Computational Study of Network and Type-I Functional Divergence in Alcohol Dehydrogenase Enzymes Across Species Using Molecular Dynamics Simulation
Abstract
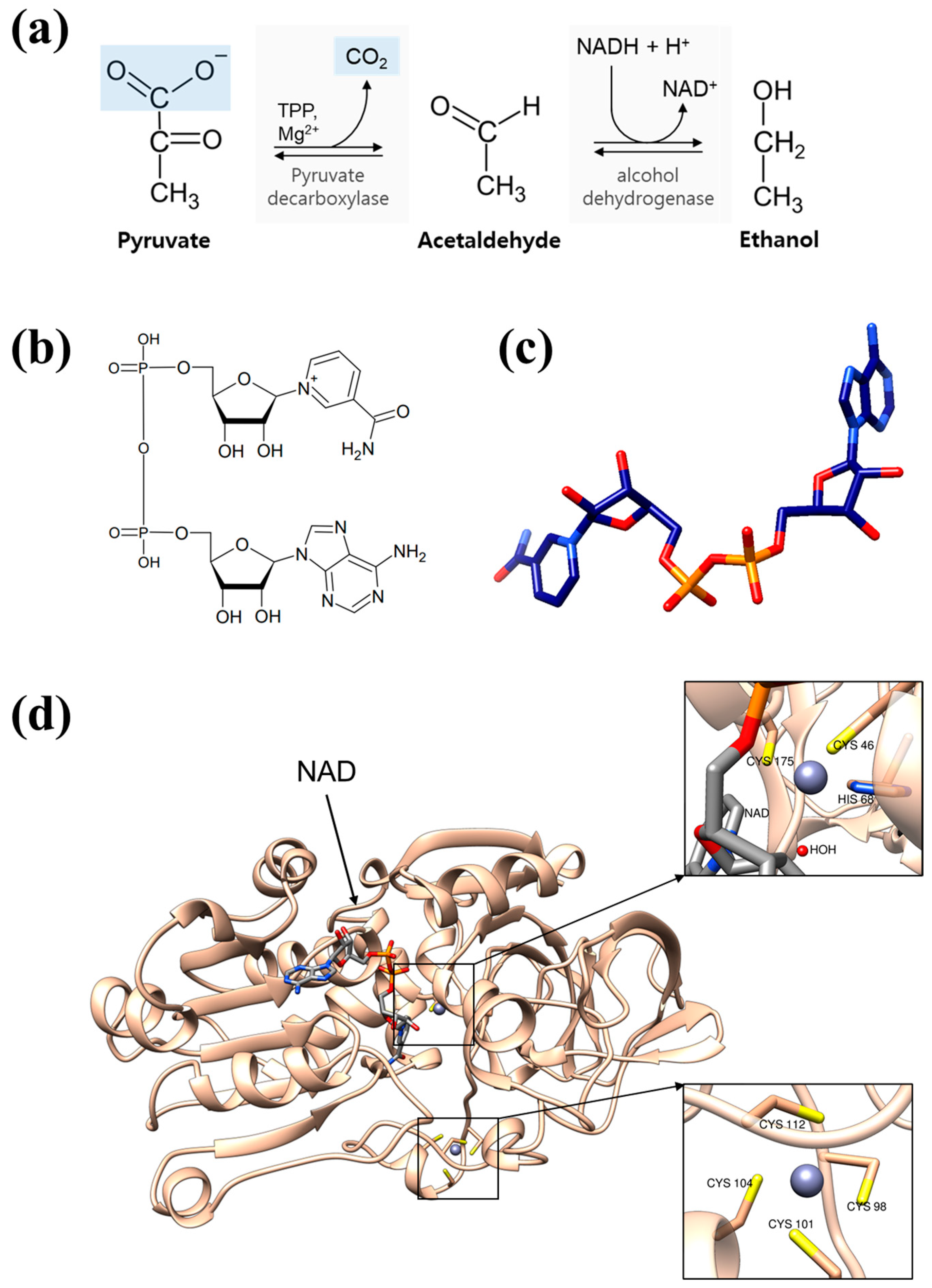
1. Introduction
2. Materials and Methods
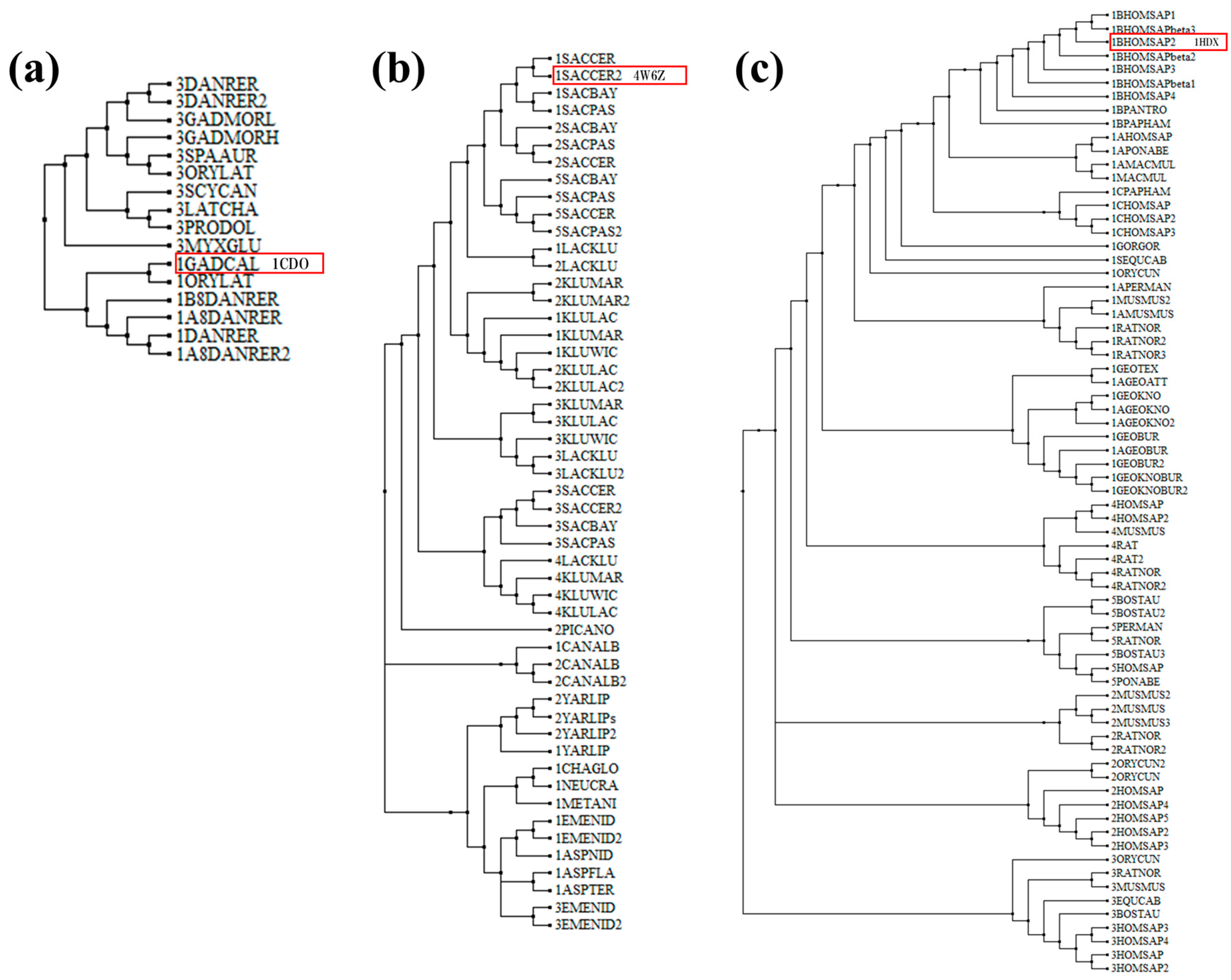
2.1. Phylogenetic Analysis
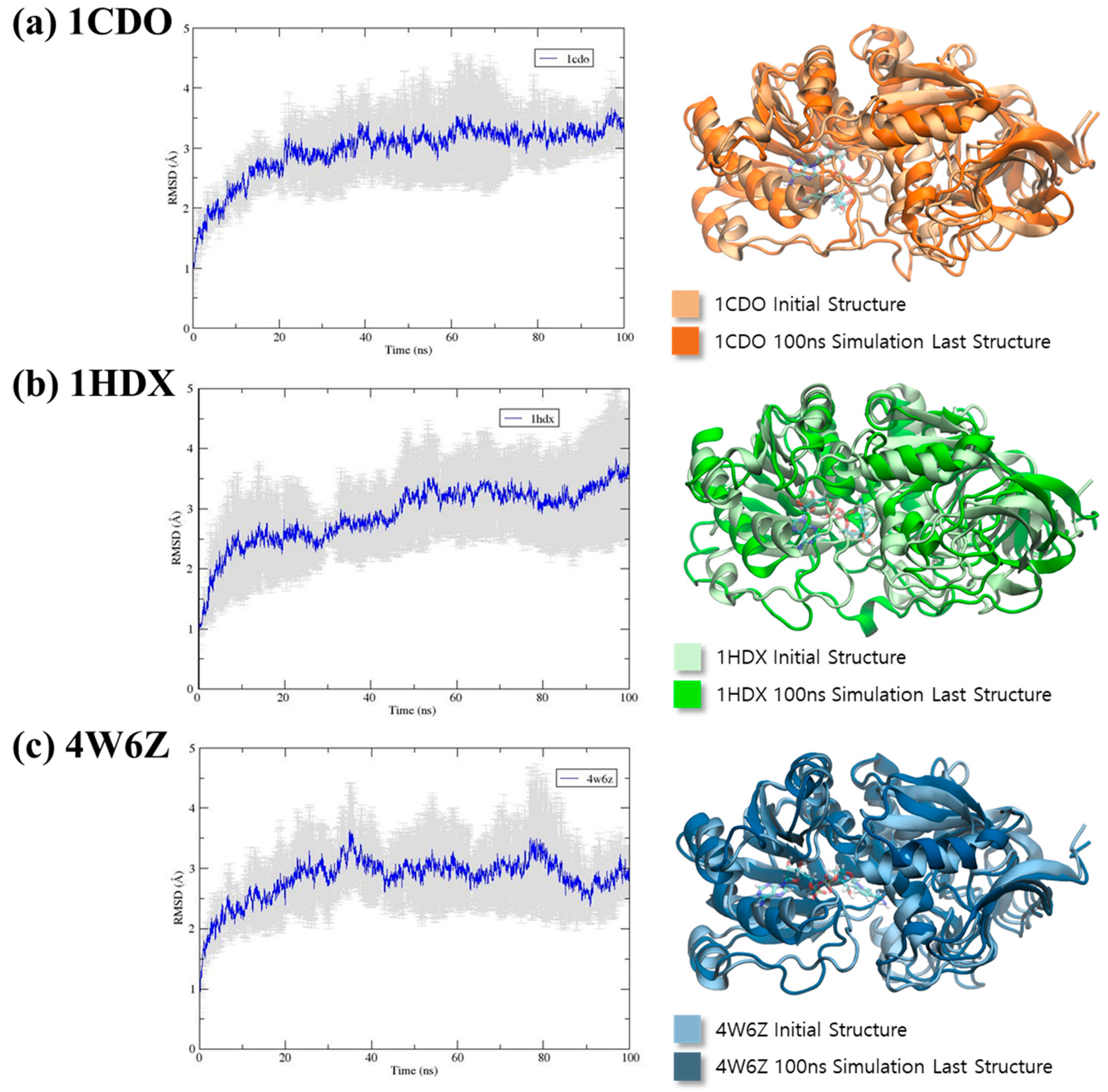
2.2. Molecular Dynamics Simulation
2.3. Network Analysis
2.4. Posterior Probability
3. Results
3.1. Phylogenetic Analysis
3.2. RMSD Analysis
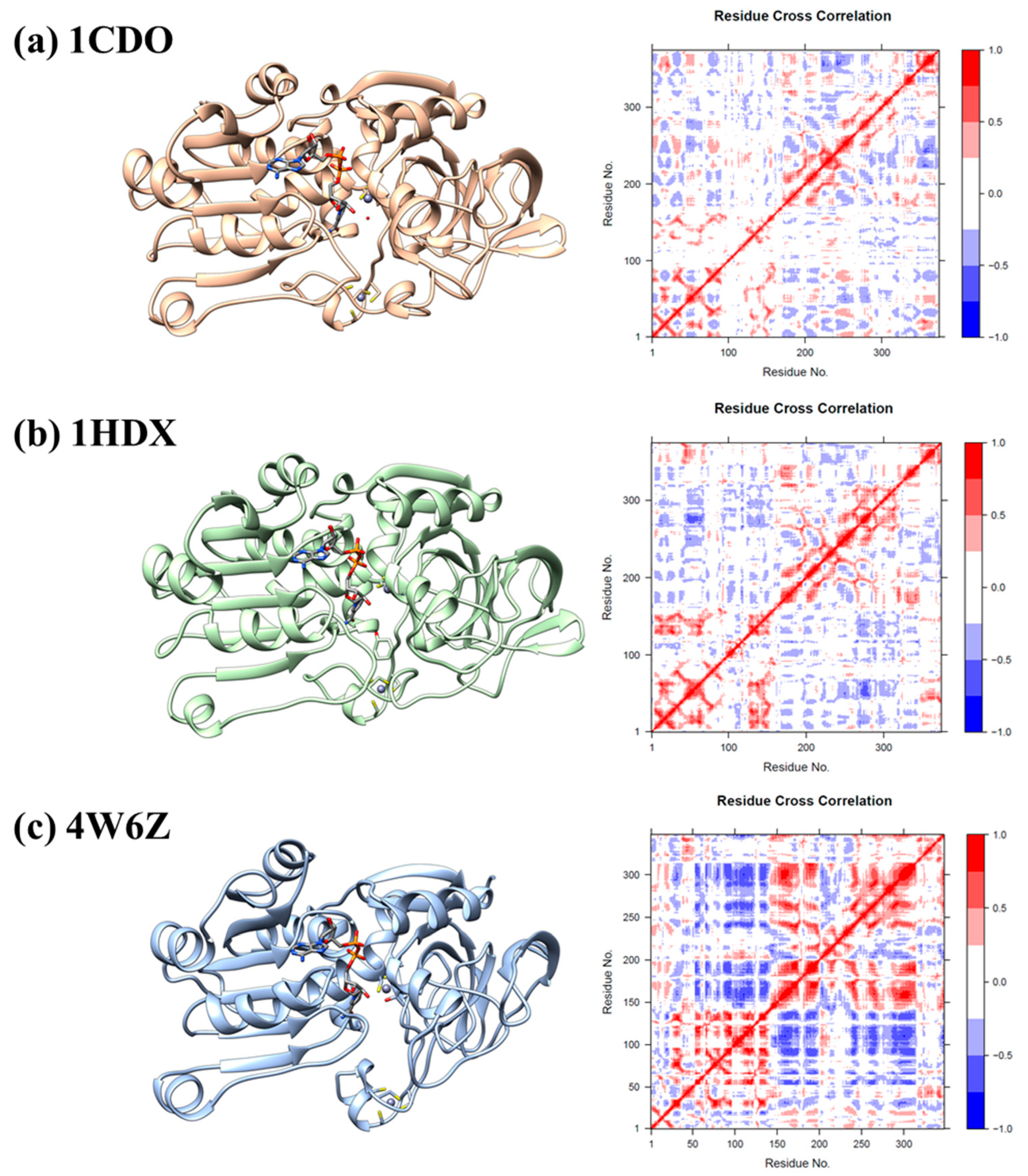
3.3. Dynamic Cross-Correlation Analysis
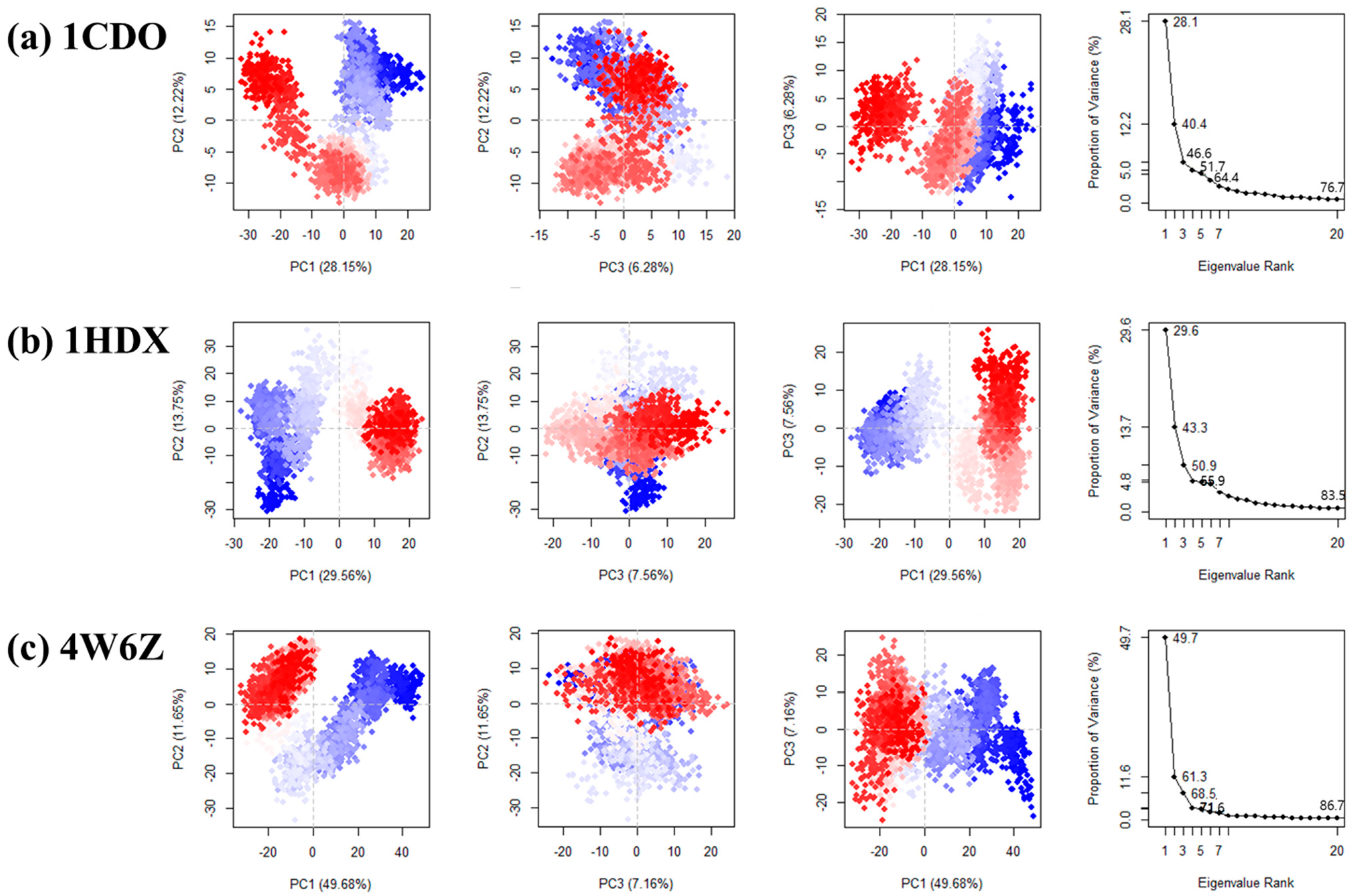
3.4. Principal Component Analysis
3.5. Centrality Measures
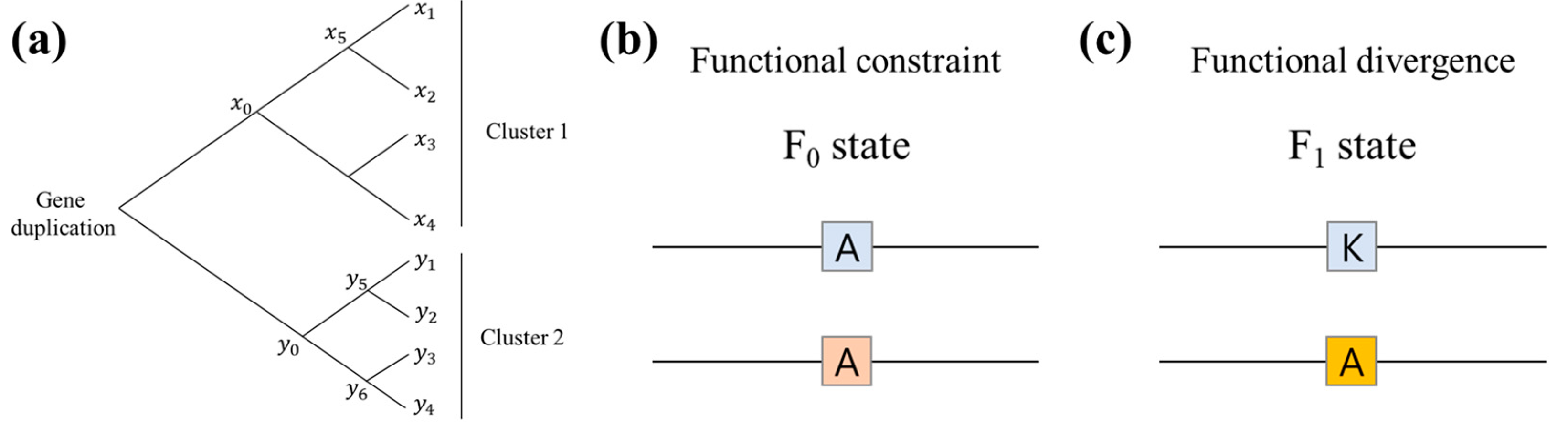
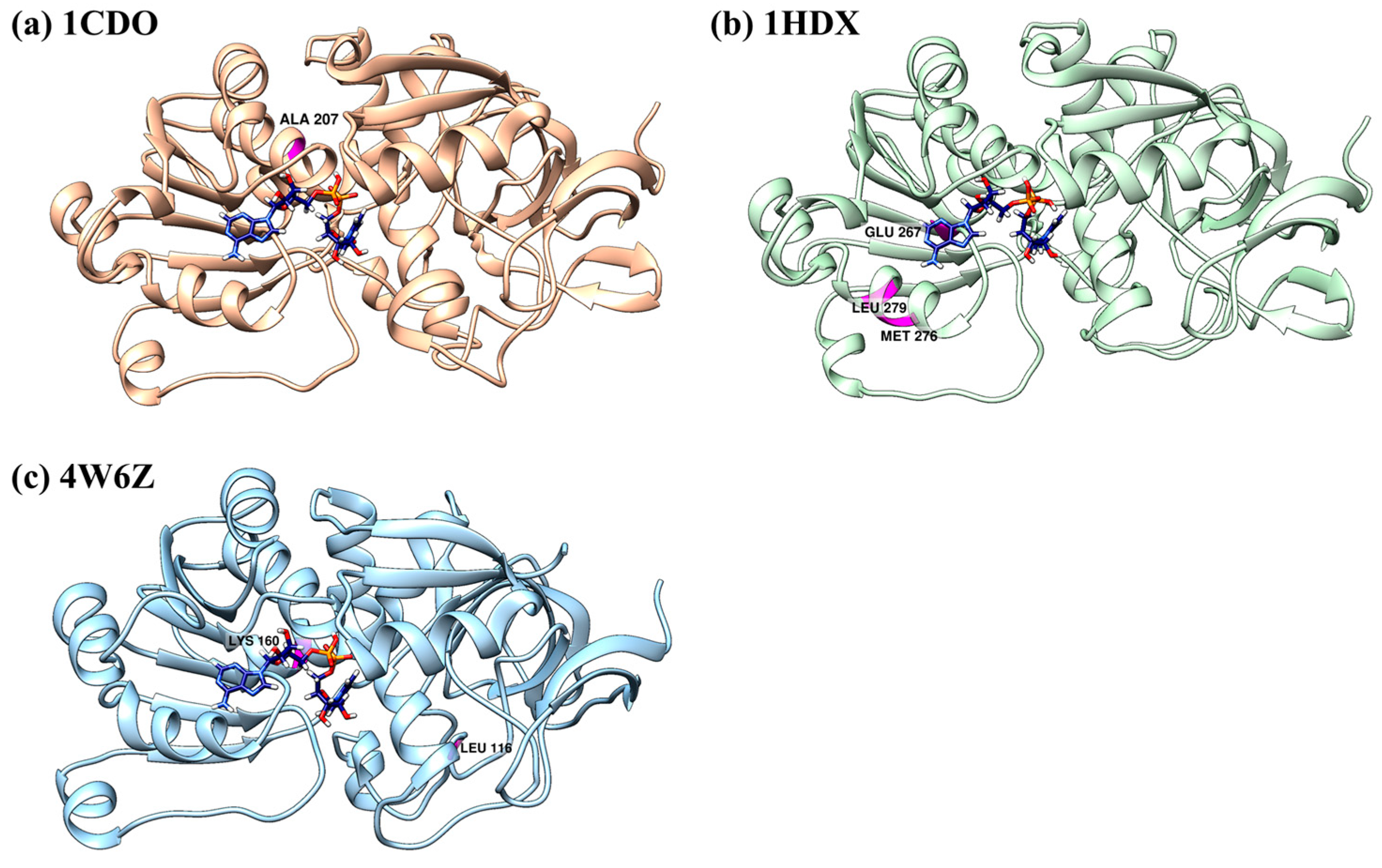
3.6. Type-I Functional Divergence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yin, S.-J.; Han, C.-L.; Lee, A.-I.; Wu, C.-W. Enzymology and Molecular Biology of Carbonyl Metabolism 7; Weiner, H., Maser, E., Crabb, D.W., Lindahl, R., Eds.; Springer: New York, NY, USA, 1999; p. 265. [Google Scholar]
- Di, L.; Balesano, A.; Jordan, S.; Shi, S.M. The Role of Alcohol Dehydrogenase in Drug Metabolism: Beyond Ethanol Oxidation. AAPS J. 2021, 23, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Zakhari, S. Alcohol metabolism and epigenetics changes. Alcohol Res. Curr. Rev. 2013, 35, 6–16. [Google Scholar]
- McQuillan, M.A.; Ranciaro, A.; Hansen, M.E.; Fan, S.; Beggs, W.; Belay, G.; Woldemeskel, D.; Tishkoff, S.A. Signatures of convergent evolution and natural selection at the alcohol dehydrogenase gene region are correlated with agriculture in ethnically diverse Africans. Mol. Biol. Evol. 2022, 39, msac183. [Google Scholar] [CrossRef] [PubMed]
- Plapp, B.V.; Green, D.W.; Sun, H.-W.; Park, D.-H.; Kim, K. Substrate Specificity of Alcohol Dehydrogenases. In Enzymology and Molecular Biology of Carbonyl Metabolism 4; Weiner, H., Crabb, D.W., Flynn, T.G., Eds.; Springer: Boston, MA, USA, 1993; pp. 391–400. [Google Scholar]
- Persson, B.; Hedlund, J.; Jornvall, H. Medium–and short-chain dehydrogenase/reductase gene and protein families: The MDR superfamily. Cell. Mol. Life Sci. 2008, 65, 3879–3894. [Google Scholar] [CrossRef]
- Kimura, M.; Yokoyama, A.; Matsushita, S.; Higuchi, S. Enzymatic Aspects of Alcoholism-ADH and ALDH. In Textbook of Addiction Treatment: International Perspectives; el-Guebaly, N., Carrà, G., Galanter, M., Eds.; Springer: Milan, Italy, 2015; pp. 333–342. [Google Scholar]
- Karabec, M.; Łyskowski, A.; Tauber, K.C.; Steinkellner, G.; Kroutil, W.; Grogan, G.; Gruber, K. Structural insights into substrate specificity and solvent tolerance in alcohol dehydrogenase ADH-‘A’from Rhodococcus ruber DSM 44541. Chem. Commun. 2010, 46, 6314–6316. [Google Scholar] [CrossRef]
- Liu, X.; Li, T.; Kong, D.; You, H.; Kong, F.; Tang, R. Prognostic implications of alcohol dehydrogenases in hepatocellular carcinoma. BMC Cancer 2020, 20, 1204. [Google Scholar] [CrossRef]
- Szalai, G.; Veres, M.; Duester, G.; Lawther, R.; Lockhart, M.; Felder, M.R. Tissue expression pattern of class II and class V genes found in the Adh complex on mouse chromosome 3. Biochem. Genet. 2008, 46, 685–695. [Google Scholar] [CrossRef]
- Nguyen Ba, A.N.; Strome, B.; Hua, J.J.; Desmond, J.; Gagnon-Arsenault, I.; Weiss, E.L.; Landry, C.R.; Moses, A.M. Detecting functional divergence after gene duplication through evolutionary changes in posttranslational regulatory sequences. PLoS Comput. Biol 2014, 10, e1003977. [Google Scholar] [CrossRef]
- Carrijo de Oliveira, L.; Figueiredo Costa, M.A.; Gonçalves Pedersolli, N.; Heleno Batista, F.A.; Migliorini Figueira, A.C.; Salgado Ferreira, R.; Alves Pinto Nagem, R.; Alves Nahum, L.; Bleicher, L. Reenacting the Birth of a Function: Functional Divergence of HIUases and Transthyretins as Inferred by Evolutionary and Biophysical Studies. J. Mol. Evol. 2021, 89, 370–383. [Google Scholar] [CrossRef]
- Thompson, C.E.; Salzano, F.M.; de Souza, O.N.; Freitas, L.B. Sequence and structural aspects of the functional diversification of plant alcohol dehydrogenases. Gene 2007, 396, 108–115. [Google Scholar] [CrossRef]
- Thompson, C.E.; Fernandes, C.L.; de Souza, O.N.; de Freitas, L.B.; Salzano, F.M. Evaluation of the impact of functional diversification on Poaceae, Brassicaceae, Fabaceae, and Pinaceae alcohol dehydrogenase enzymes. J. Mol. Model. 2010, 16, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.E.; Freitas, L.B.; Salzano, F.M. Molecular evolution and functional divergence of alcohol dehydrogenases in animals, fungi and plants. Genet. Mol. Biol. 2018, 41, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Wollenberg Valero, K.C. Aligning functional network constraint to evolutionary outcomes. BMC Evol. Biol. 2020, 20, 58. [Google Scholar] [CrossRef] [PubMed]
- Raj, S.B.; Ramaswamy, S.; Plapp, B.V. Yeast alcohol dehydrogenase structure and catalysis. Biochemistry 2014, 53, 5791–5803. [Google Scholar] [CrossRef]
- Ramaswamy, S.; Ahmad, M.E.; Danielsson, O.; Jörnvall, H.; Eklund, H. Crystal structure of cod liver class I alcohol dehydrogenase: Substrate pocket and structurally variable segments. Protein Sci. 1996, 5, 663–671. [Google Scholar] [CrossRef]
- Hurley, T.D.; Bosron, W.F.; Stone, C.L.; Amzel, L.M. Structures of three human β alcohol dehydrogenase variants: Correlations with their functional differences. J. Mol. Biol. 1994, 239, 415–429. [Google Scholar] [CrossRef]
- Israr, J.; Alam, S.; Siddiqui, S.; Misra, S.; Singh, I.; Kumar, A. Advances in Structural Bioinformatics. In Advances in Bioinformatics; Singh, V., Kumar, A., Eds.; Springer: Singapore, 2024; pp. 35–70. [Google Scholar]
- Gu, X. Functional divergence in protein (family) sequence evolution. Genetica 2003, 118, 133–141. [Google Scholar] [CrossRef]
- Lei, T.; Liao, X.; Liang, X.; Sun, L.; Xia, M.; Xia, Y.; Zhao, T.; Chen, X.; Men, W.; Wang, Y.; et al. Functional network modules overlap and are linked to interindividual connectome differences during human brain development. PLoS Biol. 2024, 22, e3002653. [Google Scholar] [CrossRef]
- Chiem, B.; Crevecoeur, F.; Delvenne, J.C. Structure-informed functional connectivity driven by identifiable and state-specific control regions. Netw. Neurosci. 2021, 5, 591–613. [Google Scholar] [CrossRef]
- Zheng, X.; Hu, C.; Spooner, D.; Liu, J.; Cao, J.; Teng, Y. Molecular evolution of Adh and LEAFY and the phylogenetic utility of their introns in Pyrus (Rosaceae). BMC Evol. Biol. 2011, 11, 255. [Google Scholar] [CrossRef]
- Fukuda, T.; Yokoyama, J.; Nakamura, T.; Song, I.J.; Ito, T.; Ochiai, T.; Kanno, A.; Kameya, T.; Maki, M. Molecular phylogeny and evolution of alcohol dehydrogenase (Adh) genes in legumes. BMC Plant Biol. 2005, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Vander Velden, K. Diverge: Phylogeny-based analysis for functional–structural divergence of a protein family. Bioinformatics 2002, 18, 500–501. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zou, Y.; Su, Z.; Huang, W.; Zhou, Z.; Arendsee, Z.; Zeng, Y. An update of DIVERGE software for functional divergence analysis of protein family. Mol. Biol. Evol. 2013, 30, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- MacKerell Jr, A.D.; Banavali, N.; Foloppe, N. Development and current status of the CHARMM force field for nucleic acids. Biopolym. Orig. Res. Biomol. 2000, 56, 257–265. [Google Scholar] [CrossRef]
- Buck, M.; Bouguet-Bonnet, S.; Pastor, R.W.; MacKerell, A.D. Importance of the CMAP correction to the CHARMM22 protein force field: Dynamics of hen lysozyme. Biophys. J. 2006, 90, L36–L38. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N· log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Hünenberger, P.; Mark, A.; Van Gunsteren, W. Fluctuation and cross-correlation analysis of protein motions observed in nanosecond molecular dynamics simulations. J. Mol. Biol. 1995, 252, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Ichiye, T.; Karplus, M. Collective motions in proteins: A covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins Struct. Funct. Bioinform. 1991, 11, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, K.; Fukuda, I.; Nakamura, H. A novel approach of dynamic cross correlation analysis on molecular dynamics simulations and its application to Ets1 dimer–DNA complex. PLoS ONE 2014, 9, e112419. [Google Scholar] [CrossRef] [PubMed]
- Sethi, A.; Eargle, J.; Black, A.A.; Luthey-Schulten, Z. Dynamical networks in tRNA: Protein complexes. Proc. Natl. Acad. Sci. USA 2009, 106, 6620–6625. [Google Scholar] [CrossRef]
- VanWart, A.T.; Eargle, J.; Luthey-Schulten, Z.; Amaro, R.E. Exploring residue component contributions to dynamical network models of allostery. J. Chem. Theory Comput. 2012, 8, 2949–2961. [Google Scholar] [CrossRef]
- Newman, M.E. A measure of betweenness centrality based on random walks. Soc. Netw. 2005, 27, 39–54. [Google Scholar] [CrossRef]
- Freeman, L.C. Centrality in social networks: Conceptual clarification. In Social Network: Critical Concepts in Sociology; Routledge: London, UK, 2002; Volume 1, pp. 238–263. [Google Scholar]
- Mason, O.; Verwoerd, M. Graph theory and networks in biology. IET Syst. Biol. 2007, 1, 89–119. [Google Scholar] [CrossRef]
- Gu, X. Maximum-likelihood approach for gene family evolution under functional divergence. Mol. Biol. Evol. 2001, 18, 453–464. [Google Scholar] [CrossRef]
- Gu, X. Statistical methods for testing functional divergence after gene duplication. Mol. Biol. Evol. 1999, 16, 1664–1674. [Google Scholar] [CrossRef]
- Kitao, A. Principal component analysis and related methods for investigating the dynamics of biological macromolecules. J 2022, 5, 298–317. [Google Scholar] [CrossRef]
- Hess, B. Similarities between principal components of protein dynamics and random diffusion. Phys. Rev. E 2000, 62, 8438. [Google Scholar] [CrossRef] [PubMed]
- Hess, B. Convergence of sampling in protein simulations. Phys. Rev. E 2002, 65, 031910. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.; Kirschner, K.N. Principal component and clustering analysis on molecular dynamics data of the ribosomal L11· 23S subdomain. J. Mol. Model. 2013, 19, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, D.; Power, A.; Chapman, J. Interpreting and reporting principal component analysis in food science analysis and beyond. Food Anal. Methods 2019, 12, 2469–2473. [Google Scholar] [CrossRef]
- Saxena, A.; Iyengar, S. Centrality measures in complex networks: A survey. arXiv 2020, arXiv:2011.07190. [Google Scholar]
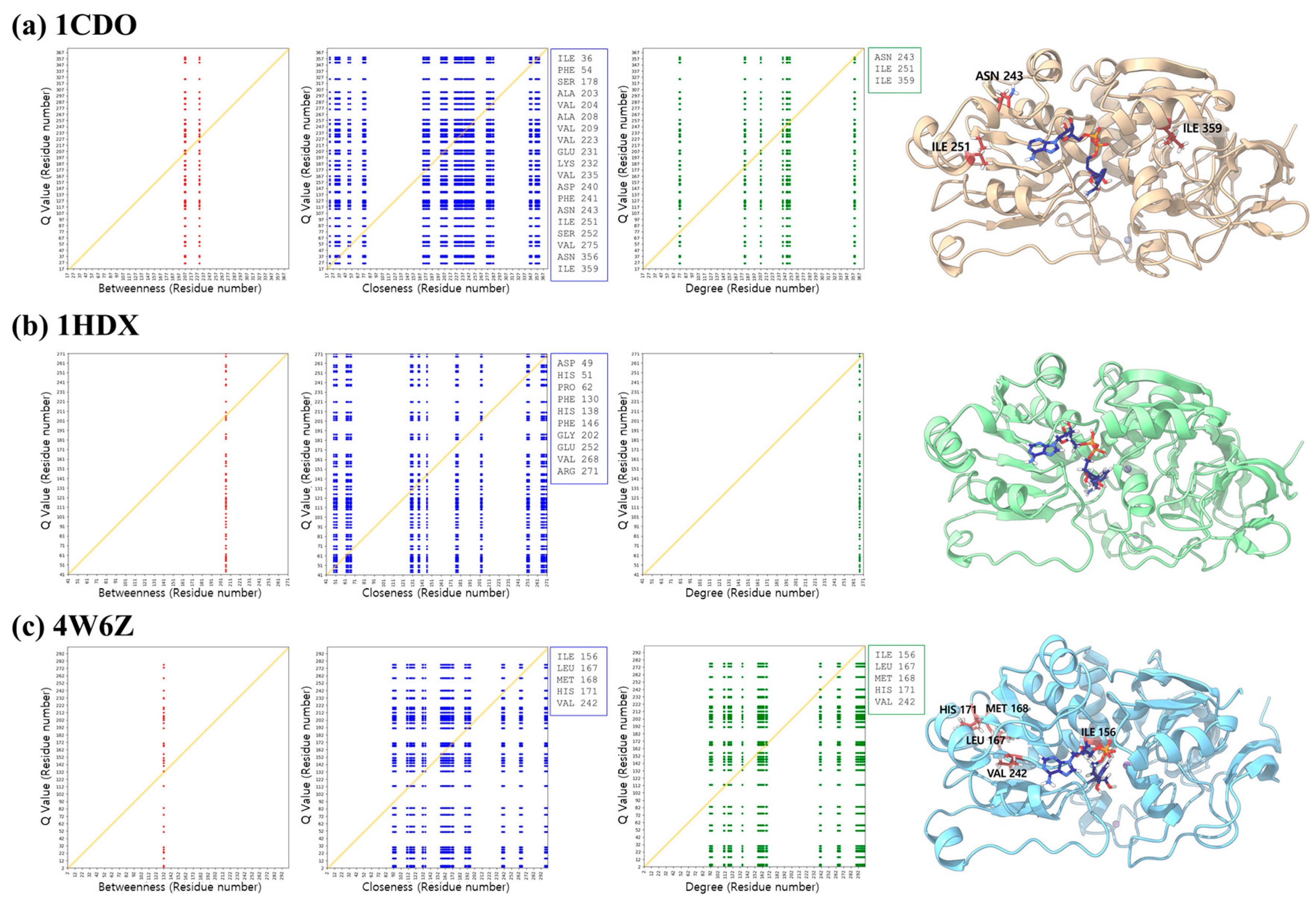
| Betweenness | Closeness | Degree | |
|---|---|---|---|
| 1 | ALA 207 | ALA 207 | SER 76 |
| 2 | PHE 230 | PHE 181 | PRO 250 |
| 3 | LEU 206 | LYS 366 | LEU 361 |
| 4 | CYS 104 | LEU 361 | ILE 251 |
| 5 | ASP 337 | LEU 206 | VAL 77 |
| 6 | LYS 285 | PHE 230 | PHE 181 |
| 7 | THR 347 | MET 362 | ASN 243 |
| 8 | GLU 27 | MET 231 | GLY 182 |
| 9 | ASN 278 | ASN 243 | GLU 249 |
| 10 | GLU 35 | ASN 278 | ALA 207 |
| 11 | LEU 172 | GLY 182 | ILE 359 |
| 12 | ARG 130 | ALA 358 | ALA 358 |
| 13 | LEU 298 | LYS 232 | VAL 254 |
| 14 | ILE 156 | GLY 365 | LYS 366 |
| 15 | GLU 281 | ASP 357 | VAL 30 |
| Betweenness | Closeness | Degree | |
|---|---|---|---|
| 1 | LEU 279 | MET 275 | GLU 267 |
| 2 | SER 206 | MET 276 | THR 274 |
| 3 | PHE 335 | THR 274 | MET 276 |
| 4 | SER 289 | LEU 279 | ARG 129 |
| 5 | ARG 37 | SER 278 | MET 275 |
| 6 | ASN 118 | ALA 277 | ALA 12 |
| 7 | GLU 267 | GLU 267 | GLY 66 |
| 8 | GLY 66 | LEU 280 | GLY 270 |
| 9 | THR 131 | ASP 273 | SER 278 |
| 10 | ARG 129 | LEU 272 | LEU 279 |
| 11 | HIS 51 | ARG 271 | ASP 49 |
| 12 | GLY 117 | PHE 266 | PHE 130 |
| 13 | ILE 137 | SER 289 | PHE 146 |
| 14 | ARG 47 | VAL 13 | PHE 266 |
| 15 | MET 276 | CYS 282 | THR 131 |
| Betweenness | Closeness | Degree | |
|---|---|---|---|
| 1 | ASP 132 | LEU 93 | ASN 297 |
| 2 | LEU 116 | LEU 116 | THR 301 |
| 3 | ASN 94 | TRP 92 | LYS 160 |
| 4 | HIS 240 | ASN 94 | VAL 266 |
| 5 | LEU 93 | LYS 163 | GLY 296 |
| 6 | ARG 196 | LEU 162 | ASP 300 |
| 7 | ILE 288 | HIS 240 | ARG 302 |
| 8 | CYS 111 | LYS 160 | CYS 111 |
| 9 | TYR 189 | SER 117 | ILE 156 |
| 10 | LYS 160 | LEU 167 | TYR 294 |
| 11 | VAL 266 | TYR 159 | ARG 298 |
| 12 | GLU 67 | ASP 115 | ALA 161 |
| 13 | ASN 110 | ALA 161 | ALA 299 |
| 14 | TYR 195 | HIS 171 | TYR 159 |
| 15 | TRP 92 | VAL 173 | LEU 116 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.; Jebamani, P.; Seo, Y.G.; Wu, S. Computational Study of Network and Type-I Functional Divergence in Alcohol Dehydrogenase Enzymes Across Species Using Molecular Dynamics Simulation. Biomolecules 2024, 14, 1473. https://doi.org/10.3390/biom14111473
Park S, Jebamani P, Seo YG, Wu S. Computational Study of Network and Type-I Functional Divergence in Alcohol Dehydrogenase Enzymes Across Species Using Molecular Dynamics Simulation. Biomolecules. 2024; 14(11):1473. https://doi.org/10.3390/biom14111473
Chicago/Turabian StylePark, Suhyun, Petrina Jebamani, Yeon Gyo Seo, and Sangwook Wu. 2024. "Computational Study of Network and Type-I Functional Divergence in Alcohol Dehydrogenase Enzymes Across Species Using Molecular Dynamics Simulation" Biomolecules 14, no. 11: 1473. https://doi.org/10.3390/biom14111473
APA StylePark, S., Jebamani, P., Seo, Y. G., & Wu, S. (2024). Computational Study of Network and Type-I Functional Divergence in Alcohol Dehydrogenase Enzymes Across Species Using Molecular Dynamics Simulation. Biomolecules, 14(11), 1473. https://doi.org/10.3390/biom14111473






