Wiggle and Shake: Managing and Exploiting Conformational Dynamics during Proteasome Biogenesis
Abstract
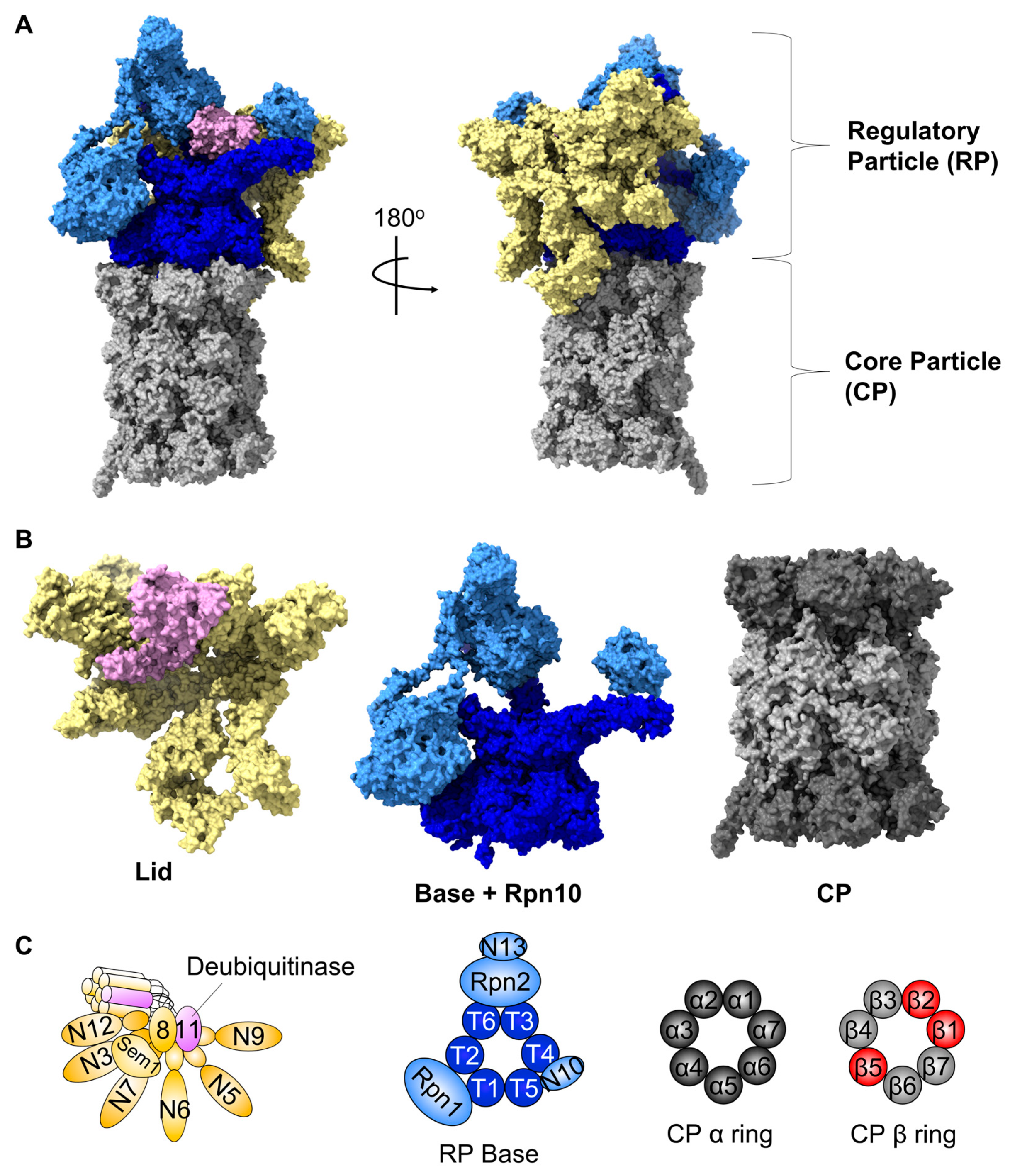
1. The 26S Proteasome: A Central Protease for Regulated Intracellular Protein Degradation
2. An Overview of Proteasome Biogenesis
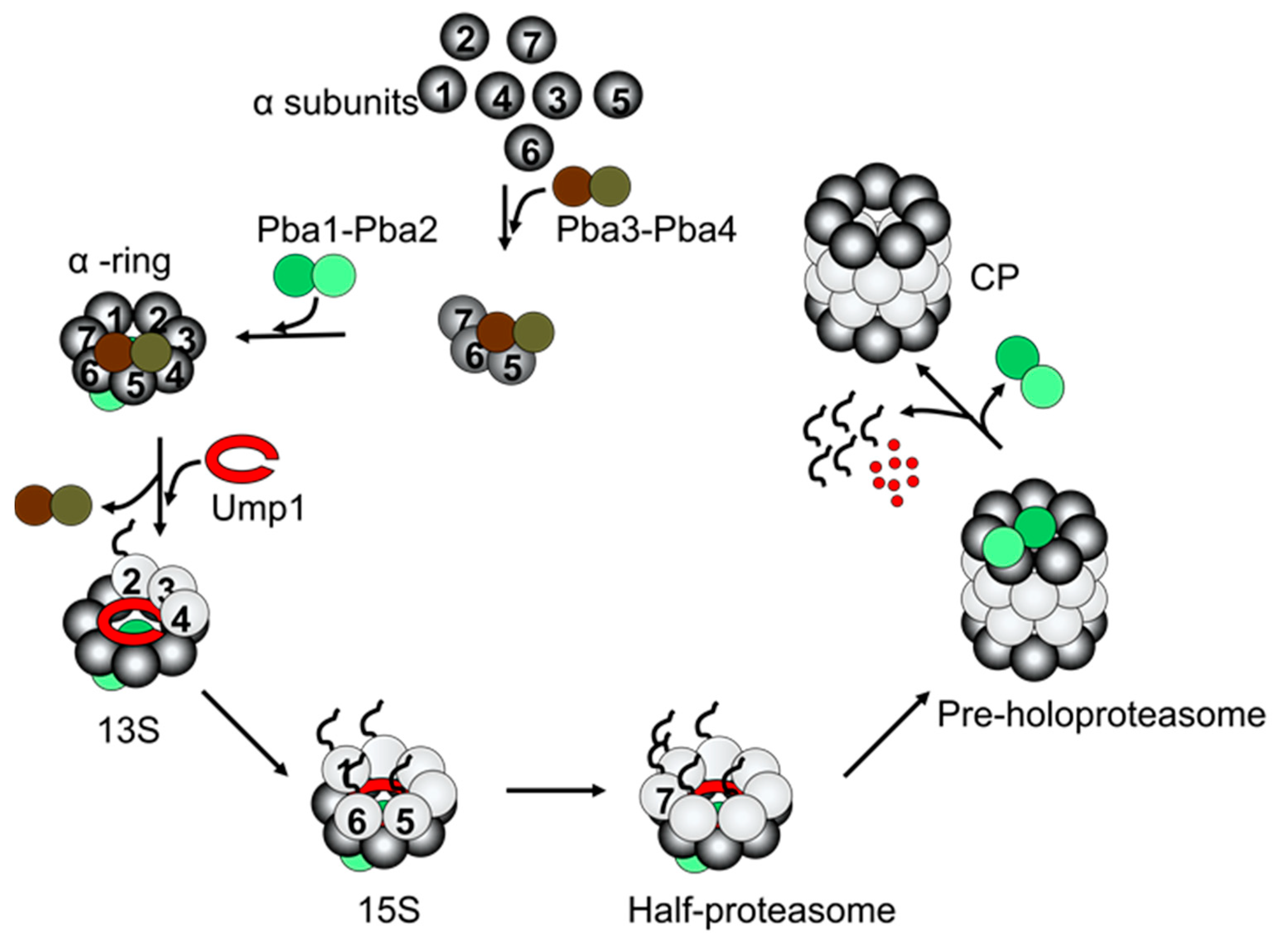
2.1. CP Assembly
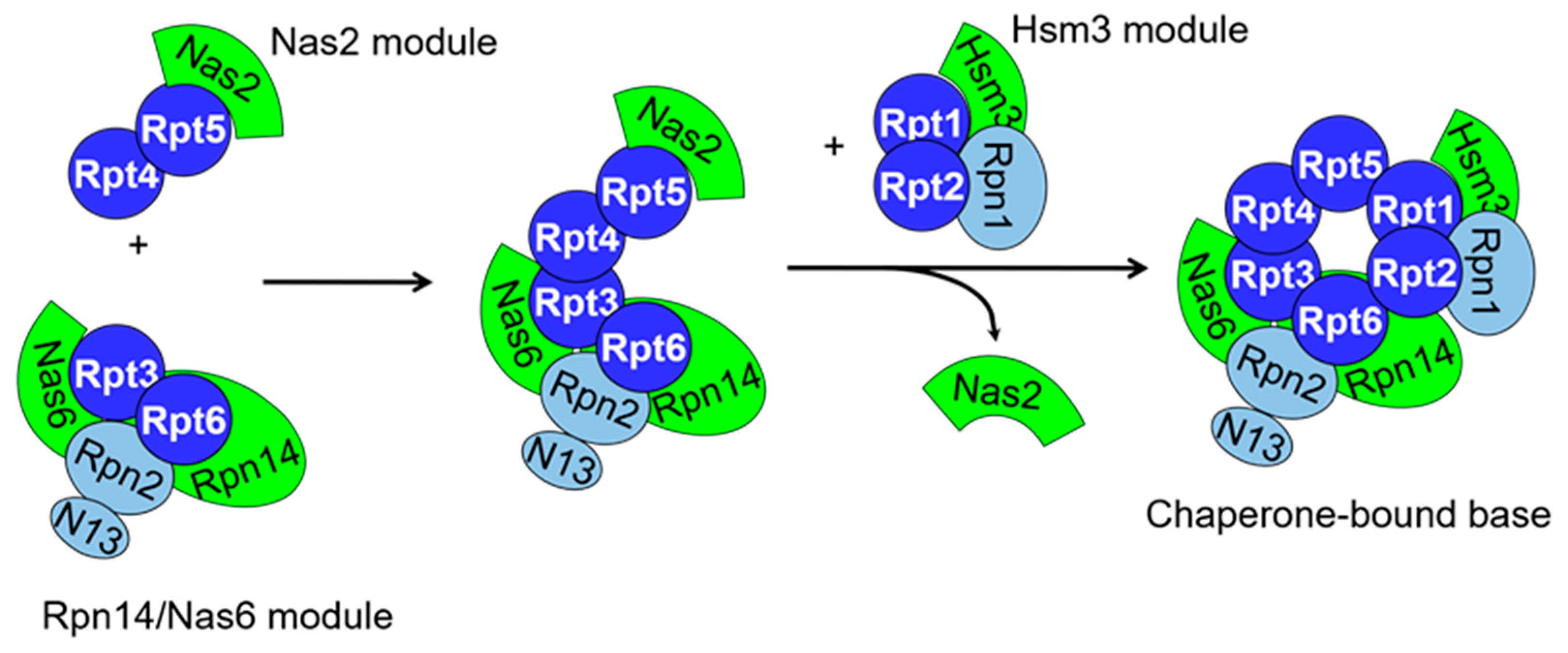
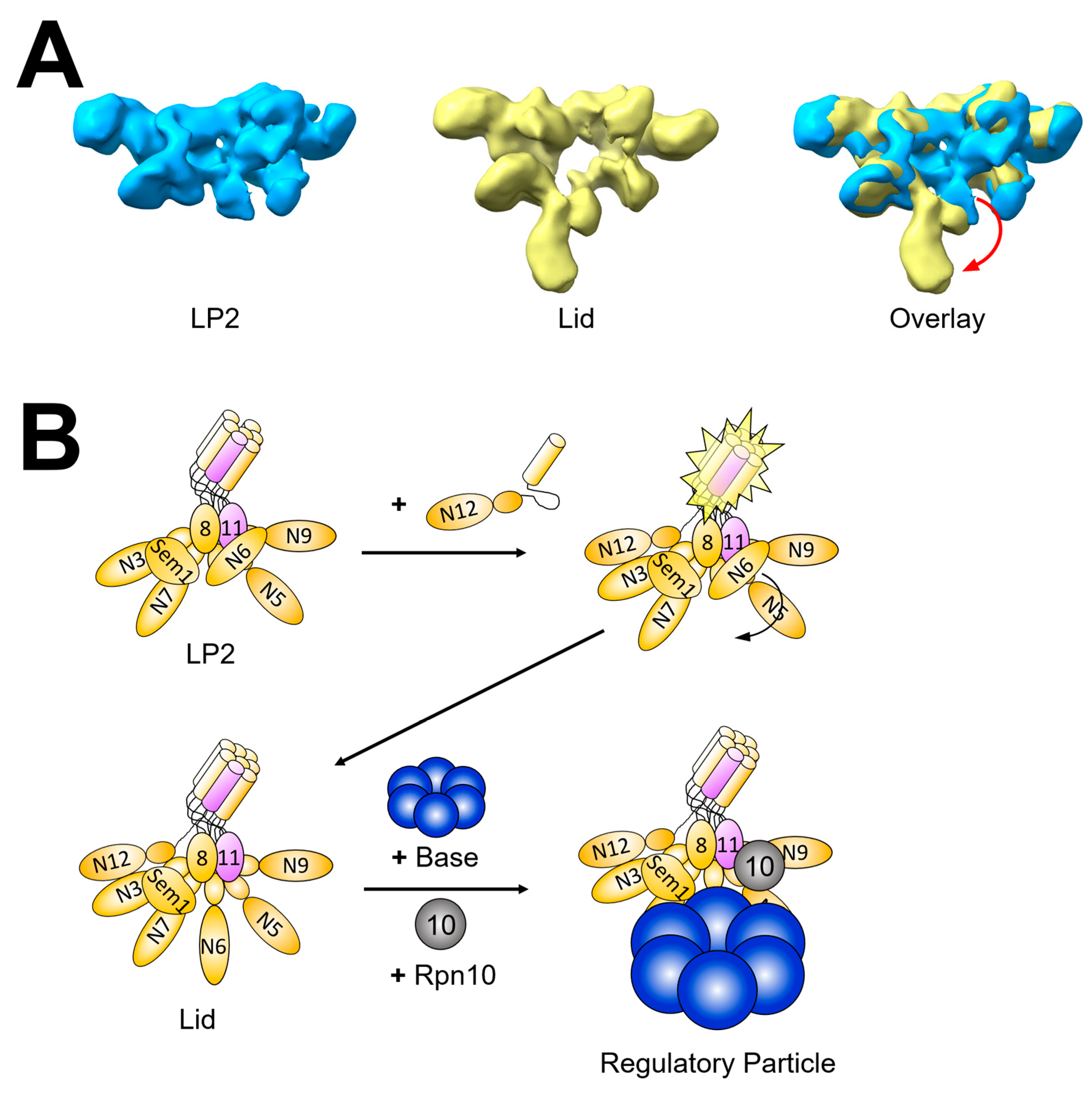
2.2. RP Assembly
3. Conformational Dynamics Provide Challenges and Opportunities during Proteasome Assembly
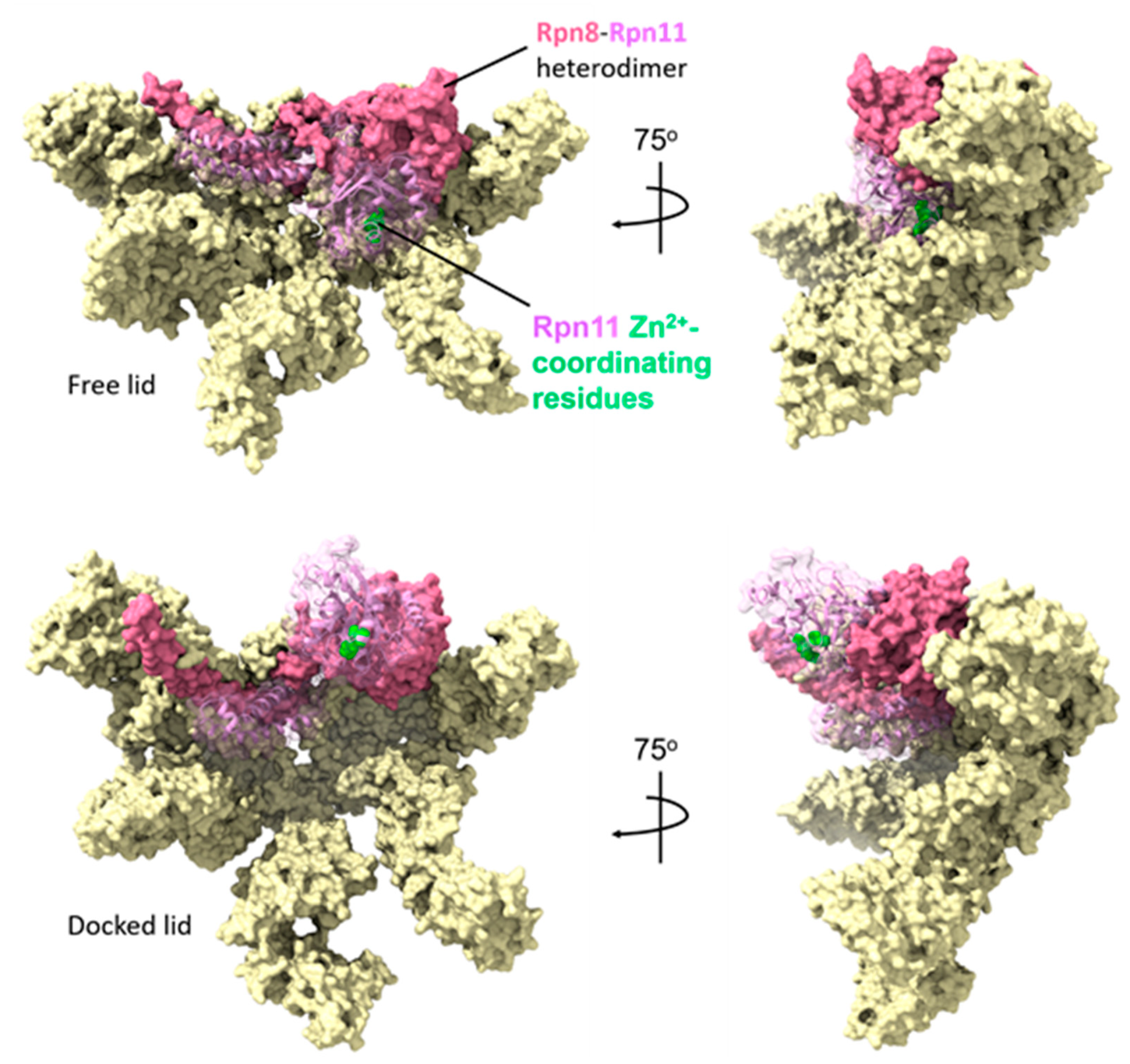
3.1. Conformational Matching as A Requirement for Association of Assembly Intermediates
3.2. Conformational Restriction of Catalytic Activity during Proteasome Assembly
3.3. Conformational Dynamics as a Means to Evict Assembly Chaperones from Nascent Complexes
3.4. Conformational Dynamics to Signal Dysfunctional Proteasomes
4. Summary and Perspective
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bard, J.A.M.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef]
- Howell, L.A.; Tomko, R.J.; Kusmierczyk, A.R. Putting it all together: Intrinsic and extrinsic mechanisms governing proteasome biogenesis. Front. Biol. 2017, 12, 19–48. [Google Scholar] [CrossRef]
- Glickman, M.H.; Rubin, D.M.; Coux, O.; Wefes, I.; Pfeifer, G.; Cjeka, Z.; Baumeister, W.; Fried, V.A.; Finley, D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 1998, 94, 615–623. [Google Scholar] [CrossRef]
- Verma, R.; Aravind, L.; Oania, R.; McDonald, W.H.; Yates, J.R., III; Koonin, E.V.; Deshaies, R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 2002, 298, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 2002, 419, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Tomko, R.J., Jr.; Funakoshi, M.; Schneider, K.; Wang, J.; Hochstrasser, M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: Implications for proteasome structure and assembly. Mol. Cell 2010, 38, 393–403. [Google Scholar] [CrossRef]
- Elsasser, S.; Chandler-Militello, D.; Müller, B.; Hanna, J.; Finley, D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 2004, 279, 26817–26822. [Google Scholar] [CrossRef] [PubMed]
- Husnjak, K.; Elsasser, S.; Zhang, N.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef]
- Schreiner, P.; Chen, X.; Husnjak, K.; Randles, L.; Zhang, N.; Elsasser, S.; Finley, D.; Dikic, I.; Walters, K.J.; Groll, M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 2008, 453, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Chen, X.; Elsasser, S.; Stocks, B.B.; Tian, G.; Lee, B.-H.; Shi, Y.; Zhang, N.; de Poot, S.A.H.; Tuebing, F.; et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 2016, 351, aad9421. [Google Scholar] [CrossRef]
- Waxman, L.; Fagan, J.M.; Goldberg, A.L. Demonstration of two distinct high molecular weight proteases in rabbit reticulocytes, one of which degrades ubiquitin conjugates. J. Biol. Chem. 1987, 262, 2451–2457. [Google Scholar] [CrossRef]
- Hough, R.; Pratt, G.; Rechsteiner, M. Ubiquitin-lysozyme conjugates. Identification and characterization of an ATP-dependent protease from rabbit reticulocyte lysates. J. Biol. Chem. 1986, 261, 2400–2408. [Google Scholar] [CrossRef]
- Budenholzer, L.; Cheng, C.L.; Li, Y.; Hochstrasser, M. Proteasome Structure and Assembly. J. Mol. Biol. 2017, 429, 3500–3524. [Google Scholar] [CrossRef]
- Marsh, J.A.; Hernández, H.; Hall, Z.; Ahnert, S.E.; Perica, T.; Robinson, C.V.; Teichmann, S.A. Protein complexes are under evolutionary selection to assemble via ordered pathways. Cell 2013, 153, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Saeki, Y.; Toh-e, A.; Kudo, T.; Kawamura, H.; Tanaka, K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell 2009, 137, 900–913. [Google Scholar] [CrossRef] [PubMed]
- Roelofs, J.; Park, S.; Haas, W.; Tian, G.; McAllister, F.E.; Huo, Y.; Lee, B.-H.; Zhang, F.; Shi, Y.; Gygi, S.P.; et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 2009, 459, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Roelofs, J.; Kim, W.; Robert, J.; Schmidt, M.; Gygi, S.P.; Finley, D. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature 2009, 459, 866–870. [Google Scholar] [CrossRef]
- Le Tallec, B.; Barrault, M.B.; Guerois, R.; Carre, T.; Peyroche, A. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol. Cell 2009, 33, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Hamazaki, J.; Iemura, S.-I.; Sasaki, K.; Furuyama, K.; Natsume, T.; Tanaka, K.; Murata, S. Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell 2009, 137, 914–925. [Google Scholar] [CrossRef]
- Funakoshi, M.; Tomko, R.J., Jr.; Kobayashi, H.; Hochstrasser, M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell 2009, 137, 887–899. [Google Scholar] [CrossRef]
- Kusmierczyk, A.R.; Kunjappu, M.J.; Funakoshi, M.; Hochstrasser, M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat. Struct. Mol. Biol. 2008, 15, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Le Tallec, B.; Barrault, M.-B.; Courbeyrette, R.; Guérois, R.; Marsolier-Kergoat, M.-C.; Peyroche, A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol. Cell 2007, 27, 660–674. [Google Scholar] [CrossRef] [PubMed]
- Hirano, Y.; Hayashi, H.; Iemura, S.-I.; Hendil, K.B.; Niwa, S.-I.; Kishimoto, T.; Kasahara, M.; Natsume, T.; Tanaka, K.; Murata, S. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol. Cell 2006, 24, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Hirano, Y.; Hendil, K.B.; Yashiroda, H.; Iemura, S.-I.; Nagane, R.; Hioki, Y.; Natsume, T.; Tanaka, K.; Murata, S. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature 2005, 437, 1381–1385. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Livnat-Levanon, N.; Kleifeld, O.; Mansour, W.; Nakasone, M.A.; Castaneda, C.A.; Dixon, E.K.; Fushman, D.; Reis, N.; Pick, E.; et al. Base-CP proteasome can serve as a platform for stepwise lid formation. Biosci. Rep. 2015, 35, e00194. [Google Scholar] [CrossRef]
- Thompson, D.; Hakala, K.; DeMartino, G.N. Subcomplexes of PA700, the 19 S regulator of the 26 S proteasome, reveal relative roles of AAA subunits in 26 S proteasome assembly and activation and ATPase activity. J. Biol. Chem. 2009, 284, 24891–24903. [Google Scholar] [CrossRef]
- Hendil, K.B.; Kriegenburg, F.; Tanaka, K.; Murata, S.; Lauridsen, A.-M.B.; Johnsen, A.H.; Hartmann-Petersen, R. The 20S proteasome as an assembly platform for the 19S regulatory complex. J. Mol. Biol. 2009, 394, 320–328. [Google Scholar] [CrossRef]
- Sharon, M.; Witt, S.; Glasmacher, E.; Baumeister, W.; Robinson, C.V. Mass spectrometry reveals the missing links in the assembly pathway of the bacterial 20 S proteasome. J. Biol. Chem. 2007, 282, 18448–18457. [Google Scholar] [CrossRef]
- Kwon, Y.D.; Nagy, I.; Adams, P.D.; Baumeister, W.; Jap, B.K. Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: Implications for the role of the pro-peptide in proteasome assembly. J. Mol. Biol. 2004, 335, 233–245. [Google Scholar] [CrossRef]
- Zühl, F.; Seemüller, E.; Golbik, R.; Baumeister, W. Dissecting the assembly pathway of the 20S proteasome. FEBS Lett. 1997, 418, 189–194. [Google Scholar] [CrossRef]
- Mayr, J.; Seemuller, E.; Muller, S.A.; Engel, A.; Baumeister, W. Late events in the assembly of 20S proteasomes. J. Struct. Biol. 1998, 124, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Zwickl, P.; Kleinz, J.; Baumeister, W. Critical elements in proteasome assembly. Nat. Struct. Biol. 1994, 1, 765–770. [Google Scholar] [CrossRef]
- Schmidtke, G.; Schmidt, M.; Kloetzel, P.M. Maturation of mammalian 20 S proteasome: Purification and characterization of 13 S and 16 S proteasome precursor complexes. J. Mol. Biol. 1997, 268, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Hirano, Y.; Kaneko, T.; Okamoto, K.; Bai, M.; Yashiroda, H.; Furuyama, K.; Kato, K.; Tanaka, K.; Murata, S. Dissecting β-ring assembly pathway of the mammalian 20S proteasome. EMBO J. 2008, 27, 2204–2213. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kusmierczyk, A.R.; Wong, P.; Emili, A.; Hochstrasser, M. β-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 2007, 26, 2339–2349. [Google Scholar] [CrossRef]
- Chen, P.; Hochstrasser, M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell 1996, 86, 961–972. [Google Scholar] [CrossRef]
- Takagi, K.; Saeki, Y.; Yashiroda, H.; Yagi, H.; Kaiho, A.; Murata, S.; Yamane, T.; Tanaka, K.; Mizushima, T.; Kato, K. Pba3–Pba4 heterodimer acts as a molecular matchmaker in proteasome α-ring formation. Biochem. Biophys. Res. Commun. 2014, 450, 1110–1114. [Google Scholar] [CrossRef]
- Gerards, W.L.; de Jong, W.W.; Bloemendal, H.; Boelens, W. The human proteasomal subunit HsC8 induces ring formation of other α-type subunits. J. Mol. Biol. 1998, 275, 113–121. [Google Scholar] [CrossRef]
- Gerards, W.L.; Enzlin, J.; Haner, M.; Hendriks, I.L.; Aebi, U.; Bloemendal, H.; Boelens, W. The human α-type proteasomal subunit HsC8 forms a double ringlike structure, but does not assemble into proteasome-like particles with the β-type subunits HsDelta or HsBPROS26. J. Biol. Chem. 1997, 272, 10080–10086. [Google Scholar] [CrossRef]
- Yashiroda, H.; Mizushima, T.; Okamoto, K.; Kameyama, T.; Hayashi, H.; Kishimoto, T.; Niwa, S.-I.; Kasahara, M.; Kurimoto, E.; Sakata, E.; et al. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat. Struct. Mol. Biol. 2008, 15, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Sahara, K.; Hirayama, S.; Zhao, X.; Watanabe, A.; Hamazaki, J.; Yashiroda, H.; Murata, S. PAC1-PAC2 proteasome assembly chaperone retains the core α4–α7 assembly intermediates in the cytoplasm. Genes Cells 2018, 23, 839–848. [Google Scholar] [CrossRef]
- Hammack, L.J.; Kusmierczyk, A.R. Assembly of proteasome subunits into non-canonical complexes in vivo. Biochem. Biophys. Res. Commun. 2017, 482, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Hammack, L.J.; Kusmierczyk, A.R. Data on the identity of non-canonical complexes formed from proteasome subunits in vivo. Data Brief 2016, 9, 1130–1137. [Google Scholar] [CrossRef]
- Schnell, H.M.; Walsh, R.M., Jr.; Rawson, S.; Kaur, M.; Bhanu, M.K.; Tian, G.; Prado, M.A.; Guerra-Moreno, A.; Paulo, J.A.; Gygi, S.P.; et al. Structures of chaperone-associated assembly intermediates reveal coordinated mechanisms of proteasome biogenesis. Nat. Struct. Mol. Biol. 2021, 28, 418–425. [Google Scholar] [CrossRef]
- Kock, M.; Nunes, M.M.; Hemann, M.; Kube, S.; Jürgen Dohmen, R.; Herzog, F.; Ramos, P.C.; Wendler, P. Proteasome assembly from 15S precursors involves major conformational changes and recycling of the Pba1–Pba2 chaperone. Nat. Commun. 2015, 6, 6123. [Google Scholar] [CrossRef]
- Padmanabhan, A.; Vuong, S.A.-T.; Hochstrasser, M. Assembly of an Evolutionarily Conserved Alternative Proteasome Isoform in Human Cells. Cell Rep. 2016, 14, 2962–2974. [Google Scholar] [CrossRef]
- Howell, L.A.; Peterson, A.K.; Tomko, R.J., Jr. Proteasome subunit α1 overexpression preferentially drives canonical proteasome biogenesis and enhances stress tolerance in yeast. Sci. Rep. 2019, 9, 12418. [Google Scholar] [CrossRef]
- Ramos, P.C.; Höckendorff, J.; Johnson, E.S.; Varshavsky, A.; Jürgen Dohmen, R. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell 1998, 92, 489–499. [Google Scholar] [CrossRef]
- Tomko, R.J., Jr.; Taylor, D.W.; Chen, Z.A.; Wang, H.-W.; Rappsilber, J.; Hochstrasser, M. A Single α Helix Drives Extensive Remodeling of the Proteasome Lid and Completion of Regulatory Particle Assembly. Cell 2015, 163, 432–444. [Google Scholar] [CrossRef]
- Tomko, R.J., Jr.; Hochstrasser, M. The intrinsically disordered Sem1 protein functions as a molecular tether during proteasome lid biogenesis. Mol. Cell 2014, 53, 433–443. [Google Scholar] [CrossRef]
- Tomko, R.J., Jr.; Hochstrasser, M. Incorporation of the Rpn12 subunit couples completion of proteasome regulatory particle lid assembly to lid-base joining. Mol. Cell 2011, 44, 907–917. [Google Scholar] [CrossRef]
- Estrin, E.; Lopez-Blanco, J.R.; Chacón, P.; Martin, A. Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. Structure 2013, 21, 1624–1635. [Google Scholar] [CrossRef]
- Fukunaga, K.; Kudo, T.; Toh-e, A.; Tanaka, K.; Saeki, Y. Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2010, 396, 1048–1053. [Google Scholar] [CrossRef]
- Sharon, M.; Taverner, T.; Ambroggio, X.I.; Deshaies, R.J.; Robinson, C.V. Structural organization of the 19S proteasome lid: Insights from MS of intact complexes. PLoS Biol. 2006, 4, e267. [Google Scholar] [CrossRef] [PubMed]
- Isono, E.; Saito, N.; Kamata, N.; Saeki, Y.; Toh-e, A. Functional analysis of Rpn6p, a lid component of the 26 S proteasome, using temperature-sensitive rpn6 mutants of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 6537–6547. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Zhao, X.; Sahara, K.; Ohte, Y.; Hirano, Y.; Kaneko, T.; Yashiroda, H.; Murata, S. In-depth Analysis of the Lid Subunits Assembly Mechanism in Mammals. Biomolecules 2019, 9, 213. [Google Scholar] [CrossRef] [PubMed]
- Matyskiela, M.E.; Lander, G.C.; Martin, A. Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 2013, 20, 781–788. [Google Scholar] [CrossRef]
- Śledź, P.; Unverdorben, P.; Beck, F.; Pfeifer, G.; Schweitzer, A.; Förster, F.; Baumeister, W. Structure of the 26S proteasome with ATP-γS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc. Natl. Acad. Sci. USA 2013, 110, 7264–7269. [Google Scholar] [CrossRef]
- Takeuchi, J.; Tamura, T. Recombinant ATPases of the yeast 26S proteasome activate protein degradation by the 20S proteasome. FEBS Lett. 2004, 565, 39–42. [Google Scholar] [CrossRef]
- Hanssum, A.; Zhong, Z.; Rousseau, A.; Krzyzosiak, A.; Sigurdardottir, A.; Bertolotti, A. An inducible chaperone adapts proteasome assembly to stress. Mol. Cell 2014, 55, 566–577. [Google Scholar] [CrossRef]
- Singh, K.; Graf, B.; Linden, A.; Sautner, V.; Urlaub, H.; Tittmann, K.; Stark, H.; Chari, A. Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase. Cell 2020, 180, 1130–1143.e20. [Google Scholar] [CrossRef]
- Barrault, M.-B.; Richet, N.; Godard, C.; Murciano, B.; Le Tallec, B.; Rousseau, E.; Legrand, P.; Charbonnier, J.-B.; Le Du, M.-H.; Guérois, R.; et al. Dual functions of the Hsm3 protein in chaperoning and scaffolding regulatory particle subunits during the proteasome assembly. Proc. Natl. Acad. Sci. USA 2012, 109, E1001–E1010. [Google Scholar] [CrossRef] [PubMed]
- Tomko, R.J., Jr.; Hochstrasser, M. Order of the proteasomal ATPases and eukaryotic proteasome assembly. Cell Biochem. Biophys. 2011, 60, 13–20. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Satoh, T.; Saeki, Y.; Hiromoto, T.; Wang, Y.-H.; Uekusa, Y.; Yagi, H.; Yoshihara, H.; Yagi-Utsumi, M.; Mizushima, T.; Tanaka, K.; et al. Structural basis for proteasome formation controlled by an assembly chaperone nas2. Structure 2014, 22, 731–743. [Google Scholar] [CrossRef]
- Lee, S.Y.-C.; De La Mota-Peynado, A.; Roelofs, J. Loss of Rpt5 protein interactions with the core particle and Nas2 protein causes the formation of faulty proteasomes that are inhibited by Ecm29 protein. J. Biol. Chem. 2011, 286, 36641–36651. [Google Scholar] [CrossRef] [PubMed]
- Rabl, J.; Smith, D.M.; Yu, Y.; Chang, S.-C.; Goldberg, A.L.; Cheng, Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 2008, 30, 360–368. [Google Scholar] [CrossRef]
- Smith, D.M.; Chang, S.-C.; Park, S.; Finley, D.; Cheng, Y.; Goldberg, A.L. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s α ring opens the gate for substrate entry. Mol. Cell 2007, 27, 731–744. [Google Scholar] [CrossRef]
- Park, S.; Li, X.; Kim, H.M.; Singh, C.R.; Tian, G.; Hoyt, M.A.; Lovell, S.; Battaile, K.P.; Zolkiewski, M.; Coffino, P.; et al. Reconfiguration of the proteasome during chaperone-mediated assembly. Nature 2013, 497, 512–516. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, S.; Wu, Z.; Li, X.; Wang, W.L.; Zhu, Y.; Stoilova-McPhie, S.; Lu, Y.; Finley, D.; Mao, Y. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 2019, 565, 49–55. [Google Scholar] [CrossRef]
- Ding, Z.; Fu, Z.; Xu, C.; Wang, Y.; Wang, Y.; Li, J.; Kong, L.; Chen, J.; Li, N.; Zhang, R.; et al. High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx. Cell Res. 2017, 27, 373–385. [Google Scholar] [CrossRef]
- Schweitzer, A.; Aufderheide, A.; Rudack, T.; Beck, F.; Pfeifer, G.; Plitzko, J.M.; Sakata, E.; Schulten, K.; Förster, F.; Baumeister, W. Structure of the human 26S proteasome at a resolution of 3.9 Å. Proc. Natl. Acad. Sci. USA 2016, 113, 7816–7821. [Google Scholar] [CrossRef]
- Huang, X.; Luan, B.; Wu, J.; Shi, Y. An atomic structure of the human 26S proteasome. Nat. Struct. Mol. Biol. 2016, 23, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Greene, E.R.; Goodall, E.A.; de la Peña, A.H.; Matyskiela, M.E.; Lander, G.C.; Martin, A. Specific lid-base contacts in the 26s proteasome control the conformational switching required for substrate degradation. Elife 2019, 8, e49806. [Google Scholar] [CrossRef] [PubMed]
- De la Peña, A.H.; Goodall, E.A.; Gates, S.N.; Lander, G.C.; Martin, A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 2018, 362, eaav0725. [Google Scholar] [CrossRef]
- Bashore, C.; Dambacher, C.M.; Goodall, E.A.; Matyskiela, M.E.; Lander, G.C.; Martin, A. Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat. Struct. Mol. Biol. 2015, 22, 712–719. [Google Scholar] [CrossRef]
- Eisele, M.R.; Reed, R.G.; Rudack, T.; Schweitzer, A.; Beck, F.; Nagy, I.; Pfeifer, G.; Plitzko, J.M.; Baumeister, W.; Tomko, R.J., Jr.; et al. Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating. Cell Rep. 2018, 24, 1301–1315.e5. [Google Scholar] [CrossRef]
- Wehmer, M.; Rudack, T.; Beck, F.; Aufderheide, A.; Pfeifer, G.; Plitzko, J.M.; Förster, F.; Schulten, K.; Baumeister, W.; Sakata, E. Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc. Natl. Acad. Sci. USA 2017, 114, 1305–1310. [Google Scholar] [CrossRef]
- Aufderheide, A.; Beck, F.; Stengel, F.; Hartwig, M.; Schweitzer, A.; Pfeifer, G.; Goldberg, A.L.; Sakata, E.; Baumeister, W.; Förster, F. Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc. Natl. Acad. Sci. USA 2015, 112, 8626–8631. [Google Scholar] [CrossRef]
- Unverdorben, P.; Beck, F.; Śledź, P.; Schweitzer, A.; Pfeifer, G.; Plitzko, J.M.; Baumeister, W.; Förster, F. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc. Natl. Acad. Sci. USA 2014, 111, 5544–5549. [Google Scholar] [CrossRef]
- Fisette, O.; Lagüe, P.; Gagné, S.; Morin, S. Synergistic applications of MD and NMR for the study of biological systems. J. Biomed. Biotechnol. 2012, 2012, 254208. [Google Scholar] [CrossRef]
- Bard, J.A.M.; Bashore, C.; Dong, K.C.; Martin, A. The 26S Proteasome Utilizes a Kinetic Gateway to Prioritize Substrate Degradation. Cell 2019, 177, 286–298.e15. [Google Scholar] [CrossRef]
- Jonsson, E.; Htet, Z.M.; Bard, J.A.M.; Dong, K.C.; Martin, A. Ubiquitin modulates 26S proteasome conformational dynamics and promotes substrate degradation. Sci. Adv. 2022, 8, eadd9520. [Google Scholar] [CrossRef]
- Dambacher, C.M.; Worden, E.J.; Herzik, M.A.; Martin, A.; Lander, G.C. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. Elife 2016, 5, e13027. [Google Scholar] [CrossRef]
- Nakamura, Y.; Umehara, T.; Tanaka, A.; Horikoshi, M.; Padmanabhan, B.; Yokoyama, S. Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26S proteasome. Biochem. Biophys. Res. Commun. 2007, 359, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Wendler, P.; Ciniawsky, S.; Kock, M.; Kube, S. Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta 2012, 1823, 2–14. [Google Scholar] [CrossRef]
- Li, F.; Tian, G.; Langager, D.; Sokolova, V.; Finley, D.; Park, S. Nucleotide-dependent switch in proteasome assembly mediated by the Nas6 chaperone. Proc. Natl. Acad. Sci. USA 2017, 114, 1548–1553. [Google Scholar] [CrossRef]
- Nemec, A.A.; Peterson, A.K.; Warnock, J.L.; Reed, R.G.; Tomko, R.J., Jr. An Allosteric Interaction Network Promotes Conformation State-Dependent Eviction of the Nas6 Assembly Chaperone from Nascent 26S Proteasomes. Cell Rep. 2019, 26, 483–495.e5. [Google Scholar] [CrossRef] [PubMed]
- Worden, E.J.; Dong, K.C.; Martin, A. An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome. Mol. Cell 2017, 67, 799–811.e8. [Google Scholar] [CrossRef] [PubMed]
- Worden, E.J.; Padovani, C.; Martin, A. Structure of the Rpn11–Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat. Struct. Mol. Biol. 2014, 21, 220–227. [Google Scholar] [CrossRef]
- Pathare, G.R.; Nagy, I.; Śledź, P.; Anderson, D.J.; Zhou, H.-J.; Pardon, E.; Steyaert, J.; Förster, F.; Bracher, A.; Baumeister, W. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc. Natl. Acad. Sci. USA 2014, 111, 2984–2989. [Google Scholar] [CrossRef]
- Nahar, A.; Sokolova, V.; Sekaran, S.; Orth, J.D.; Park, S. Assembly checkpoint of the proteasome regulatory particle is activated by coordinated actions of proteasomal ATPase chaperones. Cell Rep. 2022, 39, 110918. [Google Scholar] [CrossRef] [PubMed]
- Beckwith, R.; Estrin, E.; Worden, E.J.; Martin, A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nat. Struct. Mol. Biol. 2013, 20, 1164–1172. [Google Scholar] [CrossRef] [PubMed]
- Ehlinger, A.; Park, S.; Fahmy, A.; Lary, J.W.; Cole, J.L.; Finley, D.; Walters, K.J. Conformational dynamics of the Rpt6 ATPase in proteasome assembly and Rpn14 binding. Structure 2013, 21, 753–765. [Google Scholar] [CrossRef]
- Kim, S.; Nishide, A.; Saeki, Y.; Takagi, K.; Tanaka, K.; Kato, K.; Mizushima, T. New crystal structure of the proteasome-dedicated chaperone Rpn14 at 1.6 Å resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68, 517–521. [Google Scholar] [CrossRef]
- Kim, S.; Saeki, Y.; Fukunaga, K.; Suzuki, A.; Takagi, K.; Yamane, T.; Tanaka, K.; Mizushima, T.; Kato, K. Crystal structure of yeast rpn14, a chaperone of the 19 S regulatory particle of the proteasome. J. Biol. Chem. 2010, 285, 15159–15166. [Google Scholar] [CrossRef] [PubMed]
- Takagi, K.; Kim, S.; Yukii, H.; Ueno, M.; Morishita, R.; Endo, Y.; Kato, K.; Tanaka, K.; Saeki, Y.; Mizushima, T. Structural basis for specific recognition of Rpt1p, an ATPase subunit of 26 S proteasome, by proteasome-dedicated chaperone Hsm3p. J. Biol. Chem. 2012, 287, 12172–12182. [Google Scholar] [CrossRef]
- Wani, P.S.; Rowland, M.A.; Ondracek, A.; Deeds, E.J.; Roelofs, J. Maturation of the proteasome core particle induces an affinity switch that controls regulatory particle association. Nat. Commun. 2015, 6, 6384. [Google Scholar] [CrossRef]
- Stadtmueller, B.M.; Kish-Trier, E.; Ferrell, K.; Petersen, C.N.; Robinson, H.; Myszka, D.G.; Eckert, D.M.; Formosa, T.; Hill, C.P. Structure of a Proteasome Pba1-Pba2 Complex: Implications for Proteasome Assembly, Activation, and Biological Function. J. Biol. Chem. 2012, 287, 37371–37382. [Google Scholar] [CrossRef]
- Lu, Y.; Wu, J.; Dong, Y.; Chen, S.; Sun, S.; Ma, Y.-B.; Ouyang, Q.; Finley, D.; Kirschner, M.W.; Mao, Y. Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle. Mol. Cell 2017, 67, 322–333.e6. [Google Scholar] [CrossRef]
- Warnock, J.L.; Jobin, G.W.; Kumar, S.; Tomko, R.J., Jr. Assembly chaperone Nas6 selectively destabilizes 26S proteasomes with defective regulatory particle-core particle interfaces. J. Biol. Chem. 2023, 299, 102894. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, W.L.; Yu, D.; Ouyang, Q.; Lu, Y.; Mao, Y. Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome. Nat. Commun. 2018, 9, 1360. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Betancourt, D.; Lawal, T.; Tomko, R.J., Jr. Wiggle and Shake: Managing and Exploiting Conformational Dynamics during Proteasome Biogenesis. Biomolecules 2023, 13, 1223. https://doi.org/10.3390/biom13081223
Betancourt D, Lawal T, Tomko RJ Jr. Wiggle and Shake: Managing and Exploiting Conformational Dynamics during Proteasome Biogenesis. Biomolecules. 2023; 13(8):1223. https://doi.org/10.3390/biom13081223
Chicago/Turabian StyleBetancourt, Daniel, Tomiwa Lawal, and Robert J. Tomko, Jr. 2023. "Wiggle and Shake: Managing and Exploiting Conformational Dynamics during Proteasome Biogenesis" Biomolecules 13, no. 8: 1223. https://doi.org/10.3390/biom13081223
APA StyleBetancourt, D., Lawal, T., & Tomko, R. J., Jr. (2023). Wiggle and Shake: Managing and Exploiting Conformational Dynamics during Proteasome Biogenesis. Biomolecules, 13(8), 1223. https://doi.org/10.3390/biom13081223






