deepBBQ: A Deep Learning Approach to the Protein Backbone Reconstruction
Abstract
1. Introduction
2. Methods
2.1. Protein Backbone Reconstruction
2.2. deepBBQ Neural Network
- (a)
- 21 binary values for one-hot-encoded residue type representation, corresponding to 20 canonical amino acids and the ’X’ symbol representing unknown residue types
- (b)
- 6 floating point values corresponding to distances between C and C for
- (c)
- 4 integer values for the number of C atoms present within 4, 4.5, 5 and 6 Å from C
- (d)
- 3 binary values for one-hot-encoded secondary structure information (either helix, sheet or loop)
- (e)
- One binary value as a flag for the cis/trans classification of the peptide bond between amino acids i and
- (f)
- 2 integer values corresponding to the number of hydrogen bonds involved in helices and strands
2.3. Training Data and Tools
2.4. Testing Set
2.5. De Novo Testing Set
3. Results
3.1. Comparison with Other Methods
3.2. Reconstruction of De Novo Proteins
3.3. Accuracy of the Two Steps of the Reconstruction Process
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Levitt, M.; Warshel, A. Computer Simulation of Protein Folding. Nature 1975, 253, 694–698. [Google Scholar] [CrossRef] [PubMed]
- Purisima, E.O.; Scheraga, H.A. Conversion from a Virtual-bond Chain to a Complete Polypeptide Backbone Chain. Biopolymers 1984, 23, 1207–1224. [Google Scholar] [CrossRef] [PubMed]
- Lubecka, E.A.; Liwo, A. ESCASA: Analytical estimation of atomic coordinates from coarse-grained geometry for nuclear-magnetic-resonance-assisted protein structure modeling. I. Backbone and and Hβ Protons. J. Comput. Chem. 2021, 42, 1579–1589. [Google Scholar] [CrossRef] [PubMed]
- Adcock, S.A. Peptide Backbone Reconstruction Using Dead-End Elimination and a Knowledge-Based Forcefield. J. Comput. Chem. 2004, 25, 16–27. [Google Scholar] [CrossRef]
- Holm, L.; Sander, C. Database Algorithm for Generating Protein Backbone and Side-Chain Co-Ordinates from a CCLTrace. J. Mol. Biol. 1991, 218, 183–194. [Google Scholar] [CrossRef]
- Heo, L.; Feig, M. One Bead per Residue Can Describe All-Atom Protein Structures. Structure 2024, 32, 97–111.e6. [Google Scholar] [CrossRef]
- Payne, P.W. Reconstruction of Protein Conformations from Estimated Positions of the Cα Coordinates. Protein Sci. 1993, 2, 315–324. [Google Scholar] [CrossRef]
- Kaźmierkiewicz, R.; Liwo, A.; Scheraga, H.A. Energy-based Reconstruction of a Protein Backbone from Its A-carbon Trace by a Monte-Carlo Method. J. Comput. Chem. 2002, 23, 715–723. [Google Scholar] [CrossRef]
- Moore, B.L.; Kelley, L.A.; Barber, J.; Murray, J.W.; MacDonald, J.T. High–Quality Protein Backbone Reconstruction from Alpha Carbons Using Gaussian Mixture Models. J. Comput. Chem. 2013, 34, 1881–1889. [Google Scholar] [CrossRef]
- Iwata, Y.; Kasuya, A.; Miyamoto, S. An Efficient Method for Reconstructing Protein Backbones from α-Carbon Coordinates. J. Mol. Graph. Model. 2002, 21, 119–128. [Google Scholar] [CrossRef]
- Etchebest, C.; Benros, C.; Hazout, S.; De Brevern, A.G. A Structural Alphabet for Local Protein Structures: Improved Prediction Methods. Proteins Struct. Funct. Bioinform. 2005, 59, 810–827. [Google Scholar] [CrossRef] [PubMed]
- Rooman, M.J.; Rodriguez, J.; Wodak, S.J. Automatic Definition of Recurrent Local Structure Motifs in Proteins. J. Mol. Biol. 1990, 213, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Pandini, A.; Fornili, A.; Kleinjung, J. Structural Alphabets Derived from Attractors in Conformational Space. BMC Bioinform. 2010, 11, 97. [Google Scholar] [CrossRef]
- Park, B.H.; Levitt, M. The Complexity and Accuracy of Discrete State Models of Protein Structure. J. Mol. Biol. 1995, 249, 493–507. [Google Scholar] [CrossRef]
- Maupetit, J.; Gautier, R.; Tuffery, P. SABBAC: Online Structural Alphabet-based Protein BackBone Reconstruction from Alpha-Carbon Trace. Nucleic Acids Res. 2006, 34, W147–W151. [Google Scholar] [CrossRef][Green Version]
- Jones, T.; Thirup, S. Using Known Substructures in Protein Model Building and Crystallography. Embo J. 1986, 5, 819–822. [Google Scholar] [CrossRef]
- Claessens, M.; Van Cutsem, E.; Lasters, I.; Wodak, S. Modelling the Polypeptide Backbone with `Spare Parts’ from Known Protein Structures. Protein Eng. Des. Sel. 1989, 2, 335–345. [Google Scholar] [CrossRef]
- Reid, L.S.; Thornton, J.M. Rebuilding Flavodoxin from Cα Coordinates: A Test Study. Proteins Struct. Funct. Bioinform. 1989, 5, 170–182. [Google Scholar] [CrossRef]
- Levitt, M. Accurate Modeling of Protein Conformation by Automatic Segment Matching. J. Mol. Biol. 1992, 226, 507–533. [Google Scholar] [CrossRef]
- Milik, M.; Kolinski, A.; Skolnick, J. Algorithm for Rapid Reconstruction of Protein Backbone from Alpha Carbon Coordinates. J. Comput. Chem. 1997, 18, 80–85. [Google Scholar] [CrossRef]
- Gront, D.; Kmiecik, S.; Kolinski, A. Backbone Building from Quadrilaterals: A Fast and Accurate Algorithm for Protein Backbone Reconstruction from Alpha Carbon Coordinates. J. Comput. Chem. 2007, 28, 1593–1597. [Google Scholar] [CrossRef] [PubMed]
- Rotkiewicz, P.; Skolnick, J. Fast Procedure for Reconstruction of Full-Atom Protein Models from Reduced Representations. J. Comput. Chem. 2008, 29, 1460–1465. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y. REMO: A New Protocol to Refine Full Atomic Protein Models from C-Alpha Traces by Optimizing Hydrogen-Bonding Networks. Proteins Struct. Funct. Bioinform. 2009, 76, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Kmiecik, S.; Gront, D.; Kolinski, M.; Wieteska, L.; Dawid, A.E.; Kolinski, A. Coarse-Grained Protein Models and Their Applications. Chem. Rev. 2016, 116, 7898–7936. [Google Scholar] [CrossRef] [PubMed]
- Saqib, M.N.; Kryś, J.D.; Gront, D. Automated Protein Secondary Structure Assignment from Cα Positions Using Neural Networks. Biomolecules 2022, 12, 841. [Google Scholar] [CrossRef]
- Kryś, J.D.; Gront, D. Coarse-Grained Potential for Hydrogen Bond Interactions. J. Mol. Graph. Model. 2023, 124, 108507. [Google Scholar] [CrossRef]
- Liljas, A.; Liljas, L.; Piskur, J.; Lindblom, G.; Nissen, P.; Kjeldgaard, M. Textbook of Structural Biology; World Scientific: Singapore, 2009. [Google Scholar] [CrossRef]
- Godzik, A.; Kolinski, A.; Skolnick, J. Lattice Representations of Globular Proteins: How Good Are They? J. Comput. Chem. 1993, 14, 1194–1202. [Google Scholar] [CrossRef]
- Wang, G.; Dunbrack, R.L. PISCES: A Protein Sequence Culling Server. Bioinformatics 2003, 19, 1589–1591. [Google Scholar] [CrossRef]
- Wang, G.; Dunbrack, R.L. PISCES: Recent Improvements to a PDB Sequence Culling Server. Nucleic Acids Res. 2005, 33, W94–W98. [Google Scholar] [CrossRef]
- Macnar, J.M.; Szulc, N.A.; Kryś, J.D.; Badaczewska-Dawid, A.E.; Gront, D. BioShell 3.0: Library for Processing Structural Biology Data. Biomolecules 2020, 10, 461. [Google Scholar] [CrossRef]
- Developers, T. TensorFlow. Zenodo. 2024. [Google Scholar] [CrossRef]
- Hermann, T. Frugally-Deep. Available online: https://github.com/Dobiasd/frugally-deep (accessed on 10 October 2024).
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov Model Speed Heuristic and Iterative HMM Search Procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- wwPDB Consortium. Protein Data Bank: The Single Global Archive for 3D Macromolecular Structure Data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef] [PubMed]
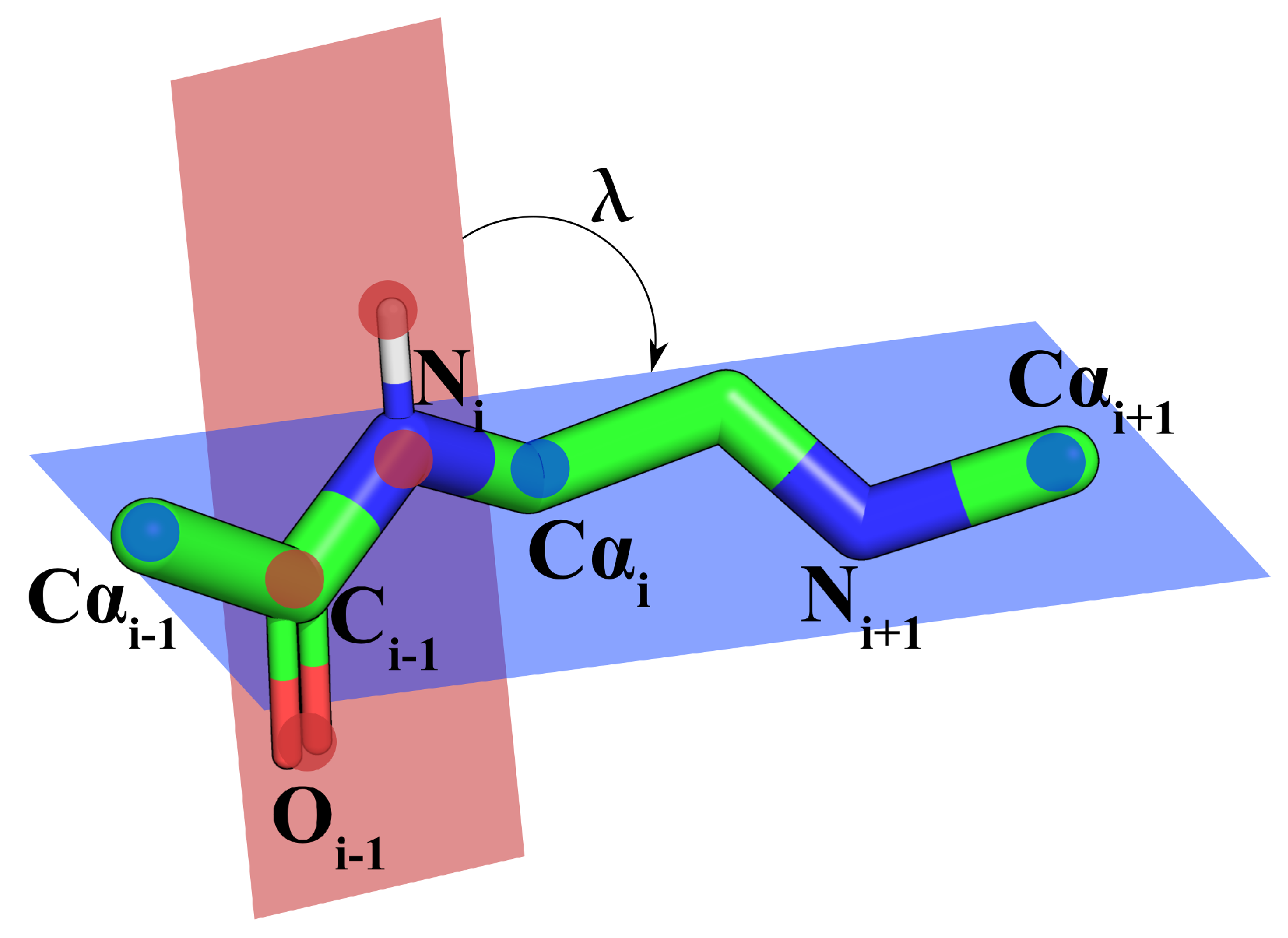


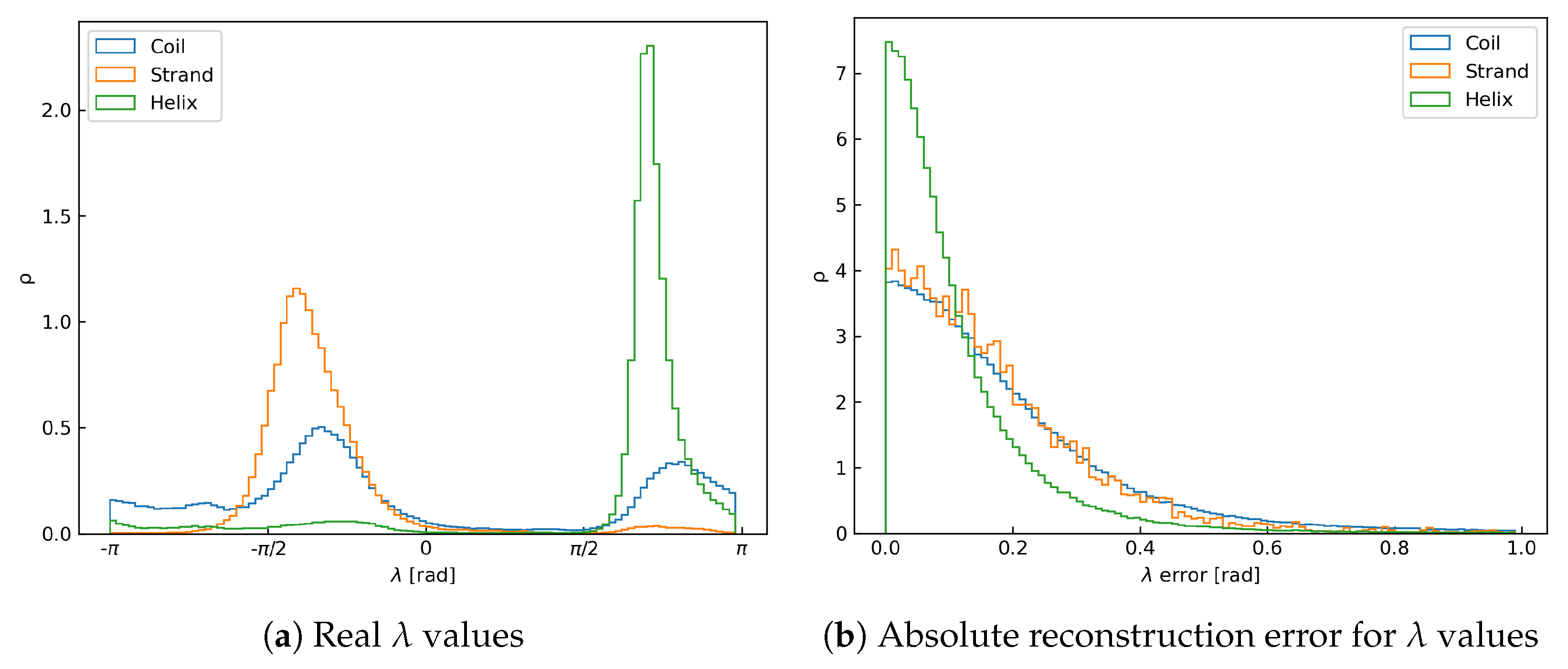
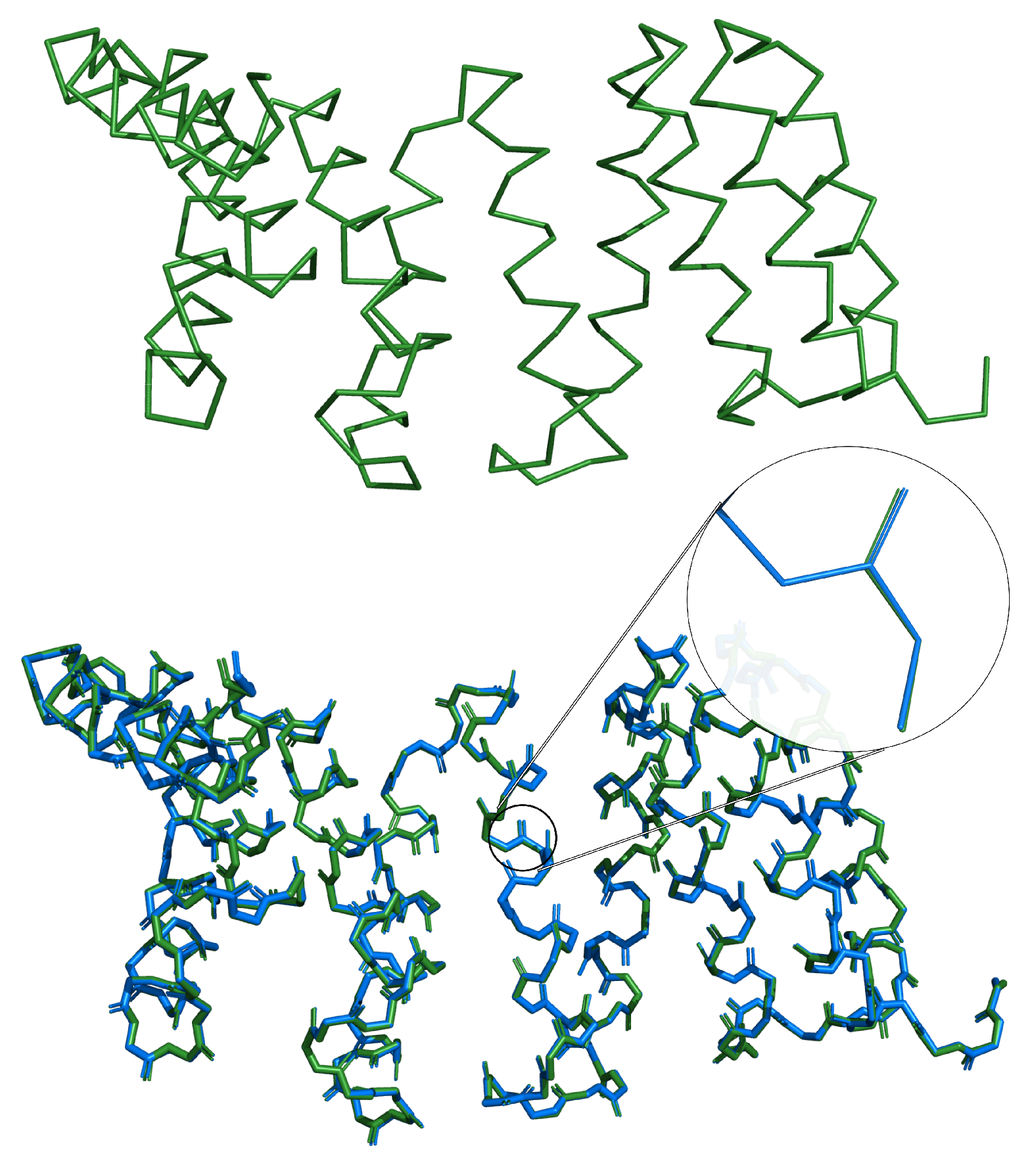

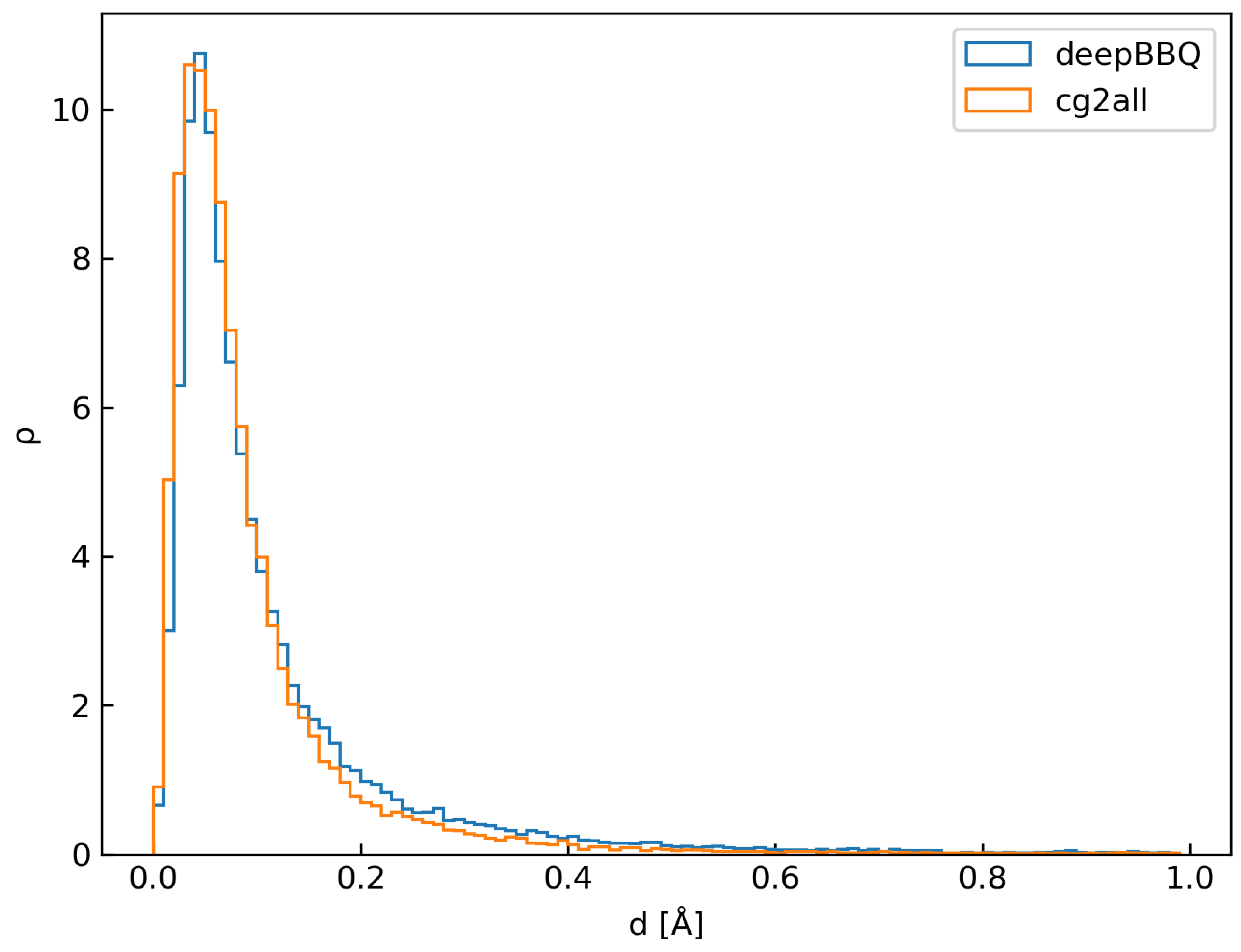
| Atom | Mean [Å] | Mode [Å] | SSE | Mean [Å] | Mode [Å] | |
|---|---|---|---|---|---|---|
| C | 0.12 ± 0.18 | 0.045 | Helix | 0.12 ± 0.20 | 0.042 | |
| N | 0.11 ± 0.15 | 0.039 | Strand | 0.18 ± 0.23 | 0.057 | |
| O | 0.33 ± 0.46 | 0.069 | Coil | 0.23 ± 0.38 | 0.056 |
| PDB Code | PD2 + Min | BBQ | MaxSprout | PULCHRA | SABBAC | REMO | deepBBQ | cg2all |
|---|---|---|---|---|---|---|---|---|
| 4EO0 | 0.211 | 0.203 | 0.392 | 0.348 | 0.286 | 0.437 | 0.229 | 0.147 |
| 4F7H | 0.293 | 0.356 | 0.505 | 0.471 | 0.480 | 0.551 | 0.318 | 0.232 |
| 4EXO | 0.264 | 0.289 | 0.447 | 0.459 | 0.436 | 0.530 | 0.235 | 0.190 |
| 4F7V | 0.444 | 0.410 | 0.436 | 0.617 | 0.450 | 0.643 | 0.377 | 0.199 |
| 4FAK | 0.285 | 0.325 | 0.461 | 0.461 | 0.366 | 0.514 | 0.184 | 0.207 |
| 4ANN | 0.238 | 0.292 | 0.405 | 0.452 | 0.445 | 0.514 | 0.320 | 0.221 |
| 4FFK | 0.355 | 0.447 | 0.390 | 0.604 | 0.441 | 0.607 | 0.361 | 0.264 |
| 4FD5 | 0.318 | 0.319 | 0.426 | 0.421 | 0.406 | 0.515 | 0.253 | 0.131 |
| 4AVX | 0.227 | 0.314 | 0.360 | 0.402 | 0.318 | 0.474 | 0.228 | 0.169 |
| 4EV1 | 0.239 | 0.304 | 0.396 | 0.411 | 0.385 | 0.504 | 0.219 | 0.159 |
| 4EG9 | 0.317 | 0.427 | 0.478 | 0.504 | 0.350 | 0.531 | 0.318 | 0.209 |
| 4EIU | 0.334 | 0.339 | 0.468 | 0.587 | 0.555 | 0.556 | 0.282 | 0.205 |
| 4F78 | 0.364 | 0.424 | 0.552 | 0.558 | 0.477 | 0.616 | 0.317 | 0.253 |
| 4FCU | 0.298 | 0.342 | 0.455 | 0.458 | 0.501 | 0.502 | 0.266 | 0.144 |
| 4FBR | 0.404 | 0.399 | 0.528 | 0.586 | 0.552 | 0.622 | 0.256 | 0.149 |
| 4FB7 | 0.248 | 0.335 | 0.344 | 0.461 | 0.359 | 0.506 | 0.183 | 0.193 |
| 4FIK | 0.374 | 0.356 | 0.288 | 0.568 | 0.575 | 0.563 | 0.326 | 0.134 |
| 4FAT | 0.227 | 0.413 | 0.359 | 0.540 | 0.431 | 0.632 | 0.337 | 0.124 |
| 4FE3 | 0.275 | 0.318 | 0.489 | 0.449 | 0.450 | 0.507 | 0.207 | 0.119 |
| 4FCS | 0.329 | 0.326 | 0.458 | 0.504 | 0.452 | 0.533 | 0.198 | 0.128 |
| 4E9L | 0.267 | 0.315 | 0.403 | 0.546 | 0.544 | 0.540 | 0.229 | 0.118 |
| 4F8X | 0.330 | 0.380 | 0.538 | 0.562 | 0.488 | 0.542 | 0.301 | 0.117 |
| 4FHG | 0.318 | 0.350 | 0.397 | 0.549 | 0.425 | 0.506 | 0.298 | 0.126 |
| 4EYO | 0.271 | 0.368 | 0.384 | 0.569 | 0.344 | 0.558 | 0.233 | 0.201 |
| 4F8J | 0.279 | 0.354 | 0.388 | 0.510 | 0.370 | 0.458 | 0.235 | 0.214 |
| 3VTF | 0.310 | 0.342 | 0.468 | 0.551 | 0.401 | 0.538 | 0.183 | 0.127 |
| 4FE9 | 0.392 | 0.406 | 0.433 | 0.573 | 0.534 | 0.568 | 0.254 | 0.152 |
| 4AVZ | 0.352 | 0.369 | 0.480 | 0.568 | 0.489 | 0.602 | 0.244 | 0.145 |
| mean | 0.306 | 0.351 | 0.433 | 0.510 | 0.440 | 0.542 | 0.264 | 0.171 |
| 0.058 | 0.052 | 0.062 | 0.070 | 0.076 | 0.052 | 0.055 | 0.044 |
| PDB Code | Nres | True [Å] | Predicted [Å] |
|---|---|---|---|
| 1CRN | 46 | 0.051 | 0.615 |
| 6PTI | 58 | 0.057 | 0.234 |
| 1CTF | 68 | 0.165 | 0.344 |
| 1UBQ | 76 | 0.055 | 0.432 |
| 2OZ9 | 104 | 0.043 | 0.143 |
| 4EO0 | 115 | 0.049 | 0.205 |
| 2MHR | 118 | 0.165 | 0.422 |
| 4F7H | 135 | 0.160 | 0.352 |
| 2FOX | 138 | 0.032 | 0.355 |
| 5NLL | 138 | 0.112 | 0.351 |
| 4EXO | 144 | 0.127 | 0.300 |
| 4F7V | 161 | 0.157 | 0.393 |
| 4FAK | 163 | 0.115 | 0.205 |
| 2ALP | 198 | 0.103 | 0.297 |
| 4ANN | 210 | 0.149 | 0.317 |
| 4FFK | 214 | 0.113 | 0.387 |
| 4FD5 | 216 | 0.110 | 0.252 |
| 4AVX | 223 | 0.109 | 0.250 |
| 4EV1 | 229 | 0.062 | 0.260 |
| 4EG9 | 232 | 0.141 | 0.308 |
| 4EIU | 241 | 0.134 | 0.299 |
| 4F78 | 254 | 0.127 | 0.323 |
| 4FCU | 262 | 0.120 | 0.315 |
| 4FBR | 273 | 0.061 | 0.291 |
| 4FB7 | 274 | 0.055 | 0.150 |
| 4FIK | 278 | 0.125 | 0.339 |
| 2PRK | 279 | 0.094 | 0.258 |
| 4FAT | 280 | 0.146 | 0.339 |
| 4FE3 | 295 | 0.060 | 0.235 |
| 5CPA | 307 | 0.172 | 0.391 |
| 4FCS | 315 | 0.079 | 0.195 |
| 4E9L | 318 | 0.085 | 0.234 |
| 3APP | 323 | 0.113 | 0.301 |
| 4F8X | 335 | 0.132 | 0.309 |
| 9WGA | 340 | 0.316 | 0.367 |
| 4FHG | 342 | 0.138 | 0.306 |
| 4EYO | 358 | 0.088 | 0.225 |
| 4F8J | 365 | 0.088 | 0.249 |
| 3VTF | 432 | 0.068 | 0.205 |
| 2CTS | 437 | 0.096 | 0.308 |
| 4FE9 | 450 | 0.097 | 0.250 |
| 1TIM | 494 | 0.412 | 0.465 |
| 4AVZ | 608 | 0.080 | 0.246 |
| Avarage | 0.115 | 0.303 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kryś, J.D.; Głowacki , M.; Śmieja , P.; Gront, D. deepBBQ: A Deep Learning Approach to the Protein Backbone Reconstruction. Biomolecules 2024, 14, 1448. https://doi.org/10.3390/biom14111448
Kryś JD, Głowacki M, Śmieja P, Gront D. deepBBQ: A Deep Learning Approach to the Protein Backbone Reconstruction. Biomolecules. 2024; 14(11):1448. https://doi.org/10.3390/biom14111448
Chicago/Turabian StyleKryś, Justyna D., Maksymilian Głowacki , Piotr Śmieja , and Dominik Gront. 2024. "deepBBQ: A Deep Learning Approach to the Protein Backbone Reconstruction" Biomolecules 14, no. 11: 1448. https://doi.org/10.3390/biom14111448
APA StyleKryś, J. D., Głowacki , M., Śmieja , P., & Gront, D. (2024). deepBBQ: A Deep Learning Approach to the Protein Backbone Reconstruction. Biomolecules, 14(11), 1448. https://doi.org/10.3390/biom14111448








