Biomarkers for Serious Bacterial Infections in Febrile Children
Abstract
1. Introduction
2. Hematological Biomarkers
2.1. White Blood Cell Count (WBC) and Absolute Neutrophil Count (ANC)
2.2. Platelet Indices
3. Inflammatory Biomarkers
3.1. C-Reactive Protein (CRP)
3.2. Procalcitonin
3.3. Cytokines and Chemokines
3.3.1. Interleukines (IL)
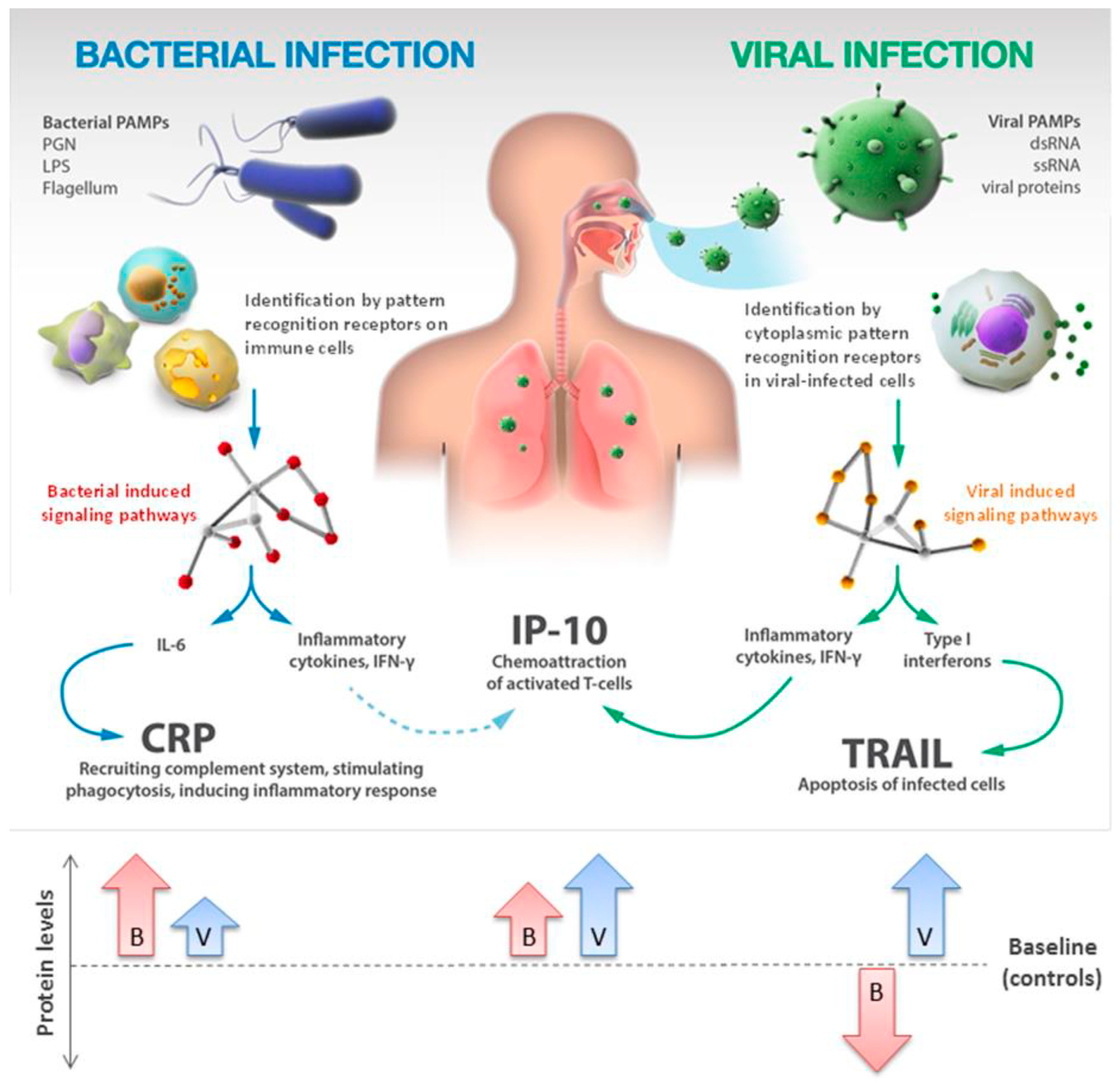
3.3.2. TRAIL and IP-10
4. Cell Adhesion Molecules
4.1. Presepsin
4.2. STREM-2
5. Future Perspective
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Armon, K.; Stephenson, T.; Gabriel, V.; MacFaul, R.; Eccleston, P.; Werneke, U.; Smith, S. Determining the common medical presenting problems to an accident and emergency department. Arch. Dis. Child. 2001, 84, 390. [Google Scholar] [CrossRef] [PubMed]
- Sands, R.; Shanmugavadivel, D.; Stephenson, T.; Wood, D. Medical problems presenting to paediatric emergency departments: 10 years on. Emerg. Med. J. 2012, 29, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Chiappini, E.; Venturini, E.; Remaschi, G.; Principi, N.; Longhi, R.; Tovo, P.A.; Becherucci, P.; Bonsignori, F.; Esposito, S.; Festini, F.; et al. 2016 Update of the Italian Pediatric Society Guidelines for Management of Fever in Children. J. Pediatr. 2017, 180, 177–183.e1. [Google Scholar] [CrossRef] [PubMed]
- National Institute for Health and Care Excellence (NICE). Fever in under 5s: Assessment and Initial Management; National Institute for Health and Care Excellence (NICE): London, UK, 2021. [Google Scholar]
- Pantell, R.H.; Roberts, K.B.; Adams, W.G.; Dreyer, B.P.; Kuppermann, N.; O’Leary, S.T.; Okechukwu, K.; Woods, C.R., Jr. Subcommittee on Febrile Infants—Evaluation and Management of Well-Appearing Febrile Infants 8 to 60 Days Old. Pediatrics 2021, 148, e2021052228. [Google Scholar] [CrossRef] [PubMed]
- Bosis, S.; Esposito, S.; Niesters, H.G.; Crovari, P.; Osterhaus, A.D.; Principi, N. Impact of human metapneumovirus in childhood: Comparison with respiratory syncytial virus and influenza viruses. J. Med. Virol. 2005, 75, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Daleno, C.; Prunotto, G.; Scala, A.; Tagliabue, C.; Borzani, I.; Fossali, E.; Pelucchi, C.; Principi, N. Impact of viral infections in children with community-acquired pneumonia: Results of a study of 17 respiratory viruses. Influenza Other Respir. Viruses 2013, 7, 18–26. [Google Scholar] [CrossRef]
- Principi, N.; Daleno, C.; Esposito, S. Human rhinoviruses and severe respiratory infections: Is it possible to identify at-risk patients early? Expert Rev. Anti Infect. Ther. 2014, 12, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Stol, K.; Nijman, R.G.; van Herk, W.; van Rossum, A.M.C. Biomarkers for Infection in Children: Current Clinical Practice and Future Perspectives. Pediatr. Infect. Dis. J. 2019, 38 (Suppl. S1), S7–S13. [Google Scholar] [CrossRef]
- Boscarino, G.; Migliorino, R.; Carbone, G.; Davino, G.; Dell’Orto, V.G.; Perrone, S.; Principi, N.; Esposito, S. Biomarkers of Neonatal Sepsis: Where We Are and Where We Are Going. Antibiotics 2023, 26, 1233. [Google Scholar]
- Nauseef, W.M. How human neutrophils kill and degrade microbes: An integrated view. Immunol. Rev. 2007, 219, 88–102. [Google Scholar] [CrossRef]
- Yo, C.H.; Hsieh, P.S.; Lee, S.H.; Wu, J.Y.; Chang, S.S.; Tasi, K.C.; Lee, C.C. Comparison of the test characteristics of procalcitonin to C-reactive protein and leukocytosis for the detection of serious bacterial infections in children presenting with fever without source: A systematic review and meta-analysis. Ann. Emerg. Med. 2012, 60, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Pratt, A.; Attia, M.W. Duration of fever and markers of serious bacterial infection in young febrile children. Pediatr. Int. 2007, 49, 31–35. [Google Scholar] [CrossRef]
- Van den Bruel, A.; Thompson, M.J.; Haj-Hassan, T.; Stevens, R.; Moll, H.; Lakhanpaul, M.; Mant, D. Diagnostic value of laboratory tests in identifying serious infections in febrile children: Systematic review. BMJ 2011, 342, d3082. [Google Scholar] [CrossRef] [PubMed]
- Semple, J.W.; Italiano, J.E., Jr.; Freedman, J. Platelets and the immune continuum. Nat. Rev. Immunol. 2011, 11, 264–274. [Google Scholar] [CrossRef]
- Vassiliou, A.G.; Mastora, Z.; Orfanos, S.E.; Jahaj, E.; Maniatis, N.A.; Koutsoukou, A.; Armaganidis, A.; Kotanidou, A. Elevated biomarkers of endothelial dysfunction/activation at ICU admission are associated with sepsis development. Cytokine 2014, 69, 240–247. [Google Scholar] [CrossRef]
- Zonneveld, R.; Martinelli, R.; Shapiro, N.I.; Kuijpers, T.W.; Plötz, F.B.; Carman, C.V. Soluble adhesion molecules as markers for sepsis and the potential pathophysiological discrepancy in neonates, children and adults. Crit. Care 2014, 18, 204. [Google Scholar] [CrossRef] [PubMed]
- Feketea, G.; Vlacha, V.; Pop, R.M.; Bocsan, I.C.; Stanciu, L.A.; Buzoianu, A.D.; Zdrenghea, M. Relationship Between Vitamin D Level and Platelet Parameters in Children with Viral Respiratory Infections. Front. Pediatr. 2022, 10, 824959. [Google Scholar] [CrossRef]
- Heijnen, H.; van der Sluijs, P. Platelet secretory behaviour: As diverse as the granules… or not? J. Thromb. Haemost. 2015, 13, 2141–2151. [Google Scholar] [CrossRef]
- Pociute, A.; Kottilingal Farook, M.F.; Dagys, A.; Kevalas, R.; Laucaityte, G.; Jankauskaite, L. Platelet-Derived Biomarkers: Potential Role in Early Pediatric Serious Bacterial Infection and Sepsis Diagnostics. J. Clin. Med. 2022, 11, 6475. [Google Scholar] [CrossRef]
- Esposito, S.; Tremolati, E.; Begliatti, E.; Bosis, S.; Gualtieri, L.; Principi, N. Evaluation of a rapid bedside test for the quantitative determination of C-reactive protein. Clin. Chem. Lab. Med. 2005, 43, 438–440. [Google Scholar] [CrossRef]
- Jaye, D.L.; Waites, K.B. Clinical applications of C-reactive protein in pediatrics. Pediatr. Infect. Dis. J. 1997, 16, 735–746. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, S.; Riordan, A. How to use: C-reactive protein. Arch. Dis. Child. Educ. Pract. Ed. 2010, 95, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Marnell, L.; Mold, C.; Du Clos, T.W. C-reactive protein: Ligands, receptors and role in inflammation. Clin. Immunol. 2005, 117, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Pepys, M.B.; Hirschfield, G.M. C-reactive protein: A critical update. J. Clin. Investig. 2003, 111, 1805–1812, Erratum in J. Clin. Investig. 2003, 112, 299. [Google Scholar] [CrossRef]
- Dyer, E.M.; Waterfield, T.; Baynes, H. How to use C-reactive protein. Arch. Dis. Child. Educ. Pract. Ed. 2019, 104, 150–153. [Google Scholar] [CrossRef] [PubMed]
- Verbakel, J.Y.; Lemiengre, M.B.; De Burghgraeve, T.; De Sutter, A.; Aertgeerts, B.; Bullens, D.M.A.; Shinkins, B.; Van den Bruel, A.; Buntinx, F. Point-of-care C reactive protein to identify serious infection in acutely ill children presenting to hospital: Prospective cohort study. Arch. Dis. Child. 2018, 103, 420–426. [Google Scholar] [CrossRef]
- Fernández Lopez, A.; Luaces Cubells, C.; García García, J.J.; Fernández Pou, J.; Spanish Society of Pediatric Emergencies. Procalcitonin in pediatric emergency departments for the early diagnosis of invasive bacterial infections in febrile infants: Results of a multicenter study and utility of a rapid qualitative test for this marker. Pediatr. Infect. Dis. J. 2003, 22, 895–903. [Google Scholar] [PubMed]
- Segal, I.; Ehrlichman, M.; Urbach, J.; Bar-Meir, M. Use of time from fever onset improves the diagnostic accuracy of C-reactive protein in identifying bacterial infections. Arch. Dis. Child. 2014, 99, 974–978. [Google Scholar] [CrossRef]
- Gómez, B.; Mintegi, S.; Benito, J.; Egireun, A.; Garcia, D.; Astobiza, E. Blood culture and bacteremia predictors in infants less than three months of age with fever without source. Pediatr. Infect. Dis. J. 2010, 29, 43–47. [Google Scholar] [CrossRef]
- Benitz, W.E.; Han, M.Y.; Madan, A.; Ramachandra, P. Serial serum C-reactive protein levels in the diagnosis of neonatal infection. Pediatrics 1998, 102, e41. [Google Scholar] [CrossRef]
- Samraj, R.S.; Zingarelli, B.; Wong, H.R. Role of biomarkers in sepsis care. Shock 2013, 40, 358–365. [Google Scholar] [CrossRef]
- Casado-Flores, J.; Blanco-Quirós, A.; Asensio, J.; Arranz, E.; Garrote, J.A.; Nieto, M. Serum procalcitonin in children with suspected sepsis: A comparison with C-reactive protein and neutrophil count. Pediatr. Crit. Care Med. 2003, 4, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Müller, B.; White, J.C.; Nylén, E.S.; Snider, R.H.; Becker, K.L.; Habener, J.F. Ubiquitous expression of the calcitonin-I gene in multiple tissues in response to sepsis. J. Clin. Endocrinol. Metab. 2001, 86, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Linscheid, P.; Seboek, D.; Schaer, D.J.; Zulewski, H.; Keller, U.; Müller, B. Expression and secretion of procalcitonin and calcitonin geneYrelated peptide by adherent monocytes and by macrophage-activated adipocytes. Crit. Care Med. 2004, 32, 1715–1721. [Google Scholar] [CrossRef] [PubMed]
- Principi, N.; Esposito, S. Biomarkers in Pediatric Community-Acquired Pneumonia. Int. J. Mol. Sci. 2017, 18, 447. [Google Scholar] [CrossRef]
- Arkader, R.; Troster, E.J.; Lopes, M.R.; Júnior, R.R.; Carcillo, J.A.; Leone, C.; Okay, T.S. Procalcitonin does discriminate between sepsis and systemic inflammatory response syndrome. Arch. Dis. Child. 2006, 91, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Milcent, K.; Faesch, S.; Gras-Le Guen, C.; Dubos, F.; Poulalhon, C.; Badier, I.; Marc, E.; Laguille, C.; de Pontual, L.; Mosca, A.; et al. Use of Procalcitonin Assays to Predict Serious Bacterial Infection in Young Febrile Infants. JAMA Pediatr. 2016, 170, 62–69. [Google Scholar] [CrossRef]
- England, J.T.; Del Vecchio, M.T.; Aronoff, S.C. Use of serum procalcitonin in evaluation of febrile infants: A meta-analysis of 2317 patients. J. Emerg. Med. 2014, 47, 682–688. [Google Scholar] [CrossRef]
- Pontrelli, G.; De Crescenzo, F.; Buzzetti, R.; Jenkner, A.; Balduzzi, S.; Calò Carducci, F.; Amodio, D.; De Luca, M.; Chiurchiù, S.; Davies, E.H.; et al. Accuracy of serum procalcitonin for the diagnosis of sepsis in neonates and children with systemic inflammatory syndrome: A meta-analysis. BMC Infect. Dis. 2017, 17, 302. [Google Scholar] [CrossRef]
- Waterfield, T.; Maney, J.A.; Lyttle, M.D.; McKenna, J.P.; Roland, D.; Corr, M.; Patenall, B.; Shields, M.D.; Woolfall, K.; Fairley, D.; et al. Diagnostic test accuracy of point-of-care procalcitonin to diagnose serious bacterial infections in children. BMC Pediatr. 2020, 20, 487. [Google Scholar] [CrossRef]
- Jacobs, D.M.; Holsen, M.; Chen, S.; Fusco, N.M.; Hassinger, A.B. Procalcitonin to Detect Bacterial Infections in Critically Ill Pediatric Patients. Clin. Pediatr. 2017, 56, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Cies, J.J.; Chopra, A. Procalcitonin use in a pediatric intensive care unit. Pediatr. Infect. Dis. J. 2014, 33, 984–986. [Google Scholar] [CrossRef] [PubMed]
- Lautz, A.J.; Dziorny, A.C.; Denson, A.R.; O’Connor, K.A.; Chilutti, M.R.; Ross, R.K.; Gerber, J.S.; Weiss, S.L. Value of Procalcitonin Measurement for Early Evidence of Severe Bacterial Infections in the Pediatric Intensive Care Unit. J. Pediatr. 2016, 179, 74–81.e2. [Google Scholar] [CrossRef] [PubMed]
- Zeng, G.; Chen, D.; Zhou, R.; Zhao, X.; Ye, C.; Tao, H.; Sheng, W.; Wu, Y. Combination of C-reactive protein, procalcitonin, IL-6, IL-8, and IL-10 for early diagnosis of hyperinflammatory state and organ dysfunction in pediatric sepsis. J. Clin. Lab. Anal. 2022, 36, e24505. [Google Scholar] [CrossRef] [PubMed]
- Jarczak, D.; Nierhaus, A. Cytokine Storm—Definition, Causes, and Implications. Int. J. Mol. Sci. 2022, 23, 11740. [Google Scholar] [CrossRef]
- Jarczak, D.; Kluge, S.; Nierhaus, A. Sepsis-Pathophysiology and Therapeutic Concepts. Front. Med. 2021, 8, 609. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Monneret, G.; Payen, D. Sepsis-induced immunosuppression: From cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 2013, 13, 862–874. [Google Scholar] [CrossRef]
- Dagys, A.; Laucaitytė, G.; Volkevičiūtė, A.; Abramavičius, S.; Kėvalas, R.; Vitkauskienė, A.; Jankauskaitė, L. Blood biomarkers in early bacterial infection and sepsis diagnostics in feverish young children. Int. J. Med. Sci. 2022, 19, 753–761. [Google Scholar] [CrossRef]
- Biron, B.M.; Ayala, A.; Lomas-Neira, J.L. Biomarkers for Sepsis: What Is and What Might Be? Biomark. Insights 2015, 10 (Suppl. S4), 7–17. [Google Scholar] [CrossRef]
- Fuster, J.J.; Walsh, K. The good, the bad, and the ugly of interleukin-6 signaling. EMBO J. 2014, 33, 1425–1427. [Google Scholar] [CrossRef]
- Vasconcellos, Â.G.; Clarêncio, J.; Andrade, D.; Cardoso, M.A.; Barral, A.; Nascimento-Carvalho, C.M. Systemic cytokines and chemokines on admission of children hospitalized with community-acquired pneumonia. Cytokine 2018, 107, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Li, L.; Dou, Y.; Li, P.; Chen, R.; Liu, H. Diagnostic utility of neutrophil CD64 as a marker for early-onset sepsis in preterm neonates. PLoS ONE 2014, 9, e102647. [Google Scholar] [CrossRef]
- Foo, C.P.Z.; Seabrook, J.A.; Sangha, G.; Foster, J.R. Presumed Systemic Inflammatory Response Syndrome in the Pediatric Emergency Department. Pediatr. Emerg. Care 2019, 35, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Umlauf, V.N.; Dreschers, S.; Orlikowsky, T.W. Flow cytometry in the detection of neonatal sepsis. Int. J. Pediatr. 2013, 2013, 763191. [Google Scholar] [CrossRef]
- Levy, M.M.; Fink, M.P.; Marshall, J.C.; Abraham, E.; Angus, D.; Cook, D.; Cohen, J.; Opal, S.M.; Vincent, J.L.; Ramsay, G. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit. Care Med. 2003, 31, 1250–1256. [Google Scholar] [CrossRef] [PubMed]
- Urbonas, V.; Eidukaitė, A.; Tamulienė, I. Increased interleukin-10 levels correlate with bacteremia and sepsis in febrile neutropenia pediatric oncology patients. Cytokine 2012, 57, 313–315. [Google Scholar] [CrossRef] [PubMed]
- Theodosiou, A.A.; Mashumba, F.; Flatt, A. Excluding Clinically Significant Bacteremia by 24 h in Otherwise Well Febrile Children Younger Than 16 Years: A Study of More Than 50,000 Blood Cultures. Pediatr. Infect. Dis. J. 2019, 38, e203–e208. [Google Scholar] [CrossRef] [PubMed]
- Tamelytė, E.; Vaičekauskienė, G.; Dagys, A.; Lapinskas, T.; Jankauskaitė, L. Early Blood Biomarkers to Improve Sepsis/Bacteremia Diagnostics in Pediatric Emergency Settings. Medicina 2019, 55, 99. [Google Scholar] [CrossRef]
- Romagnoli, C.; Frezza, S.; Cingolani, A.; De Luca, A.; Puopolo, M.; De Carolis, M.P.; Vento, G.; Antinori, A.; Tortorolo, G. Plasma levels of interleukin-6 and interleukin-10 in preterm neonates evaluated for sepsis. Eur. J. Pediatr. 2001, 160, 345–350. [Google Scholar] [CrossRef]
- Wong, H.R.; Cvijanovich, N.Z.; Hall, M.; Allen, G.L.; Thomas, N.J.; Freishtat, R.J.; Anas, N.; Meyer, K.; Checchia, P.A.; Lin, R.; et al. Interleukin-27 is a novel candidate diagnostic biomarker for bacterial infection in critically ill children. Crit. Care 2012, 16, R213. [Google Scholar] [CrossRef]
- Hanna, W.J.; Berrens, Z.; Langner, T.; Lahni, P.; Wong, H.R. Interleukin-27: A novel biomarker in predicting bacterial infection among the critically ill. Crit. Care 2015, 19, 378. [Google Scholar] [CrossRef] [PubMed]
- Gyurkovska, V.; Ivanovska, N. Distinct roles of TNF-related apoptosis-inducing ligand (TRAIL) in viral and bacterial infections: From pathogenesis to pathogen clearance. Inflamm. Res. 2016, 65, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Unsinger, J.; Kazama, H.; McDonough, J.S.; Griffith, T.S.; Hotchkiss, R.S.; Ferguson, T.A. Sepsis—Induced apoptosis leads to active suppression of delayed-type hypersensitivity by CD8+ regulatory T cells through a TRAIL-dependent mechanism. J. Immunol. 2010, 184, 6766–6772. [Google Scholar] [CrossRef]
- Tian, Y.; Tao, T.; Zhu, J.; Zou, Y.; Wang, J.; Li, J.; Bo, L.; Deng, X. Soluble Tumor Necrosis Factor Related Apoptosis Inducing Ligand Level as a Predictor of Severity of Sepsis and the Risk of Mortality in Septic Patients. PLoS ONE 2013, 8, e82204. [Google Scholar] [CrossRef] [PubMed]
- Papan, C.; Sidorov, S.; Greiter, B.; Bühler, N.; Berger, C.; Becker, S.L.; Sauteur, P.M.M. Combinatorial host-response biomarker signature (BV score) and its subanalytes TRAIL, IP-10, and CRP in children with Mycoplasma pneumoniae community-acquired pneumonia. J. Infect. Dis. 2023, 13, jiad573. [Google Scholar] [CrossRef]
- Fröhlich, F.; Gronwald, B.; Bay, J.; Simon, A.; Poryo, M.; Geisel, J.; Tegethoff, S.A.; Last, K.; Rissland, J.; Smola, S.; et al. Expression of TRAIL, IP-10, and CRP in children with suspected COVID-19 and real-life impact of a computational signature on clinical decision-making: A prospective cohort study. Infection 2023, 51, 1349–1356. [Google Scholar] [CrossRef]
- Ashkenazi-Hoffnung, L.; Oved, K.; Navon, R.; Friedman, T.; Boico, O.; Paz, M.; Kronenfeld, G.; Etshtein, L.; Cohen, A.; Gottlieb, T.M.; et al. A host-protein signature is superior to other biomarkers for differentiating between bacterial and viral disease in patients with respiratory infection and fever without source: A prospective observational study. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1361–1371. [Google Scholar] [CrossRef]
- Ashkenazi-Hoffnung, L.; Livni, G.; Scheuerman, O.; Berger, I.; Eden, E.; Oved, K.; Shani, L.; Kronenfeld, G.; Simon, E.; Boico, O.; et al. Differential Serum and Urine CRP, IP-10, and TRAIL Levels in Pediatric Urinary Tract Infection. Front. Pediatr. 2021, 9, 771118. [Google Scholar] [CrossRef]
- Ko, T.M.; Kuo, H.C.; Chang, J.S.; Chen, S.P.; Liu, Y.M.; Chen, H.W.; Tsai, F.J.; Lee, Y.C.; Chen, C.H.; Wu, J.Y.; et al. CXCL10/IP-newly10 is a biomarker and mediator for Kawasaki disease. Circ. Res. 2015, 116, 876–883. [Google Scholar] [CrossRef]
- Azzurri, A.; Sow, O.Y.; Amedei, A.; Bah, B.; Diallo, S.; Peri, G.; Benagiano, M.; D’Elios, M.M.; Mantovani, A.; Del Prete, G. IFN-gamma-inducible protein 10 and pentraxin 3 plasma levels are tools for monitoring inflammation and disease activity in Mycobacterium tuberculosis infection. Microbes Infect. 2005, 7, 421429. [Google Scholar] [CrossRef]
- Esposito, S.; Tagliabue, C.; Picciolli, I.; Semino, M.; Sabatini, C.; Consolo, S.; Bosis, S.; Pinzani, R.; Principi, N. Procalcitonin measurements for guiding antibiotic treatment in pediatric pneumonia. Respir. Med. 2011, 105, 1939–1945. [Google Scholar] [CrossRef]
- Van Houten, C.B.; de Groot, J.A.H.; Klein, A.; Srugo, I.; Chistyakov, I.; de Waal, W.; Meijssen, C.B.; Avis, W.; Wolfs, T.F.W.; Shachor-Meyouhas, Y.; et al. A host-protein based assay to differentiate between bacterial and viral infections in preschool children (OPPORTUNITY): A double-blind, multicentre, validation study. Lancet Infect. Dis. 2017, 17, 431–440. [Google Scholar] [CrossRef]
- König, R.; Kolte, A.; Ahlers, O.; Oswald, M.; Krauss, V.; Roell, D.; Sommerfeld, O.; Dimopoulos, G.; Tsangaris, I.; Antoniadou, E.; et al. Use of IFNγ/IL10 Ratio for Stratification of Hydrocortisone Therapy in Patients with Septic Shock. Front. Immunol. 2021, 12, 607217. [Google Scholar] [CrossRef]
- Papan, C.; Argentiero, A.; Adams, O.; Porwoll, M.; Hakim, U.; Farinelli, E.; Testa, I.; Pasticci, M.B.; Mezzetti, D.; Perruccio, K.; et al. Association of viral load with TRAIL, IP-10, CRP biomarker signature and disease severity in children with respiratory tract infection or fever without source: A prospective, multicentre cohort study. J. Med. Virol. 2022, 95, e28113. [Google Scholar] [CrossRef]
- Papan, C.; Argentiero, A.; Porwoll, M.; Hakim, U.; Farinelli, E.; Testa, I.; Pasticci, M.B.; Mezzetti, D.; Perruccio, K.; Etshtein, L.; et al. A host signature based on TRAIL, IP-10, and CRP for reducing antibiotic overuse in children by differentiating bacterial from viral infections: A prospective, multicentre cohort study. Clin. Microbiol. Infect. 2022, 28, 723–730. [Google Scholar] [CrossRef]
- Velissaris, D.; Zareifopoulos, N.; Karamouzos, V.; Karanikolas, E.; Pierrakos, C.; Koniari, I.; Karanikolas, M. Presepsin as a Diagnostic and Prognostic Biomarker in Sepsis. Cureus 2021, 13, e15019. [Google Scholar] [CrossRef]
- Henriquez-Camacho, C.; Losa, J. Biomarkers for sepsis. Biomed. Res. Int. 2014, 2014, 547818. [Google Scholar] [CrossRef]
- Wu, C.C.; Lan, H.M.; Han, S.T.; Chaou, C.H.; Yeh, C.F.; Liu, S.H.; Li, C.H.; Blaney, G.N., 3rd; Liu, Z.Y.; Chen, K.F. Comparison of diagnostic accuracy in sepsis between presepsin, procalcitonin, and C-reactive protein: A systematic review and meta-analysis. Ann. Intensive Care 2017, 7, 91. [Google Scholar] [CrossRef]
- Romualdo, L.G.; Torrella, P.E.; González, M.V.; Sánchez, R.J.; Holgado, A.H.; Freire, A.O.; Acebes, S.R.; Otón, M.D. Diagnostic accuracy of presepsin (soluble CD14 subtype) for prediction of bacteremia in patients with systemic inflammatory response syndrome in the Emergency Department. Clin. Biochem. 2014, 47, 505–508. [Google Scholar] [CrossRef]
- Vouloumanou, E.K.; Plessa, E.; Karageorgopoulos, D.E.; Mantadakis, E.; Falagas, M.E. Serum procalcitonin as a diagnostic marker for neonatal sepsis: A systematic review and meta-analysis. Intensive Care Med. 2011, 37, 747–762. [Google Scholar] [CrossRef]
- Schuetz, P.; Christ-Crain, M.; Müller, B. Procalcitonin and other biomarkers to improve assessment and antibiotic stewardship in infections–hope for hype? Swiss Med. Wkly. 2009, 139, 318–326. [Google Scholar]
- Poggi, C.; Bianconi, T.; Gozzini, E.; Generoso, M.; Dani, C. Presepsin for the detection of late-onset sepsis in preterm newborns. Pediatrics 2015, 135, 68–75. [Google Scholar] [CrossRef]
- Kumar, N.; Dayal, R.; Singh, P.; Pathak, S.; Pooniya, V.; Goyal, A.; Kamal, R.; Mohanty, K.K. A Comparative Evaluation of Presepsin with Procalcitonin and CRP in Diagnosing Neonatal Sepsis. Indian J. Pediatr. 2019, 86, 177–179. [Google Scholar] [CrossRef]
- Smok, B.; Domagalski, K.; Pawłowska, M. Diagnostic and Prognostic Value of IL-6 and sTREM-1 in SIRS and Sepsis in Children. Mediat. Inflamm. 2020, 2020, 8201585. [Google Scholar] [CrossRef]
- Esposito, S.; Di Gangi, M.; Cardinale, F.; Baraldi, E.; Corsini, I.; Da Dalt, L.; Tovo, P.A.; Correra, A.; Villani, A.; Sacco, O.; et al. Sensitivity and Specificity of Soluble Triggering Receptor Expressed on Myeloid Cells-1, Midregional Proatrial Natriuretic Peptide and Midregional Proadrenomedullin for Distinguishing Etiology and to Assess Severity in Community-Acquired Pneumonia. PLoS ONE 2016, 11, e0163262. [Google Scholar] [CrossRef]
- Balanza, N.; Erice, C.; Ngai, M.; Varo, R.; Kain, K.C.; Bassat, Q. Host-Based Prognostic Biomarkers to Improve Risk Stratification and Outcome of Febrile Children in Low- and Middle-Income Countries. Front. Pediatr. 2020, 8, 552083. [Google Scholar] [CrossRef]
- Adly, A.A.; Ismail, E.A.; Andrawes, N.G.; El-Saadany, M.A. Circulating soluble triggering receptor expressed on myeloid cells-1 (sTREM-1) as diagnostic and prognostic marker in neonatal sepsis. Cytokine 2014, 65, 184–191. [Google Scholar] [CrossRef]
- Sarafidis, K.; Soubasi-Griva, V.; Piretzi, K.; Thomaidou, A.; Agakidou, E.; Taparkou, A.; Diamanti, E.; Drossou-Agakidou, V. Diagnostic utility of elevated serum soluble triggering receptor expressed on myeloid cells (sTREM)-1 in infected neonates. Intensive Care Med. 2010, 36, 864–868. [Google Scholar] [CrossRef]
- Pontrelli, G.; De Crescenzo, F.; Buzzetti, R.; Calò Carducci, F.; Jenkner, A.; Amodio, D.; De Luca, M.; Chiurchiù, S.; Davies, E.H.; Simonetti, A.; et al. Diagnostic value of soluble triggering receptor expressed on myeloid cells in paediatric sepsis: A systematic review. Ital. J. Pediatr. 2016, 42, 44. [Google Scholar] [CrossRef]
- Jiyong, J.; Tiancha, H.; Wei, C.; Huahao, S. Diagnostic value of the soluble triggering receptor expressed on myeloid cells-1 in bacterial infection: A meta-analysis. Intensive Care Med. 2009, 35, 587–595. [Google Scholar] [CrossRef]
- Goldstein, B.; Giroir, B.; Randolph, A. International pediatric sepsis consensus conference: Definitions for sepsis and organ dysfunction in pediatrics. Pediatr. Crit. Care Med. 2005, 6, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Balamuth, F.; Scott, H.F.; Weiss, S.L.; Webb, M.; Chamberlain, J.M.; Bajaj, L.; Depinet, H.; Grundmeier, R.W.; Campos, D.; Deakyne Davies, S.J.; et al. Validation of the Pediatric Sequential Organ Failure Assessment Score and Evaluation of Third International Consensus Definitions for Sepsis and Septic Shock Definitions in the Pediatric Emergency Department. JAMA Pediatr. 2022, 176, 672–678. [Google Scholar] [CrossRef]
- Esposito, S.; Rinaldi, V.E.; Argentiero, A.; Farinelli, E.; Cofini, M.; D’Alonzo, R.; Mencacci, A.; Principi, N. Approach to Neonates and Young Infants with Fever without a Source Who Are at Risk for Severe Bacterial Infection. Mediat. Inflamm. 2018, 2018, 4869329. [Google Scholar] [CrossRef]
- Blot, F.; Nitenberg, G.; Chachaty, E.; Raynard, B.; Germann, N.; Antoun, S.; Laplanche, A.; Brun-Buisson, C.; Tancrède, C. Diagnosis of catheter-related bacteraemia: A prospective comparison of the time to positivity of hub-blood versus peripheral-blood cultures. Lancet 1999, 354, 1071–1077. [Google Scholar] [CrossRef]
- Cecinati, V.; Brescia, L.; Tagliaferri, L.; Giordano, P.; Esposito, S. Catheter-related infections in pediatric patients with cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2869–2877. [Google Scholar] [CrossRef]
- McGowan, K.I.; Foster, J.A.; Coffin, S.E. Outpatient pediatric blood cultures: Time to positivity. Pediatrics 2000, 106, 251–255. [Google Scholar] [CrossRef]
- Alpern, E.R.; Alessandrini, E.A.; Bell, L.M.; Shaw, K.N.; McGowan, K.L. Occult bacteremia from a pediatric emergency department: Current prevalence, time to detection, and outcome. Pediatrics 2000, 106, 505–511. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Yi, Q.; Suo, F.; Tang, Y.; Luo, S.; Tian, X.; Zhang, G.; Chen, D.; Luo, Z. Prognostic roles of time to positivity of blood culture in children with Streptococcus pneumoniae bacteremia. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 457–465. [Google Scholar] [CrossRef]
- Kang, C.M.; Chen, X.I.; Chih, C.C.; Hsu, C.C.; Chen, P.H.; Fen Lee, T.; Teng, L.J.; Hsueh, P.R. Rapid identification of bloodstream bacterial and fungal pathogens and their antibiotic resistance determinants from positively flagged blood cultures using the BioFire FilmArray blood culture identification panel. J. Microbiol. Immunol. Infect. 2020, 53, 882–891. [Google Scholar] [CrossRef]
- She, R.C.; Bender, J.M. Advances in Rapid Molecular Blood Culture Diagnostics: Healthcare Impact, Laboratory Implications, and Multiplex Technologies. J. Appl. Lab. Med. 2019, 3, 617–630. [Google Scholar] [CrossRef]
- Matsushita, F.Y.; Jornada Krebs, V.L.; de Carvalho, W.B. Complete blood count and C-reactive protein to predict positive blood culture among neonates using machine learning algorithms. Clinics 2022, 78, 100148. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.C.; Chen, H.L.; Lin, S.Y.; Chen, T.C.; Lu, P.L. Short time to positivity of blood culture predicts mortality and septic shock in bacteremic patients: A systematic review and meta-analysis. BMC Infect. Dis. 2022, 22, 142. [Google Scholar] [CrossRef]
- Han, J.H.; Nachamkin, I.; Coffin, S.E.; Gerber, J.S.; Fuchs, B.; Garrigan, C.; Han, X.; Bilker, W.B.; Wise, J.; Tolomeo, P.; et al. Use of a Combination Biomarker Algorithm to Identify Medical Intensive Care Unit Patients with Suspected Sepsis at Very Low Likelihood of Bacterial Infection. Antimicrob. Agents Chemother. 2015, 59, 6494–6500. [Google Scholar] [CrossRef] [PubMed]
- Witting, C.S.; Simon, N.J.E.; Lorenz, D.; Murphy, J.S.; Nelson, J.; Lehnig, K.; Alpern, E.R. Sepsis Electronic Decision Support Screen in High-Risk Patients Across Age Groups in a Pediatric Emergency Department. Pediatr. Emerg. Care 2022, 38, e1479–e1484. [Google Scholar] [CrossRef]
- Narayanan, N.; Gross, A.K.; Pintens, M.; Fee, C.; Macdougall, C. Effect of an electronic medical record alert for severe sepsis among ED patients. Am. J. Emerg. Med. 2016, 34, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Lamping, F.; Jack, T.; Rübsamen, N.; Sasse, M.; Beerbaum, P.; Mikolajczyk, R.T.; Boehne, M.; Karch, A. Development and validation of a diagnostic model for early differentiation of sepsis and non-infectious SIRS in critically ill children-a data-driven approach using machine-learning algorithms. BMC Pediatr. 2018, 18, 112. [Google Scholar] [CrossRef] [PubMed]
- Komorowski, M.; Green, A.; Tatham, K.C.; Seymour, C.; Antcliffe, D. Sepsis biomarkers and diagnostic tools with a focus on machine learning. EBioMedicine 2022, 86, 104394. [Google Scholar] [CrossRef]
- Calfee, C.S.; Delucchi, K.; Parsons, P.E.; Thompson, B.T.; Ware, L.B.; Matthay, M.A. Subphenotypes in acute respiratory distress syndrome: Latent class analysis of data from two randomised controlled trials. Lancet Resp Med. 2014, 2, 611–620. [Google Scholar] [CrossRef]
- Downes, K.J.; Fitzgerald, J.C.; Schriver, E.; Boge, C.L.K.; Russo, M.E.; Weiss, S.L.; Balamuth, F.; Kubis, S.E.; Tolomeo, P.; Bilker, W.B.; et al. Implementation of a Pragmatic Biomarker-Driven Algorithm to Guide Antibiotic Use in the Pediatric Intensive Care Unit: The Optimizing Antibiotic Strategies in Sepsis (OASIS) II Study. J. Pediatr. Infect. Dis. Soc. 2020, 9, 36–43. [Google Scholar] [CrossRef]
- Bos, D.A.G.; De Burghgraeve, T.; De Sutter, A.; Buntinx, F.; Verbakel, J.Y. Clinical prediction models for serious infections in children: External validation in ambulatory care. BMC Med. 2023, 21, 151. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernardi, L.; Bossù, G.; Dal Canto, G.; Giannì, G.; Esposito, S. Biomarkers for Serious Bacterial Infections in Febrile Children. Biomolecules 2024, 14, 97. https://doi.org/10.3390/biom14010097
Bernardi L, Bossù G, Dal Canto G, Giannì G, Esposito S. Biomarkers for Serious Bacterial Infections in Febrile Children. Biomolecules. 2024; 14(1):97. https://doi.org/10.3390/biom14010097
Chicago/Turabian StyleBernardi, Luca, Gianluca Bossù, Giulia Dal Canto, Giuliana Giannì, and Susanna Esposito. 2024. "Biomarkers for Serious Bacterial Infections in Febrile Children" Biomolecules 14, no. 1: 97. https://doi.org/10.3390/biom14010097
APA StyleBernardi, L., Bossù, G., Dal Canto, G., Giannì, G., & Esposito, S. (2024). Biomarkers for Serious Bacterial Infections in Febrile Children. Biomolecules, 14(1), 97. https://doi.org/10.3390/biom14010097







