and Physiological Role in Environmental Stress Tolerance. Physiol. Mol. Biol. Plants 2017, 23, 249–268. [Google Scholar] [CrossRef]
- Saiz-Fernández, I.; Milenković, I.; Berka, M.; Černý, M.; Tomšovský, M.; Brzobohatý, B.; Kerchev, P. Integrated Proteomic and Metabolomic Profiling of Phytophthora cinnamomi Attack on Sweet Chestnut (Castanea sativa) Reveals Distinct Molecular Reprogramming Proximal to the Infection Site and Away from It. Int. J. Mol. Sci. 2020, 21, 8525. [Google Scholar] [CrossRef] [PubMed]
- MacRobbie, E.A.C. Signalling in Guard Cells and Regulation of Ion Channel Activity. J. Exp. Bot. 1997, 48, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Luan, S. Signalling Drought in Guard Cells. Plant Cell Environ. 2002, 25, 229–237. [Google Scholar] [CrossRef]
- Xu, P.; Ma, W.; Liu, J.; Hu, J.; Cai, W. Overexpression of a Small GTP-binding Protein Ran1 in Arabidopsis Leads to Promoted Elongation Growth and Enhanced Disease Resistance against P. Syringae DC3000. Plant J. 2021, 108, 977–991. [Google Scholar] [CrossRef]
- Engelhardt, S.; Trutzenberg, A.; Hückelhoven, R. Regulation and Functions of ROP GTPases in Plant–Microbe Interactions. Cells 2020, 9, 2016. [Google Scholar] [CrossRef] [PubMed]
- Ono, E. Essential Role of the Small GTPase Rac in Disease Resistance of Rice. Proc. Natl. Acad. Sci. USA 2001, 98, 759–764. [Google Scholar] [CrossRef]
- Kawano, Y.; Kaneko-Kawano, T.; Shimamoto, K. Rho Family GTPase-Dependent Immunity in Plants and Animals. Front. Plant Sci. 2014, 5, 522. [Google Scholar] [CrossRef]
- Nadimpalli, R.; Yalpani, N.; Johal, G.S.; Simmons, C.R. Prohibitins, Stomatins, and Plant Disease Response Genes Compose a Protein Superfamily That Controls Cell Proliferation, Ion Channel Regulation, and Death. J. Biol. Chem. 2000, 275, 29579–29586. [Google Scholar] [CrossRef] [PubMed]
- Gershater, M.C.; Edwards, R. Regulating Biological Activity in Plants with Carboxylesterases. Plant Sci. J. 2007, 173, 579–588. [Google Scholar] [CrossRef]
- Leinweber, F.-J. Possible Physiological Roles of Carboxylic Ester Hydrolases. Drug Metab. Rev. 1987, 18, 379–439. [Google Scholar] [CrossRef] [PubMed]
- Viswanath, K.K.; Varakumar, P.; Pamuru, R.R.; Basha, S.J.; Mehta, S.; Rao, A.D. Plant Lipoxygenases and Their Role in Plant Physiology. J. Plant Biol. 2020, 63, 83–95. [Google Scholar] [CrossRef]
- Soprano, A.S.; Smetana, J.H.C.; Benedetti, C.E. Regulation of tRNA Biogenesis in Plants and Its Link to Plant Growth and Response to Pathogens. Biochim. Biophys. Acta Gene Regul. 2018, 1861, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Duan, N.; Chen, X.; Li, H.; Zhao, X.; Duo, P.; Wang, J.; Li, Q. Metabolic Pathways Involved in the Drought Stress Response of Nitraria tangutorum as Revealed by Transcriptome Analysis. Forests 2022, 13, 509. [Google Scholar] [CrossRef]
- Gugger, P.F.; Peñaloza-Ramírez, J.M.; Wright, J.W.; Sork, V.L. Whole-Transcriptome Response to Water Stress in a California Endemic Oak, Quercus lobata. Tree Physiol. 2016, 37, 632–644. [Google Scholar] [CrossRef]
- Ren, S.; Ma, K.; Lu, Z.; Chen, G.; Cui, J.; Tong, P.; Wang, L.; Teng, N.; Jin, B. Transcriptomic and Metabolomic Analysis of the Heat-Stress Response of Populus tomentosa Carr. Forests 2019, 10, 383. [Google Scholar] [CrossRef]
- Tronchet, M.; Balagué, C.; Kroj, T.; Jouanin, L.; Roby, D. Cinnamyl Alcohol Dehydrogenases-C and D, Key Enzymes in Lignin Biosynthesis, Play an Essential Role in Disease Resistance in Arabidopsis. Mol. Plant Pathol. 2010, 11, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Madritsch, S.; Wischnitzki, E.; Kotrade, P.; Ashoub, A.; Burg, A.; Fluch, S.; Brüggemann, W.; Sehr, E.M. Elucidating Drought Stress Tolerance in European Oaks Through Cross-Species Transcriptomics. G3 2019, 9, 3181–3199. [Google Scholar] [CrossRef] [PubMed]
- Tienda-Parrilla, M.; López-Hidalgo, C.; Guerrero-Sanchez, V.M.; Infantes-González, Á.; Valderrama-Fernández, R.; Castillejo, M.-Á.; Jorrín-Novo, J.V.; Rey, M.-D. Untargeted MS-Based Metabolomics Analysis of the Responses to Drought Stress in Quercus ilex L. Leaf Seedlings and the Identification of Putative Compounds Related to Tolerance. Forests 2022, 13, 551. [Google Scholar] [CrossRef]
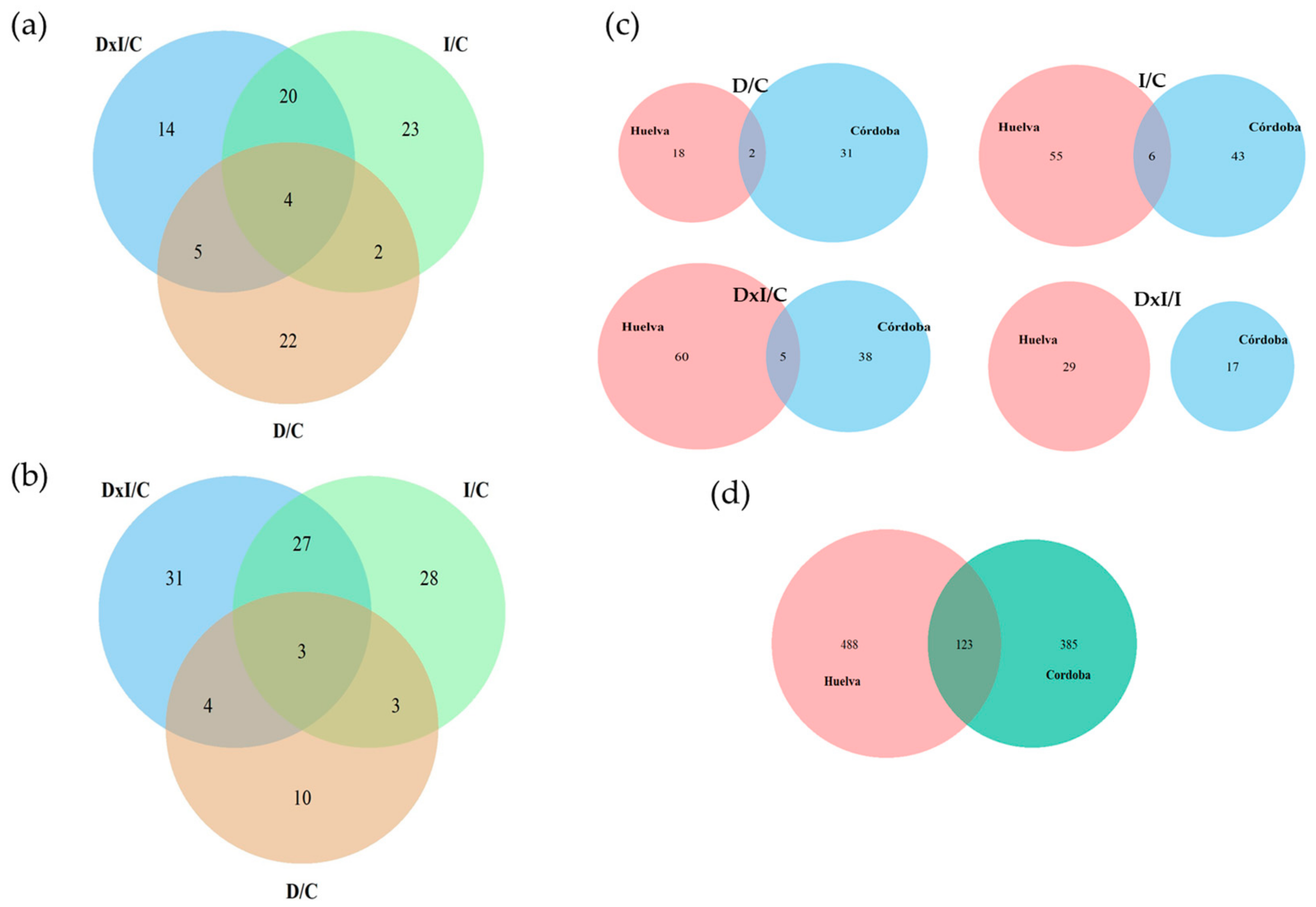
| Feature | Cordoba | Huelva | Common to Both |
|---|---|---|---|
| Raw data | 2735/10,983 | 3019/10,051 | 2540/7798 |
| ≥2 peptides | 2261 | 2451 | 2261 |
| Score ≥ 2 | 2260 | 2448 | 2203 |
| Consistent proteins | 1590 | 1444 | 1334 |
| Statistical significance (p ≤ 0.05) | 600/1298 | 525/1309 | 350/503 |
| Up-accumulated: | |||
| D/C | 33 | 20 | 2 |
| I/C | 49 | 61 | 6 |
| DxI/C | 43/385 | 65/488 | 5/123 |
| DxI/I | 17 | 29 | 0 |
| Molecular Function | Protein ID | Protein Description | Up-Accumulated | Population |
|---|---|---|---|---|
| Carbohydrate metabolism | qilexprot_70617 | Fructose-1,6-bisphosphatase, chloroplastic | D × I/D × I, I | Co/Hu |
| qilexprot_74362 | Beta-xylosidase/alpha-L-arabinofuranosidase 2 | D√I/I | Co/Hu | |
| qilexprot_27855 | D-tagatose-1,6-bisphosphate aldolase subunit gatY | D × I, I/D × I, I | Co/Hu | |
| qilexprot_67493 | Phosphoglycerate mutase (2,3-diphosphoglycerate-independent) | D × I, I/I | Co/Hu | |
| qilexprot_46285 | Phosphoenolpyruvate carboxykinase (ATP) 1 | D × I, I, D/D × I, I | Co/Hu | |
| qilexprot_11680 | Pyruvate dehydrogenase E1 component subunit alpha, mitochondrial | I/D × I | Co/Hu | |
| qilexprot_45099 | Beta-galactosidase | I/D × I, I | Co/Hu | |
| qilexprot_65736 | Phosphoglycerate kinase | D | Co | |
| qilexprot_54727 | Malate dehydrogenase | D | Co | |
| qilexprot_57046 | Malate dehydrogenase | D × I | Co | |
| qilexprot_32059 | Glucose-6-phosphate 1-dehydrogenase, cytoplasmic isoform 2 | D | Co | |
| qilexprot_19946 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | D | Co | |
| qilexprot_10831 | FGGY carbohydrate kinase domain-containing protein | D | Co | |
| qilexprot_58277 | Aconitate hydratase | D × I | Co | |
| qilexprot_73875 | Malic enzyme | D × I, D | Co | |
| qilexprot_20787 | Uridine 5’-diphosphate-sulfoquinovose synthase | D × I, I | Co | |
| qilexprot_43884 | Glyoxalase I | D × I, I | Co | |
| qilexprot_52600 | Pyruvate kinase | D × I, I | Co | |
| qilexprot_12453 | Fructose-1,6-bisphosphatase | D × I, I | Co | |
| qilexprot_32875 | Alcohol dehydrogenase 1 | D × I, I | Co | |
| qilexprot_48598 | Pyruvate decarboxylase 1 | D × I, I, D | Co | |
| qilexprot_38653 | Succinate dehydrogenase (ubiquinone) flavoprotein subunit, mitochondrial | I | Co | |
| qilexprot_18792 | Triosephosphate isomerase, cytosolic | I | Co | |
| qilexprot_38624 | ATP-dependent 6-phosphofructokinase | I | Co | |
| qilexprot_19665 | Ribokinase | I | Co | |
| qilexprot_61660 | Sorbitol dehydrogenase | I | Co | |
| qilexprot_49147 | Succinyl–CoA ligase (ADP-forming) subunit alpha | I, D | Co | |
| qilexprot_48238 | Granule-bound starch synthase 1, chloroplastic/amyloplastic | D | Hu | |
| qilexprot_27919 | Alpha 1,4-glucan phosphorylase | D × I | Hu | |
| qilexprot_55364 | Glyceraldehyde-3-phosphate dehydrogenase | D × I | Hu | |
| qilexprot_75569 | Glyceraldehyde-3-phosphate dehydrogenase | D × I | Hu | |
| qilexprot_4907 | Probable galactinol-sucrose galactosyltransferase 2 | D × I, I | Hu | |
| qilexprot_49174 | 6-Phosphogluconate dehydrogenase, decarboxylating | D × I, I, D | Hu | |
| qilexprot_16282 | Aconitase | I | Hu | |
| qilexprot_178 | Acetyltransferase component of pyruvate dehydrogenase complex | I | Hu | |
| qilexprot_50083 | Phosphoglycerate kinase | D × I | Hu | |
| qilexprot_46354 | Succinyl–CoA ligase (ADP-forming) subunit alpha | D × I | Hu | |
| qilexprot_7036 | 6-phosphofructokinase, pyrophosphate-dependent | I | Hu | |
| Amino acid metabolism | qilexprot_49163 | Aspartate aminotransferase 3, chloroplastic | D/D | Co/Hu |
| qilexprot_19886 | Phosphoserine aminotransferase 1, chloroplastic | D/D × I, I | Co/Hu | |
| qilexprot_7937 | Glutamine synthetase | D × I | Co | |
| qilexprot_968 | Ferredoxin-dependent glutamate synthase | D × I, I | Co | |
| qilexprot_73896 | Carbamoyl-phosphate synthase large chain, chloroplastic | D × I, I | Co | |
| qilexprot_49389 | Arginase 1, mitochondrial | D | Hu | |
| qilexprot_74641 | D-3-phosphoglycerate dehydrogenase 1, chloroplastic | D × I | Hu | |
| qilexprot_41437 | Glutamate synthase (ferredoxin) | D × I, I | Hu | |
| qilexprot_45599 | Glutamate synthase (ferredoxin) | D × I, I | Hu | |
| qilexprot_26928 | Glutamate synthase (ferredoxin) | D × I, I | Hu | |
| qilexprot_7419 | Serine hydroxymethyltransferase | I | Hu | |
| qilexprot_55598 | Alpha isopropylmalate synthase | I | Hu | |
| Lipid metabolism | qilexprot_55785 | Epoxide hydrolase 3 | D | Co |
| qilexprot_56587 | NADH-cytochrome b5 reductase-like protein | D | Co | |
| qilexprot_36359 | Acyl-CoA oxidase 1 | D | Co | |
| qilexprot_19906 | Acyl-CoA dehydrogenase | D | Co | |
| qilexprot_58843 | Acyl-coenzyme A oxidase | D | Co | |
| qilexprot_29429 | Acetyl-CoA acetyltransferase | D × I | Co | |
| qilexprot_66905 | CXE carboxylesterase | I | Co | |
| qilexprot_40937 | CXE1 Carboxylesterase 1 | I | Co | |
| qilexprot_72077 | Lipoxygenase * | D × I | Co * | |
| qilexprot_60909 | 12-Oxophytodienoate reductase 1 | D | Hu | |
| qilexprot_77659 | Phospholipase D alpha 1 | D × I | Hu | |
| qilexprot_61680 | 3-Hydroxybutyryl-CoA dehydratase | I | Hu | |
| qilexprot_10167 | Acetate/butyrate–CoA ligase AAE7, peroxisomal | I, D | Hu | |
| Nucleotide metabolism | qilexprot_35697 | Phosphoribosylaminoimidazolecarboxamide formyltransferase | D × I, I | Co |
| qilexprot_38489 | Dihydropyrimidine dehydrogenase (NADP(+)), chloroplastic | D × I | Hu | |
| qilexprot_29525 | Adenylosuccinate synthetase, chloroplastic | D × I, I | Hu | |
| Cellular processes | qilexprot_59288 | MPBQ/MSBQ methyltransferase | D × I | Co |
| qilexprot_72324 | Putative membrane protease, stomatin/prohibitin-like protein | D × I, D | Co | |
| qilexprot_26311 | Aluminum-induced protein with YGL and LRDR motifs | I | Co | |
| qilexprot_32288 | Dynamin-related protein 3A | D × I | Hu | |
| qilexprot_19558 | Dynamin-related protein 3A | D × I, I | Hu | |
| qilexprot_31321 | Carotenoid cleavage dioxygenase 1 | D × I | Hu | |
| qilexprot_9244 | Turgor-responsive-like protein | D × I, I | Hu | |
| qilexprot_16156 | Tublin binding cofactor C | I | Hu | |
| Development | qilexprot_67941 | Apoptosis inhibitor 5-like protein API5 | I | Hu |
| qilexprot_56133 | ATPase family AAA domain-containing protein 3 | I | Hu | |
| qilexprot_68348 | Sieve element occlusion protein 1 | I | Hu | |
| Photosynthesis | qilexprot_6646 | Chlorophyll a-b binding protein, chloroplastic | D | Co |
| qilexprot_16571 | Chlorophyll a-b binding protein, chloroplastic | D | Hu | |
| qilexprot_77212 | Cytochrome b559 subunit alpha | D | Hu | |
| Energy metabolism | qilexprot_2302 | Ferredoxin-NADP reductase | I | Hu |
| Folding, sorting and degradation | qilexprot_41199 | Small heat shock protein, chloroplastic | I/D × I, I | Co/Hu |
| qilexprot_73457 | Non-heme dioxygenase N-terminal domain-containing protein | D | Co | |
| qilexprot_24541 | Non-heme dioxygenase N-terminal domain-containing protein | D × I, D | Co | |
| qilexprot_10667 | Glycine cleavage system H protein, mitochondrial | D × I | Co | |
| qilexprot_22106 | Xaa-Pro aminopeptidase 2 | D × I | Co | |
| qilexprot_76191 | Heat shock 70 kDa protein | D × I, I | Co | |
| qilexprot_68090 | Putative heat shock protein 70 | I | Co | |
| qilexprot_42055 | Proteasome subunit beta type-1 | I | Co | |
| qilexprot_4991 | Heat shock protein 40 | D | Hu | |
| qilexprot_11703 | Chaperone protein DnaJ A6, chloroplastic | D × I | Hu | |
| qilexprot_17542 | 22.0 kDa class IV heat shock protein | D × I | Hu | |
| qilexprot_67032 | Proteasome subunit beta type 4 | D × I | Hu | |
| qilexprot_36038 | Small heat shock protein | D × I | Hu | |
| qilexprot_55307 | Small heat shock protein, chloroplastic | D × I, I | Hu | |
| qilexprot_29045 | Protein Cpn60 * | D × I | Hu/Co * | |
| qilexprot_38494 | Presequence protease 1, chloroplastic/mitochondrial | D × I, I | Hu | |
| qilexprot_31991 | Oligopeptidase A (partial) | D × I, I | Hu | |
| qilexprot_50467 | ClpB | D × I, I | Hu | |
| qilexprot_46770 | TCPB | D × I, I | Hu | |
| qilexprot_46485 | Chaperone protein DnaK | D × I, I | Hu | |
| qilexprot_55829 | T-complex protein 1 subunit beta | D × I, I, D | Hu | |
| qilexprot_68821 | Prolyl tripeptidyl peptidase | I | Hu | |
| Redox | qilexprot_41994 | Oxidoreductase | D | Co |
| qilexprot_47035 | NADH-ubiquinone reductase 75 kDa subunit | D × I | Co | |
| qilexprot_8286 | Monothiol glutaredoxin-S17 | D × I | Co | |
| qilexprot_22755 | Thioredoxin reductase | D × I, I | Co | |
| qilexprot_13916 | Glutathione reductase, chloroplastic | D × I, I | Co | |
| qilexprot_4433 | NADPH-dependent thioredoxin reductase 3 | D × I, I | Co | |
| qilexprot_27576 | Short-chain dehydrogenase/reductase SDR | I | Co | |
| qilexprot_67856 | Sulfite reductase 1 (ferredoxin), chloroplastic | I | Co | |
| qilexprot_53092 | Thioredoxin reductase 1 | I | Co | |
| qilexprot_38487 | Glutathione reductase | I | Co | |
| qilexprot_21080 | Putative uncharacterized protein PDI | D × I | Hu | |
| qilexprot_21353 | Mitochondrial peroxiredoxin IIF | D × I | Hu | |
| qilexprot_67058 | Aldo/keto reductase | D × I | Hu | |
| qilexprot_6108 | Peroxiredoxin-2B | D × I | Hu | |
| qilexprot_4209 | Polyphenol oxidase | D × I, D | Hu | |
| qilexprot_45306 | NADPH HC toxin reductase | D × I, D | Hu | |
| Secondary metabolism | qilexprot_7930 | Cinnamoyl alcohol dehydrogenase | D | Co |
| qilexprot_62609 | Clavaminate synthase-like protein | D | Co | |
| qilexprot_67426 | Cinnamyl alcohol dehydrogenase 1 | D × I, I | Co | |
| qilexprot_29542 | Chalcone synthase | I | Co | |
| qilexprot_16776 | Protein STRICTOSIDINE SYNTHASE-LIKE 3 | D | Hu | |
| qilexprot_29588 | Betaine aldehyde dehydrogenase | I | Hu | |
| qilexprot_6949 | Glutamate decarboxylase | I | Hu | |
| Stress | qilexprot_16524 | Ornithine aminotransferase | D/D × I, D | Co/Hu |
| qilexprot_66333 | Thaumatin-like protein 1a | D × I, D/D × I, I | Co/Hu | |
| qilexprot_52513 | Thaumatin-like protein 1 | D × I, D/D × I, I | Co/Hu | |
| qilexprot_62485 | Snakin-2 | D × I | Hu | |
| qilexprot_4320 | Temperature-induced lipocalin-1 | D × I | Hu | |
| qilexprot_75875 | Cysteine desulfurase 1, chloroplastic | I | Hu | |
| qilexprot_77543 | Succinate-semialdehyde dehydrogenase | I | Hu | |
| Transcription/Translation | qilexprot_49279 | Plastid lipid-associated protein 3, chloroplastic | I/D × I, I | Co/Hu |
| qilexprot_32535 | Plastid transcriptionally active 16 | D × I, I | Co | |
| qilexprot_33379 | Plastid transcriptionally active 16 | D × I, I | Co | |
| qilexprot_40661 | Histone H2A | D × I, I | Co | |
| qilexprot_59253 | Histone H2A | I, D | Hu | |
| qilexprot_31771 | EF-Tu | D × I, I | Co | |
| qilexprot_48469 | DNA-directed RNA polymerase subunit alpha | D × I, I, D | Co | |
| qilexprot_70773 | Alanine–tRNA ligase | I | Co | |
| qilexprot_67436 | 50S ribosomal protein L1, chloroplastic | I | Co | |
| qilexprot_61172 | Ubiquitin-conjugating enzyme E2 variant 1D | D | Hu | |
| qilexprot_35357 | Developmentally regulated G-protein 3 | D × I | Hu | |
| qilexprot_47135 | 50S ribosomal protein L17, chloroplastic | D × I | Hu | |
| qilexprot_28988 | Elongation factor 2 | D × I | Hu | |
| qilexprot_48468 | 30S ribosomal protein S8, chloroplastic | D × I | Hu | |
| qilexprot_22027 | Histone H4 | D × I | Hu | |
| qilexprot_71551 | Ubiquitin-60S ribosomal protein L40 | D × I | Hu | |
| qilexprot_16675 | 50S ribosomal protein L22, chloroplastic | D × I, D | Hu | |
| qilexprot_74256 | Leucine–tRNA ligase, cytoplasmic * | D × I, I | Hu * | |
| qilexprot_42234 | Glycine–tRNA ligase, chloroplastic/mitochondrial 2 | D × I, I | Hu | |
| qilexprot_49297 | 60S ribosomal protein L13-1 | I | Hu | |
| qilexprot_19368 | Polyadenylate-binding protein RBP45 | I | Hu | |
| qilexprot_50137 | 60S ribosomal protein L13a-2 | I | Hu | |
| qilexprot_38968 | 60S ribosomal protein L21-1 | I | Hu | |
| qilexprot_56118 | 60S ribosomal protein L6 | I | Hu | |
| qilexprot_46161 | Methionyl-tRNA synthetase | D × I, I | Hu | |
| Signaling | qilexprot_22041 | Mitochondrial Rho GTPase 1 | D × I | Co |
| qilexprot_41913 | NADH:ubiquinone oxidoreductase complex I intermediate-associated protein 30 | D × I, I | Co | |
| Transport | qilexprot_13099 | Porin/voltage-dependent anion-selective channel protein * | D × I, I, D | Co */Hu * |
| qilexprot_22308 | GTP-binding nuclear protein Ran-3 | I | Co | |
| qilexprot_20347 | Hexapeptide transferase family protein | I | Co | |
| qilexprot_25532 | Ferritin | D × I | Hu | |
| qilexprot_9762 | Nuclear pore complex protein NUP98A | D × I, I | Hu | |
| qilexprot_48381 | Clathrin heavy chain 2 | D × I, I, D | Hu | |
| qilexprot_19028 | Polyadenylate-binding protein 8 * | I | Hu/Co * | |
| qilexprot_23804 | Protein TOC75, chloroplastic | I | Hu | |
| qilexprot_16395/ qilexprot_4497 | V-type proton ATPase catalytic subunit A/V-type proton ATPase subunit C * | I | Hu * | |
| qilexprot_64280 | Translocon at outer membrane of chloroplasts 64 | I | Hu | |
| qilexprot_74226 | PABP | I | Hu | |
| Miscellaneous | qilexprot_29442 | Aldehyde dehydrogenase * | D | Co * |
| qilexprot_42491 | Citrate synthase | D | Co | |
| qilexprot_71234 | UDP-sulfoquinovose synthase | D × I | Co | |
| qilexprot_14071 | At2g30880/F7F1.9 * | I, D | Hu * | |
| Not assigned | qilexprot_79169 | Putative uncharacterized protein Sb03g011745 | D | Co |
| qilexprot_20701 | Putative reversibly glycosylatable polypeptide | D | Co | |
| qilexprot_48145 | Putative uncharacterized protein At5g13030 | D × I | Co | |
| qilexprot_36527 | Fruit protein pKIWI502 | I, D | Co | |
| qilexprot_45328 | NAD(P)-binding Rossmann-fold protein | D | Hu | |
| qilexprot_34141 | NAD(P)-binding Rossmann-fold protein | D × I | Hu | |
| qilexprot_72624 | Spermatogenesis-associated protein 20 | D × I, I | Hu | |
| qilexprot_63727 | Pdx1 * | D × I | Hu * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Lao, T.; Tienda-Parrilla, M.; Labella-Ortega, M.; Guerrero-Sánchez, V.M.; Rey, M.-D.; Jorrín-Novo, J.V.; Castillejo-Sánchez, M.Á. Proteomic and Metabolomic Analysis of the Quercus ilex–Phytophthora cinnamomi Pathosystem Reveals a Population-Specific Response, Independent of Co-Occurrence of Drought. Biomolecules 2024, 14, 160. https://doi.org/10.3390/biom14020160
Hernández-Lao T, Tienda-Parrilla M, Labella-Ortega M, Guerrero-Sánchez VM, Rey M-D, Jorrín-Novo JV, Castillejo-Sánchez MÁ. Proteomic and Metabolomic Analysis of the Quercus ilex–Phytophthora cinnamomi Pathosystem Reveals a Population-Specific Response, Independent of Co-Occurrence of Drought. Biomolecules. 2024; 14(2):160. https://doi.org/10.3390/biom14020160
Chicago/Turabian StyleHernández-Lao, Tamara, Marta Tienda-Parrilla, Mónica Labella-Ortega, Victor M. Guerrero-Sánchez, María-Dolores Rey, Jesús V. Jorrín-Novo, and María Ángeles Castillejo-Sánchez. 2024. "Proteomic and Metabolomic Analysis of the Quercus ilex–Phytophthora cinnamomi Pathosystem Reveals a Population-Specific Response, Independent of Co-Occurrence of Drought" Biomolecules 14, no. 2: 160. https://doi.org/10.3390/biom14020160
APA StyleHernández-Lao, T., Tienda-Parrilla, M., Labella-Ortega, M., Guerrero-Sánchez, V. M., Rey, M.-D., Jorrín-Novo, J. V., & Castillejo-Sánchez, M. Á. (2024). Proteomic and Metabolomic Analysis of the Quercus ilex–Phytophthora cinnamomi Pathosystem Reveals a Population-Specific Response, Independent of Co-Occurrence of Drought. Biomolecules, 14(2), 160. https://doi.org/10.3390/biom14020160











