Neurofilament Light Protein Rod Domain Exhibits Structural Heterogeneity
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Expression and Purification
2.2. Circular Dichroism Spectroscopy (CD)
2.3. Differential Scanning Calorimetry (DSC)
2.4. Fluorescence Experiments
2.5. Asymmetrical Flow Field-Flow Fractionation Coupled with UV/VIS Detector
2.6. Size Exclusion Chromatography (SEC) of NFL Fragments
3. Results
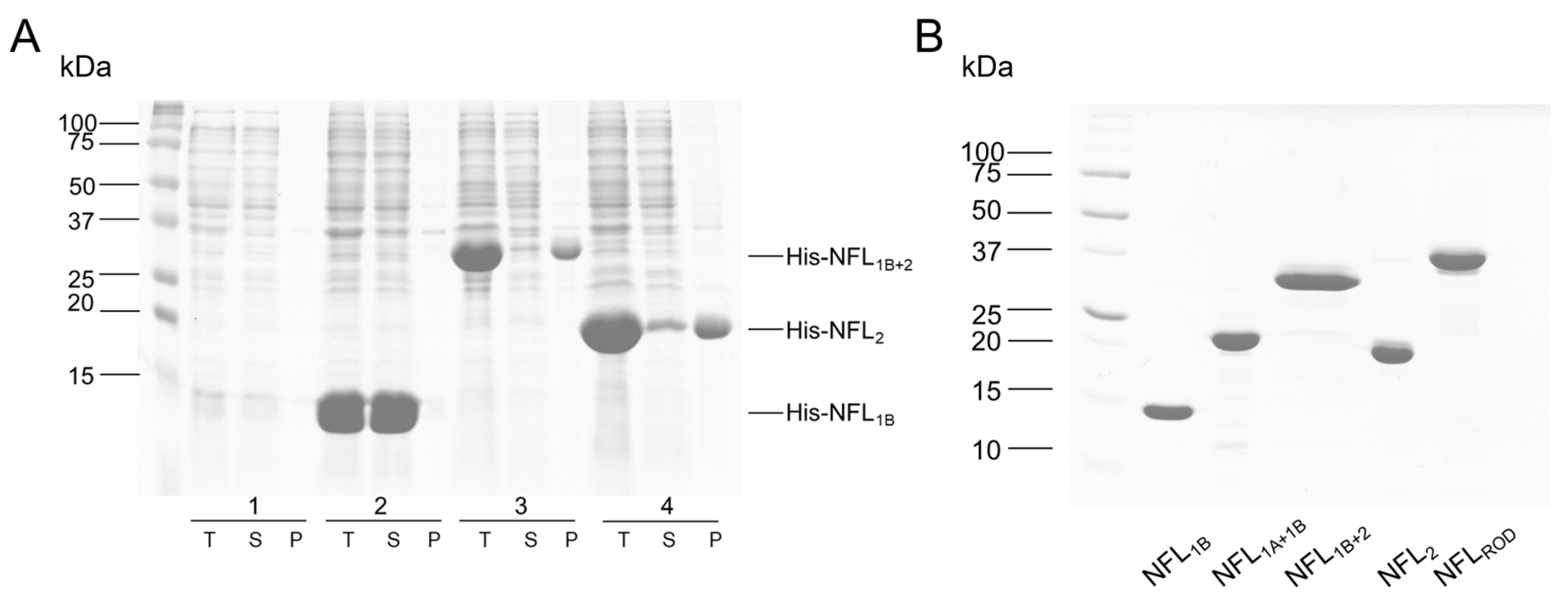
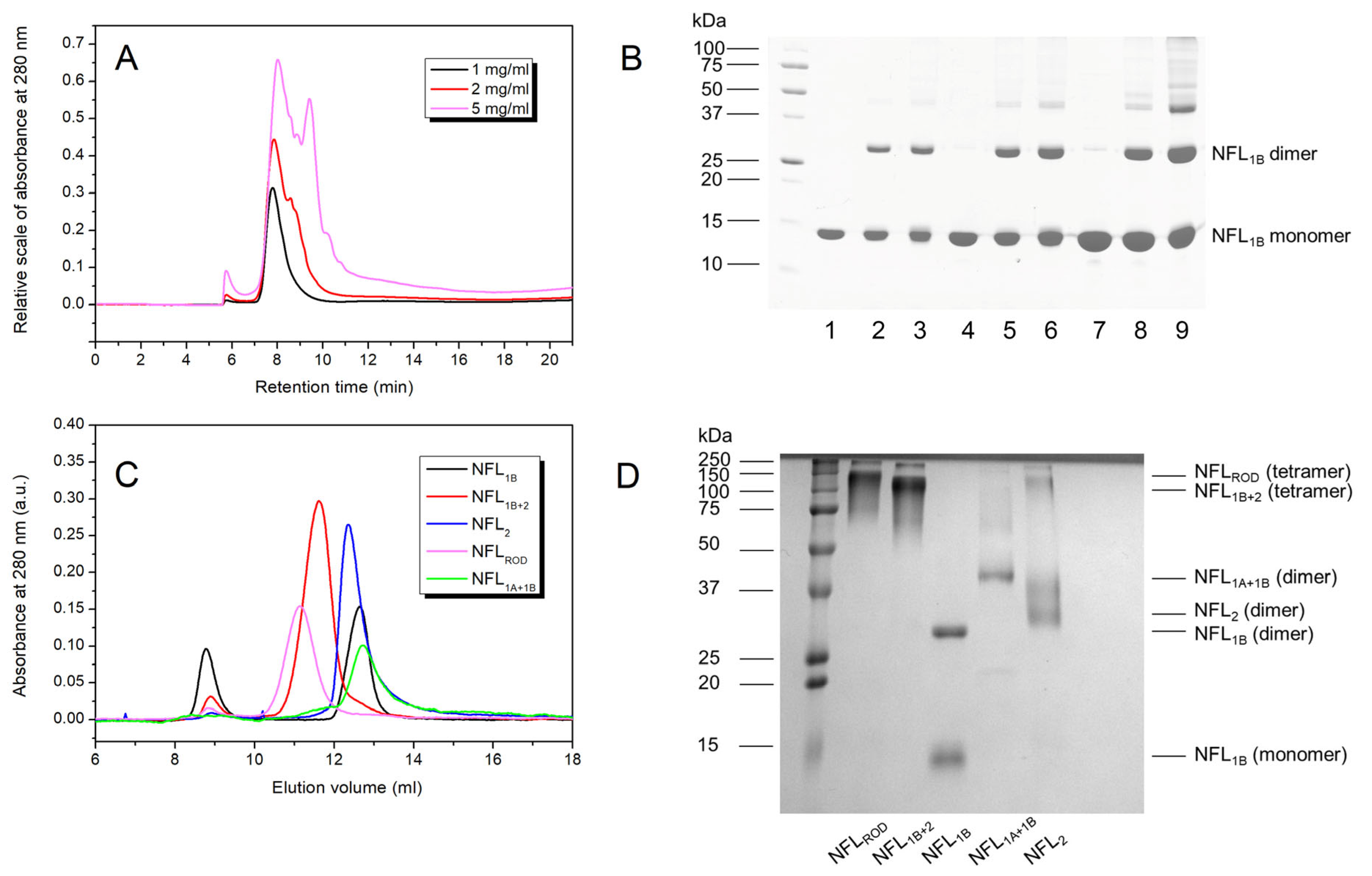
3.1. Expression and Purification of the Recombinant NFL Domains
3.2. Thermal Stability of the NFL Domains
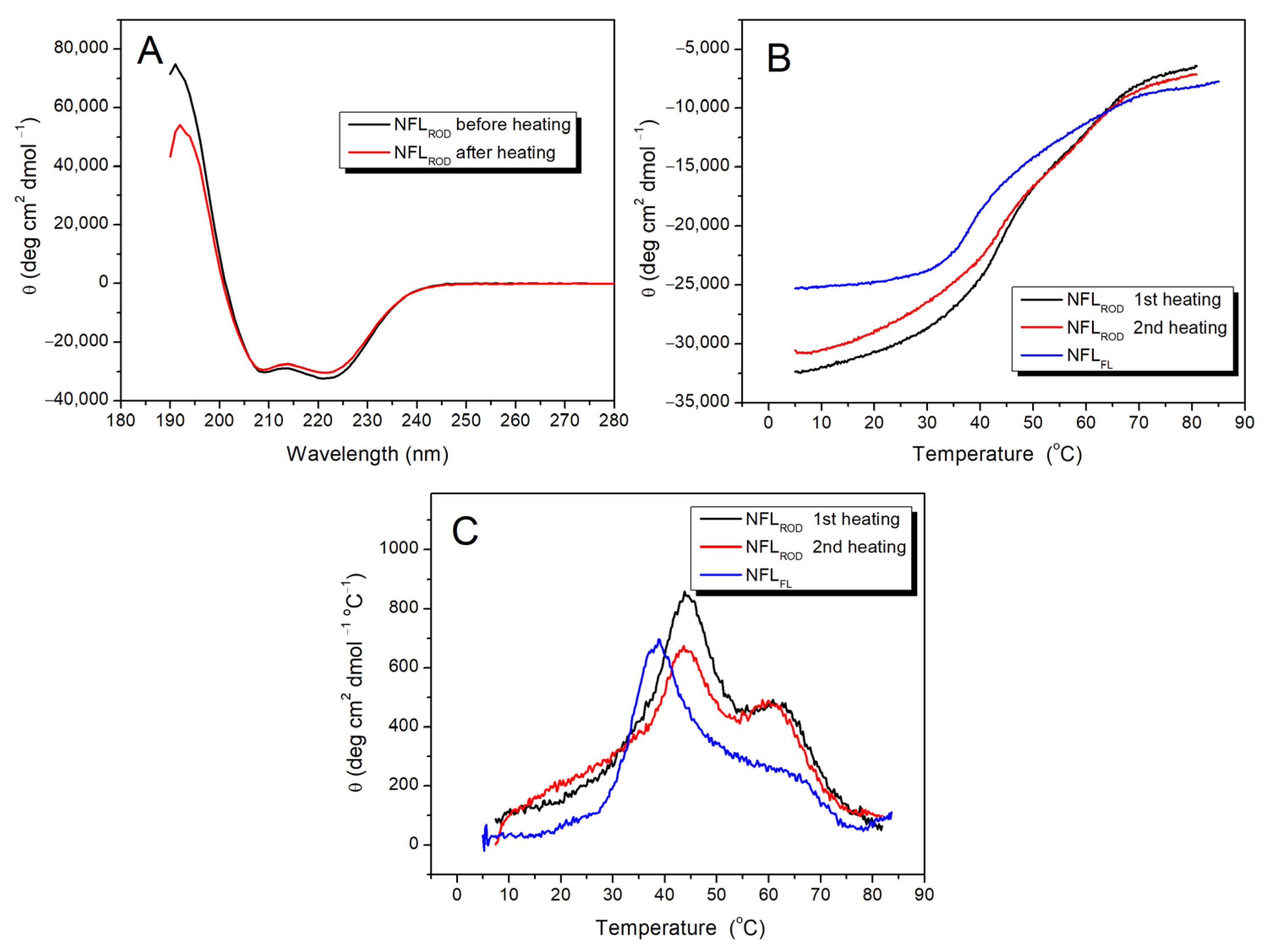
3.2.1. NFL Rod Domain
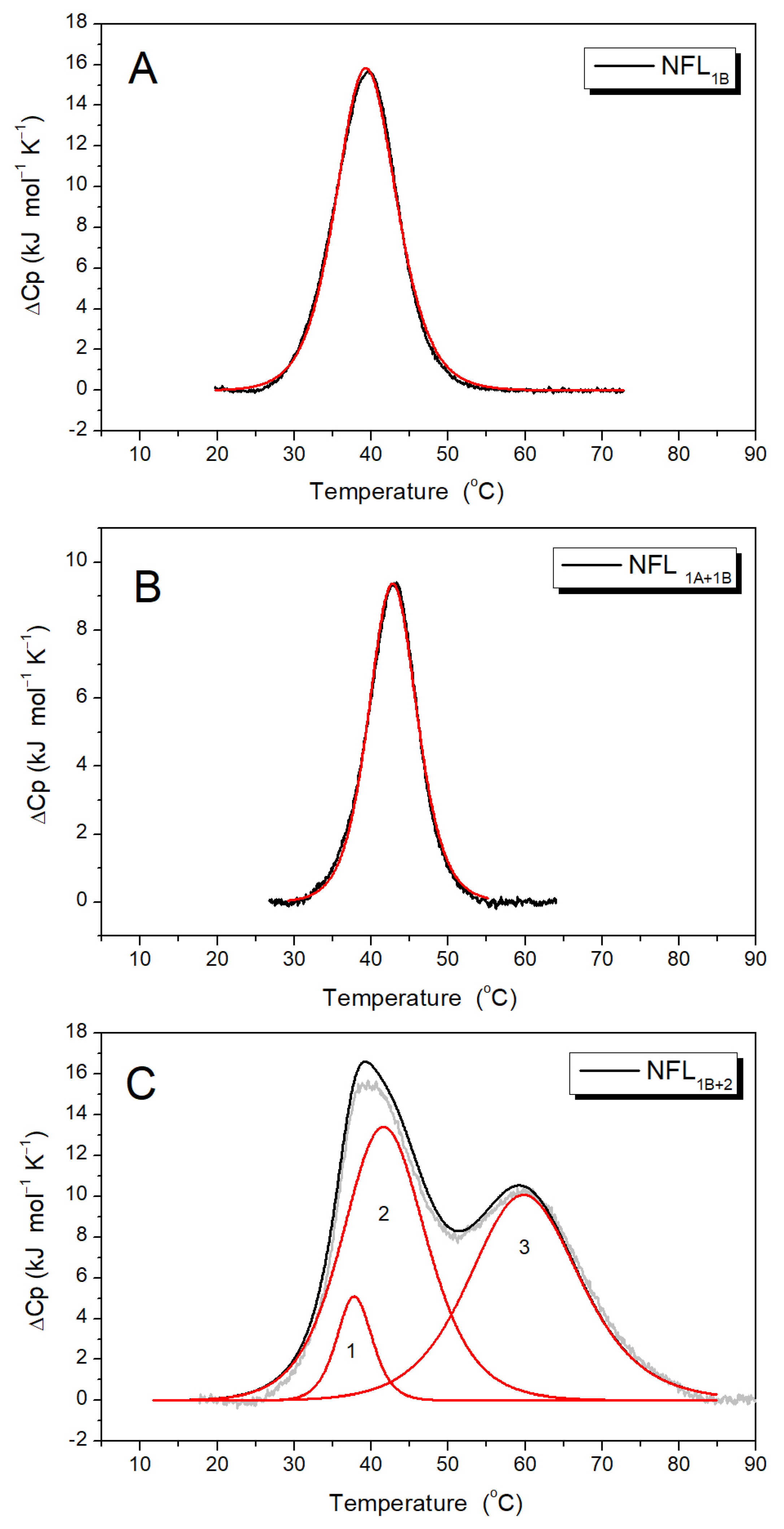
3.2.2. NFL Coiled-Coil Fragments Containing Coil 1B (NFL1B, NFL1A+1B, and NFL1B+2)
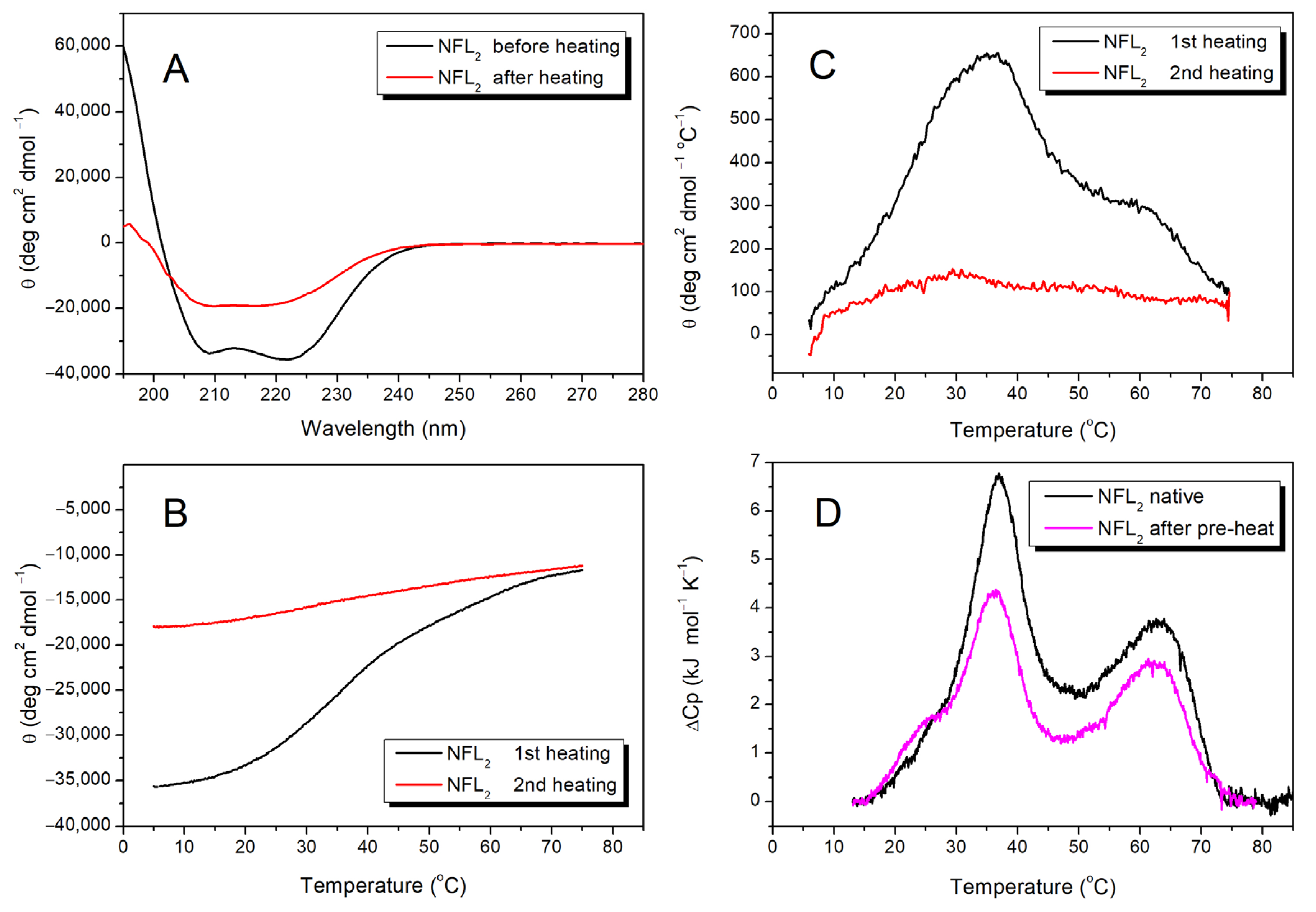
3.2.3. NFL Coil 2
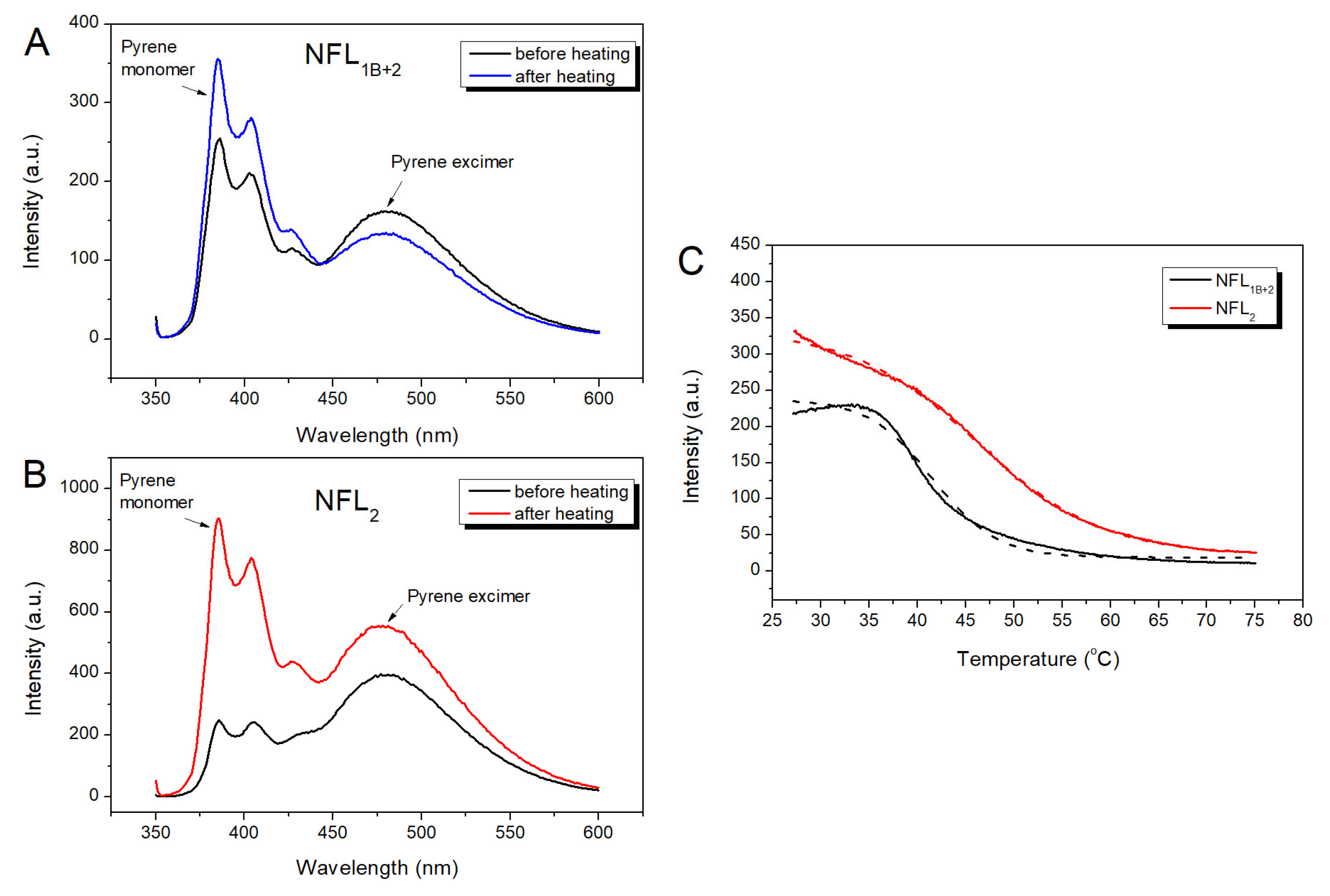
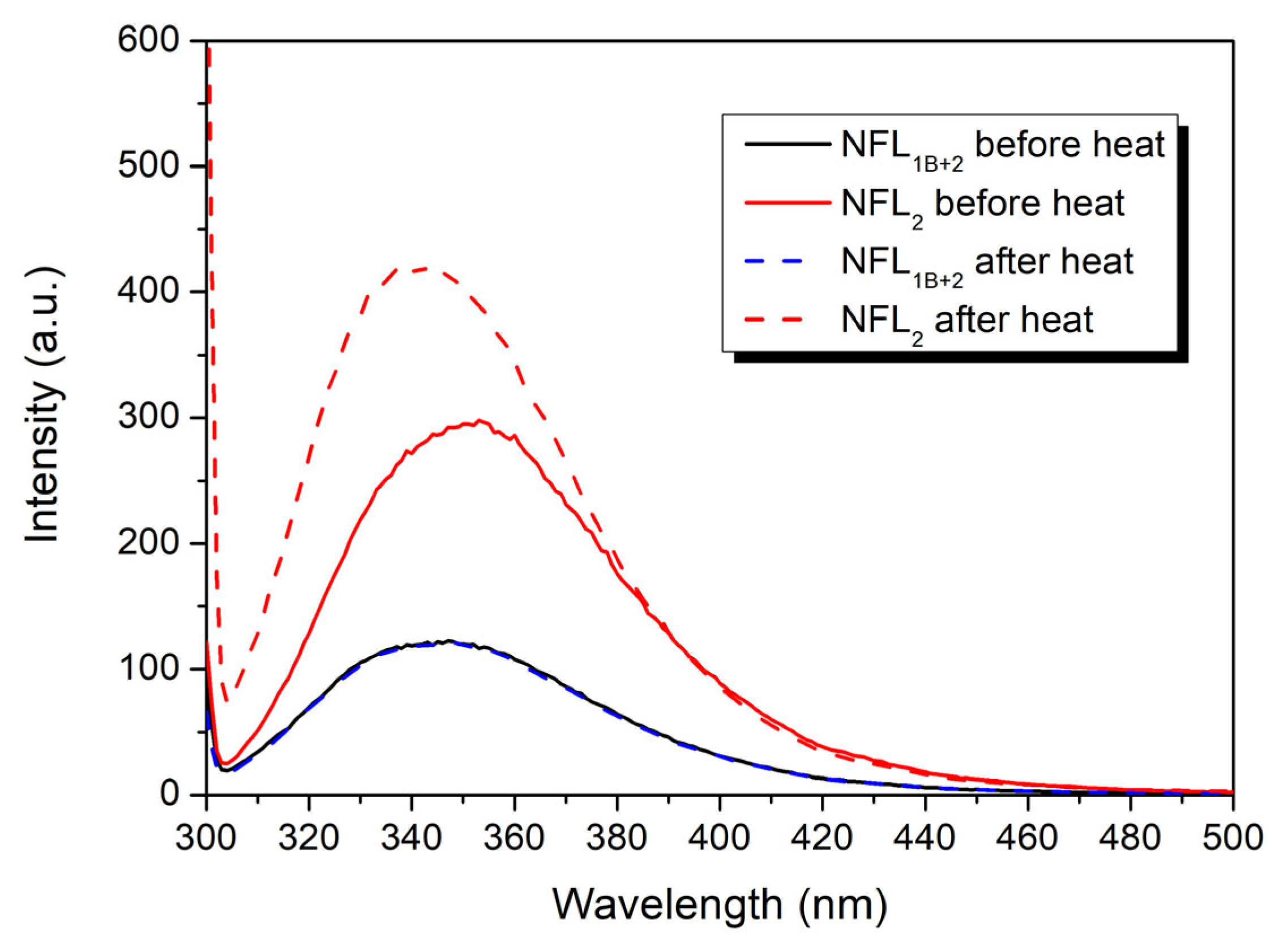
3.2.4. Fluorescence Studies of the NFL Fragments
3.2.5. Oligomerization of NFL Fragments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kornreich, M.; Avinery, R.; Malka-Gibor, E.; Laser-Azogui, A.; Beck, R. Order and disorder in intermediate filament proteins. FEBS Lett. 2015, 589 Pt A, 2464–2476. [Google Scholar] [CrossRef]
- Eldirany, S.A.; Lomakin, I.B.; Ho, M.; Bunick, C.G. Recent insight into intermediate filament structure. Curr. Opin. Cell Biol. 2020, 68, 132–143. [Google Scholar] [CrossRef]
- Etienne-Manneville, S. Cytoplasmic Intermediate Filaments in Cell Biology. Annu. Rev. Cell Dev. Biol. 2018, 34, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Vermeire, P.-J.; Stalmans, G.; Lilina, A.V.; Fiala, J.; Novak, P.; Herrmann, H.; Strelkov, S.V. Molecular interactions driving intermediate filament assembly. Cells 2021, 10, 2457. [Google Scholar] [CrossRef] [PubMed]
- Burkhard, P.; Stetefeld, J.; Strelkov, S.V. Coiled coils: A highly versatile protein folding motif. Trends Cell Biol. 2001, 11, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Mücke, N.; Wedig, T.; Bürer, A.; Marekov, L.N.; Steinert, P.M.; Langowski, J.; Aebi, U.; Herrmann, H. Molecular and biophysical characterization of assembly-starter units of human vimentin. J. Mol. Biol. 2004, 340, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Wickert, U.; Mücke, N.; Wedig, T.; Müller, S.A.; Aebi, U.; Herrmann, H. Characterization of the in vitro co-assembly process of the intermediate filament proteins vimentin and desmin: Mixed polymers at all stages of assembly. Eur. J. Cell Biol. 2005, 84, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Soellner, P.; Quinlan, R.A.; Franke, W.W. Identification of a distinct soluble subunit of an intermediate filament protein: Tetrameric vimentin from living cells. Proc. Natl. Acad. Sci. USA 1985, 82, 7929–7933. [Google Scholar] [CrossRef] [PubMed]
- Eldirany, S.; Ho, M.; Hinbest, A.J.; Lomakin, I.B.; Bunick, C.G. Human keratin 1/10-1B tetramer structures reveal a knob-pocket mechanism in intermediate filament assembly. EMBO J. 2019, 38, e100741. [Google Scholar] [CrossRef]
- Herrmann, H.; Haner, M.; Brettel, M.; Ku, N.O.; Aebi, U. Characterization of distinct early assembly units of different intermediate filament proteins. J. Mol. Biol. 1999, 286, 1403–1420. [Google Scholar] [CrossRef]
- Nakamura, Y.Y.; Takeda, M.; Angelides, K.J.; Tanaka, T.; Tada, K.; Nishimura, T. Effect of phosphorylation on 68 KDa neurofilament subunit protein assembly by the cyclic AMP dependent protein kinase in vitro. Biochem. Biophys. Res. Commun. 1990, 169, 744–750. [Google Scholar] [CrossRef]
- Yates, D.M.; Manser, C.; De Vos, K.J.; Shaw, C.E.; McLoughlin, D.M.; Miller, C.C. Neurofilament subunit (NFL) head domain phosphorylation regulates axonal transport of neurofilaments. Eur. J. Cell. Biol. 2009, 88, 193–202. [Google Scholar] [CrossRef]
- Hisanaga, S.; Gonda, Y.; Inagaki, M.; Ikai, A.; Hirokawa, N. Effects of phosphorylation of the neurofilament L protein on filamentous structures. Cell Regul. 1990, 1, 237–248. [Google Scholar] [CrossRef]
- Ahn, J.; Jeong, S.; Kang, S.-M.; Jo, I.; Park, B.-J.; Ha, N.-C. Separation of coiled-coil structures in lamin A/C is required for the elongation of the filament. Cells 2021, 10, 55. [Google Scholar] [CrossRef] [PubMed]
- Turgay, Y.; Eibauer, M.; Goldman, A.E.; Shimi, T.; Khayat, M.; Ben-Harush, K.; Dubrovsky-Gaupp, A.; Sapra, K.T.; Goldman, R.D.; Medalia, O. The molecular architecture of lamins in somatic cells. Nat. Cell Biol. 2017, 543, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-H.; Kim, M.-S.; Chung, B.M.; Leahy, D.J.; Coulombe, P.A. Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat. Struct. Mol. Biol. 2012, 19, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Nicolet, S.; Herrmann, H.; Aebi, U.; Strelkov, S.V. Atomic structure of vimentin coil 2. J. Struct. Biol. 2010, 170, 369–376. [Google Scholar] [CrossRef]
- Chernyatina, A.A.; Nicolet, S.; Aebi, U.; Herrmann, H.; Strelkov, S.V. Atomic structure of the vimentin central α-helical domain and its implications for intermediate filament assembly. Proc. Natl. Acad. Sci. USA 2012, 109, 13620–13625. [Google Scholar] [CrossRef] [PubMed]
- Bollati, M.; Barbiroli, A.; Favalli, V.; Arbustini, E.; Charron, P.; Bolognesi, M. Structures of the lamin A/C R335W and E347K mutants: Implications for dilated cardiolaminopathies. Biochem. Biophys. Res. Commun. 2012, 418, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Xu, C.; Bian, C.; Lam, R.; Wang, J.P.; Kania, J.; Min, J.R.; Zang, J.Y. Crystal structures of the coil 2B fragment and the globular tail domain of human lamin B1. FEBS Lett 2012, 586, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Leterrier, C.; Dubey, P.; Roy, S. The nano-architecture of the axonal cytoskeleton. Nat. Rev. Neurosci. 2017, 18, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Laser-Azogui, A.; Kornreich, M.; Malka-Gibor, E.; Beck, R. Neurofilament assembly and function during neuronal development. Curr. Opin. Cell Biol. 2015, 32, 92–101. [Google Scholar] [CrossRef]
- Athlan, E.S.; Mushynski, W.E. Heterodimeric associations between neuronal intermediate filament proteins. J. Biol. Chem. 1997, 272, 31073–31078. [Google Scholar] [CrossRef]
- Garden, M.J.; Eagles, P.A. Chemical cross-linking analyses of ox neurofilaments. Biochem. J. 1986, 234, 587–591. [Google Scholar] [CrossRef]
- Stone, E.J.; Uchida, A.; Brown, A. Charcot–Marie–Tooth disease type 2E/1F mutant neurofilament proteins assemble into neurofilaments. Cytoskeleton 2019, 76, 423–439. [Google Scholar] [CrossRef] [PubMed]
- Adebola, A.A.; Gastri, T.D.; He, C.-Z.; Salvatierra, L.A.; Zhao, J.; Brown, K.; Lin, C.-S.; Worman, H.J.; Liem, R.K.H. Neurofilament light polypeptide gene N98S mutation in mice leads to neurofilament network abnormalities and a Charcot–Marie–Tooth type 2E phenotype. Hum. Mol. Genet. 2014, 24, 2163–2174. [Google Scholar] [CrossRef] [PubMed]
- Perez-Olle, R.; Jones, S.T.; Liem, R.K.H. Phenotypic analysis of neurofilament light gene mutations linked to Charcot–Marie–Tooth disease in cell culture models. Hum. Mol. Genet. 2004, 13, 2207–2220. [Google Scholar] [CrossRef] [PubMed]
- Jordanova, A.; De Jonghe, P.; Boerkoel, C.F.; Takashima, H.; De Vriendt, E.; Ceuterick, C.; Martin, J.-J.; Butler, I.J.; Mancias, P.; Papasozomenos, S.C.; et al. Mutations in the neurofilament light chain gene (NEFL) cause early onset severe Charcot–Marie–Tooth disease. Brain 2003, 126, 590–597. [Google Scholar] [CrossRef]
- Meier, M.; Padilla, G.P.; Herrmann, H.; Wedig, T.; Hergt, M.; Patel, T.R.; Stetefeld, J.; Aebi, U.; Burkhard, P. Vimentin coil 1A—A molecular switch involved in the initiation of filament elongation. J. Mol. Biol. 2009, 390, 245–261. [Google Scholar] [CrossRef]
- Lilina, A.V.; Leekens, S.; Hashim, H.M.; Vermeire, P.-J.; Harvey, J.N.; Strelkov, S.V. Stability profile of vimentin rod domain. Protein Sci. 2022, 31, e4505. [Google Scholar] [CrossRef]
- Brennich, M.E.; Vainio, U.; Wedig, T.; Bauch, S.; Herrmann, H.; Köster, S. Mutation-induced alterations of intra-filament subunit organization in vimentin filaments revealed by SAXS. Soft Matter 2019, 15, 1999–2008. [Google Scholar] [CrossRef]
- Nefedova, V.V.; Yampolskaya, D.S.; Kleymenov, S.Y.; Chebotareva, N.A.; Matyushenko, A.M.; Levitsky, D.I. Effect of neurodegenerative mutations in the NEFL gene on thermal denaturation of the neurofilament light chain protein. Biochemistry 2023, 88, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Strelkov, S.V.; Herrmann, H.; Geisler, N.; Lustig, A.; Ivaninskii, S.; Zimbelmann, R.; Burkhard, P.; Aebi, U. Divide-and-conquer crystallographic approach towards an atomic structure of intermediate filaments. J. Mol. Biol. 2001, 306, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Guzenko, D.; Chernyatina, A.A.; Strelkov, S.V. Crystallographic studies of intermediate filament proteins. Subcell. Biochem. 2017, 82, 151–170. [Google Scholar] [CrossRef] [PubMed]
- Nefedova, V.V.; Sudnitsyna, M.V.; Gusev, N.B. Interaction of small heat shock proteins with light component of neurofilaments (NFL). Cell Stress Chaperones 2017, 22, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.J.; Ramalli, S.G.; Wallace, B.A. DichroWeb, a website for calculating protein secondary structure from circular dichroism spectroscopic data. Protein Sci. 2022, 31, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Freire, E.; Biltonen, R.L. Statistical mechanical deconvolution of thermal transitions in macromolecules. I. Theory and application to homogeneous systems. Biopolymers 1978, 17, 463–479. [Google Scholar] [CrossRef]
- Safenkova, I.V.; Slutskaya, E.S.; Panferov, V.G.; Zherdev, A.V.; Dzantiev, B.B. Complex analysis of concentrated antibody-gold nanoparticle conjugates’ mixtures using asymmetric flow field-flow fractionation. J. Chromatogr. A 2016, 1477, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Strelkov, S.V.; Herrmann, H.; Aebi, U. Molecular architecture of intermediate filaments. Bioessays 2003, 25, 243–251. [Google Scholar] [CrossRef]
- Chernyatina, A.A.; Strelkov, S.V. Stabilization of vimentin coil2 fragment via an engineered disulfide. J. Struct. Biol. 2012, 177, 46–53. [Google Scholar] [CrossRef]
- Herrmann, H.; Aebi, U. Intermediate Filaments: Structure and Assembly. Cold Spring Harb. Perspect. Biol. 2016, 8, a018242. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Aebi, U. Intermediate filaments: Molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu. Rev. Biochem. 2004, 73, 749–789. [Google Scholar] [CrossRef] [PubMed]
- Burstein, E.A.; Vedenkina, N.S.; Ivkova, M.N. Fluorescence and the location of tryptophan residues in protein molecules. Photochem. Photobiol. 1973, 18, 263–279. [Google Scholar] [CrossRef] [PubMed]
- Geisler, N.; Heimburg, T.; Schunemann, J.; Weber, K. Peptides from the conserved ends of the rod domain of desmin disassemble intermediate filaments and reveal unexpected structural features: A circular dichroism, Fourier transform infrared, and electron microscopic study. J. Struct. Biol. 1993, 110, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Vermeire, P.-J.; Lilina, A.V.; Hashim, H.M.; Dlabolová, L.; Fiala, J.; Beelen, S.; Kukačka, Z.; Harvey, J.N.; Novák, P.; Strelkov, S.V. Molecular structure of soluble vimentin tetramers. Sci. Rep. 2023, 13, 8841. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Kim, S.; Jin, M.S. Crystal structure of the human glial fibrillary acidic protein 1B domain. Biochem. Biophys. Res. Commun. 2018, 503, 2899–2905. [Google Scholar] [CrossRef]
- Potschka, M.; Nave, R.; Weber, K.; Geisler, N. The two coiled coils in the isolated rod domain of the intermediate filament protein desmin are staggered A hydrodynamic analysis of tetramers and dimers. Eur. J. Biochem. 1990, 190, 503–508. [Google Scholar] [CrossRef]
- Ramm, B.; Stigler, J.; Hinczewski, M.; Thirumalai, D.; Herrmann, H.; Woehlke, G.; Rief, M. Sequence-resolved free energy profiles of stress-bearing vimentin intermediate filaments. Proc. Natl. Acad. Sci. USA 2014, 111, 11359–11364. [Google Scholar] [CrossRef]
- Premchandar, A.; Mücke, N.; Poznański, J.; Wedig, T.; Kaus-Drobek, M.; Herrmann, H.; Dadlez, M. Structural dynamics of the vimentin coiled-coil contact regions involved in filament assembly as revealed by hydrogen-deuterium exchange. J. Biol. Chem. 2016, 291, 24931–24950. [Google Scholar] [CrossRef]
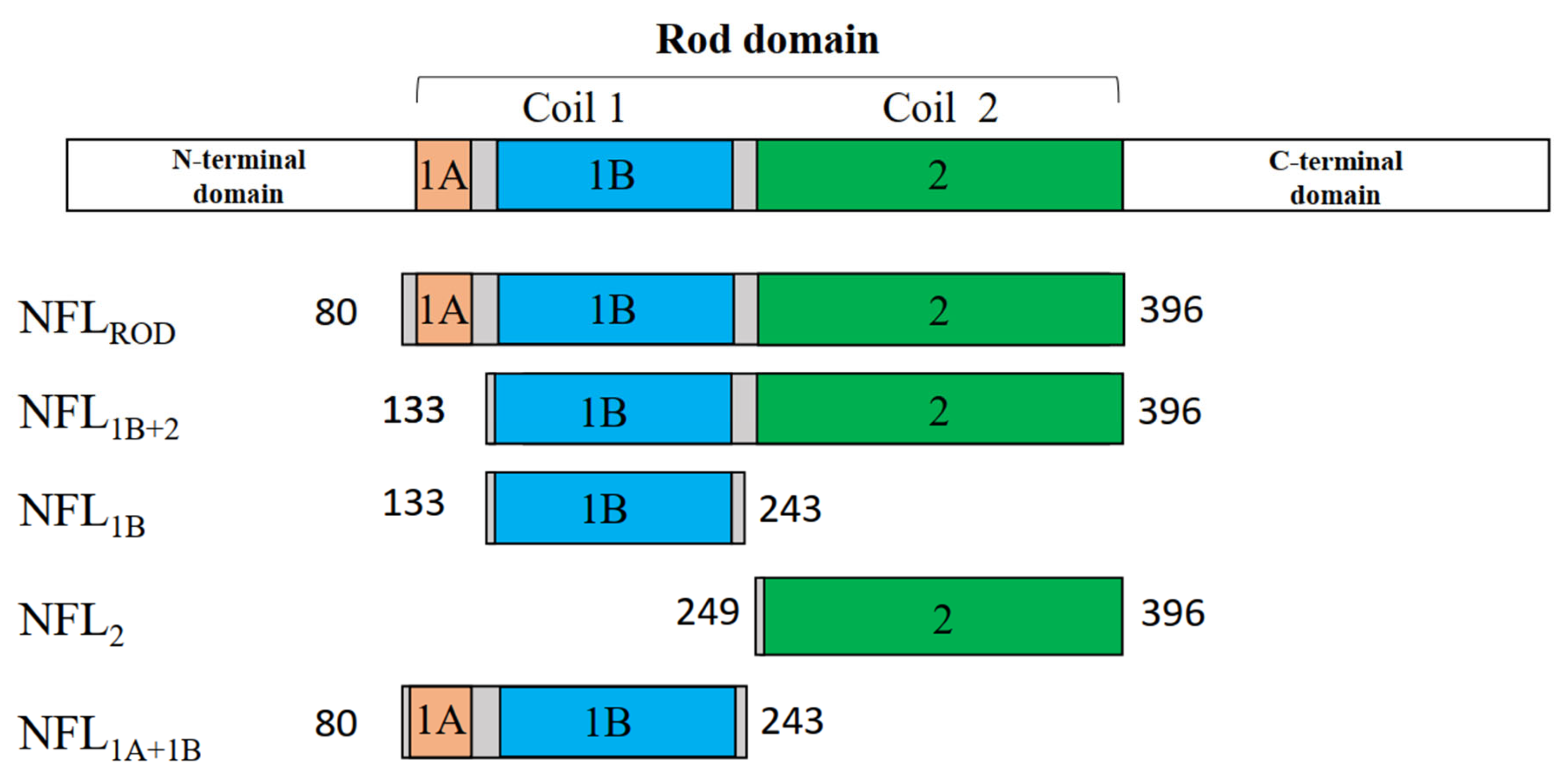


| Short Name of the Fragment | Name of the Structural Domain | Borders of the Truncated Fragment |
|---|---|---|
| NFL1B | Coil 1B | Ser133—Ile243 |
| NFL2 | Coil 2 | Val249—Glu396 |
| NFL1B+2 | Coils 1B and 2 | Ser133—Glu396 |
| NFL1A+1B | Coils 1A and 1B | Ser80—Ile243 |
| NFLROD | Coils 1A, 1B, and 2 | Ser80—Glu396 |
| Primer | Sequence |
|---|---|
| FW80 | 5′-ATATATATCATATGCATCACCATCACCATCACCTGGAAGTGCTGTTTCAGGGCCCGAGCAACGACCTCAAGTCCATCCG-3′ |
| FW133 | 5′-ATATATATCATATGCATCACCATCACCATCACCTGGAAGTGCTGTTTCAGGGCCCGTCCCGCTTCCGGGCGCT-3′ |
| FW249 | 5′-ATATATATCATATGCATCACCATCACCATCACCTGGAAGTGCTGTTTCAGGGCCCGGTGACCAAGCCCGACCTTTCC-3′ |
| REV243 | 5′-ATATATATGAATTCTTAGATCTGCGCGTACTGGATCTGCG-3′ |
| REV396 | 5′-ATATATATGAATTCTTACTCCCCCTCCAGCAGTTTACGG-3′ |
| Sample | Number of Domain | Tm # (°C) | ΔHcal § (kJ mol−1) | ΔHcal (% of Total ΔHcal) | Total ΔHcal (kJ mol−1) |
|---|---|---|---|---|---|
| NFL1B | Domain 1 | 39.4 | 165 | 100 | 165 |
| NFL1A+1B | Domain 1 | 41.9 | 80 | 100 | 80 |
| NFL1B+2 | Domain 1 Domain 2 Domain 3 | 37.8 41.5 59.6 | 30 195 190 | 7 47 46 | 415 |
| NFLROD | Domain 1 Domain 2 Domain 3 | 33.7 41.8 61.1 | 35 205 155 | 9 52 39 | 395 |
| NFLFL | Domain 1 Domain 2 Domain 3 Domain 4 | 33.2 41.7 53.6 63.2 | 15 245 215 115 | 3 42 36 19 | 590 |
| NFL2 | Domain 1 Domain 2 | 37.2 62.7 | ND | ND | 160 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nefedova, V.V.; Kleymenov, S.Y.; Safenkova, I.V.; Levitsky, D.I.; Matyushenko, A.M. Neurofilament Light Protein Rod Domain Exhibits Structural Heterogeneity. Biomolecules 2024, 14, 85. https://doi.org/10.3390/biom14010085
Nefedova VV, Kleymenov SY, Safenkova IV, Levitsky DI, Matyushenko AM. Neurofilament Light Protein Rod Domain Exhibits Structural Heterogeneity. Biomolecules. 2024; 14(1):85. https://doi.org/10.3390/biom14010085
Chicago/Turabian StyleNefedova, Victoria V., Sergey Y. Kleymenov, Irina V. Safenkova, Dmitrii I. Levitsky, and Alexander M. Matyushenko. 2024. "Neurofilament Light Protein Rod Domain Exhibits Structural Heterogeneity" Biomolecules 14, no. 1: 85. https://doi.org/10.3390/biom14010085
APA StyleNefedova, V. V., Kleymenov, S. Y., Safenkova, I. V., Levitsky, D. I., & Matyushenko, A. M. (2024). Neurofilament Light Protein Rod Domain Exhibits Structural Heterogeneity. Biomolecules, 14(1), 85. https://doi.org/10.3390/biom14010085






