CXCL10 Is Associated with Increased Cerebrospinal Fluid Immune Cell Infiltration and Disease Duration in Multiple Sclerosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient and Demographic Information
2.2. Preparation of Human CSF Samples
2.3. Preparation of Human Blood Samples
2.4. Cytokine Measurements
2.5. CSF Cell Flow Cytometry
2.6. PBMC Flow Cytometry
2.7. RNA Isolation and qPCR Analyses
2.8. Analysis
3. Results
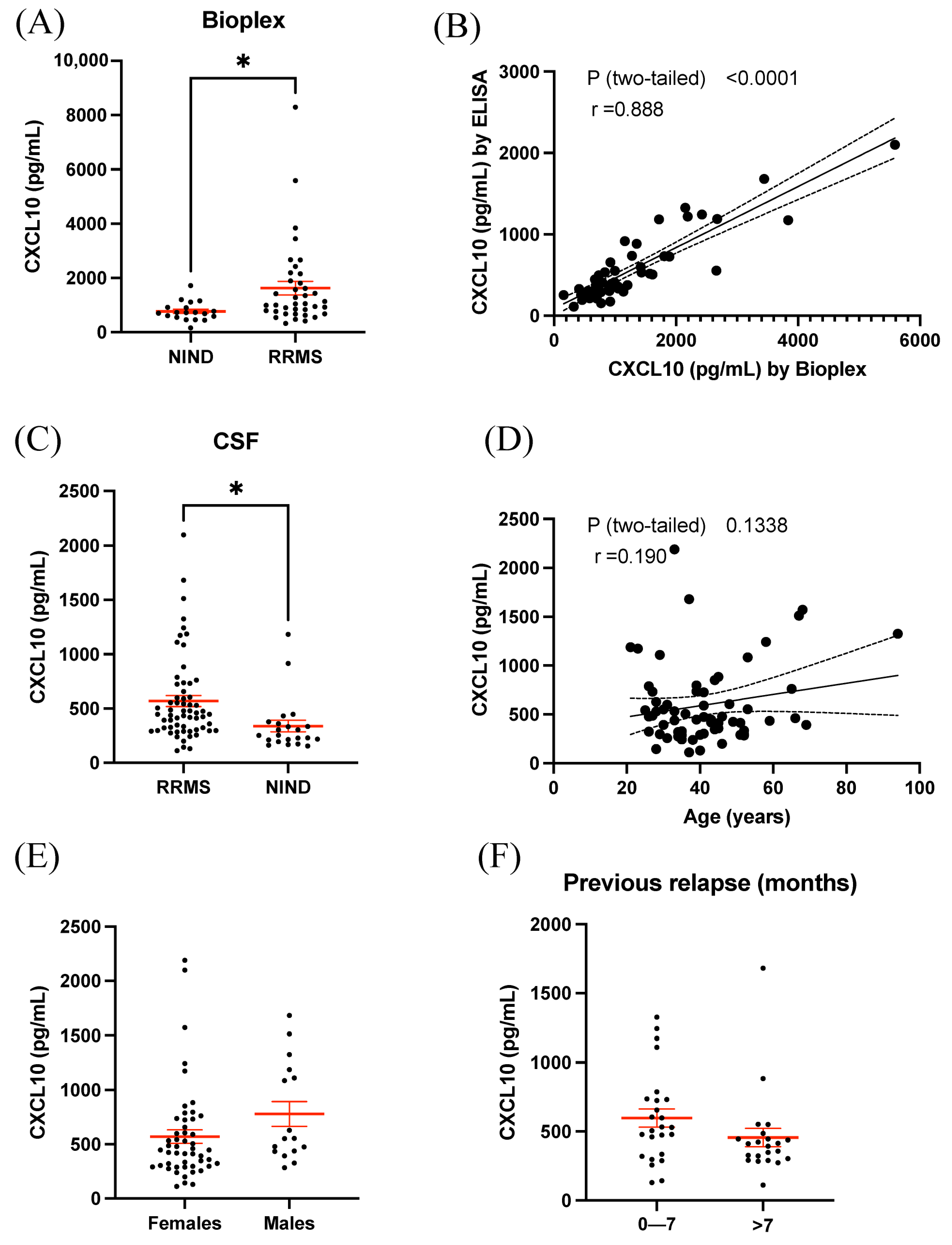
3.1. BioPlex Analysis of CSF Revealed Increased Expression of CXCL10 in RRMS
3.2. ELISA Results Validated Elevated CXCL10 Expression in CSF of RRMS
3.3. Plasma CXCL10 Is Not Increased in RRMS Compared to Controls but Is Associated with Overall Disease Duration
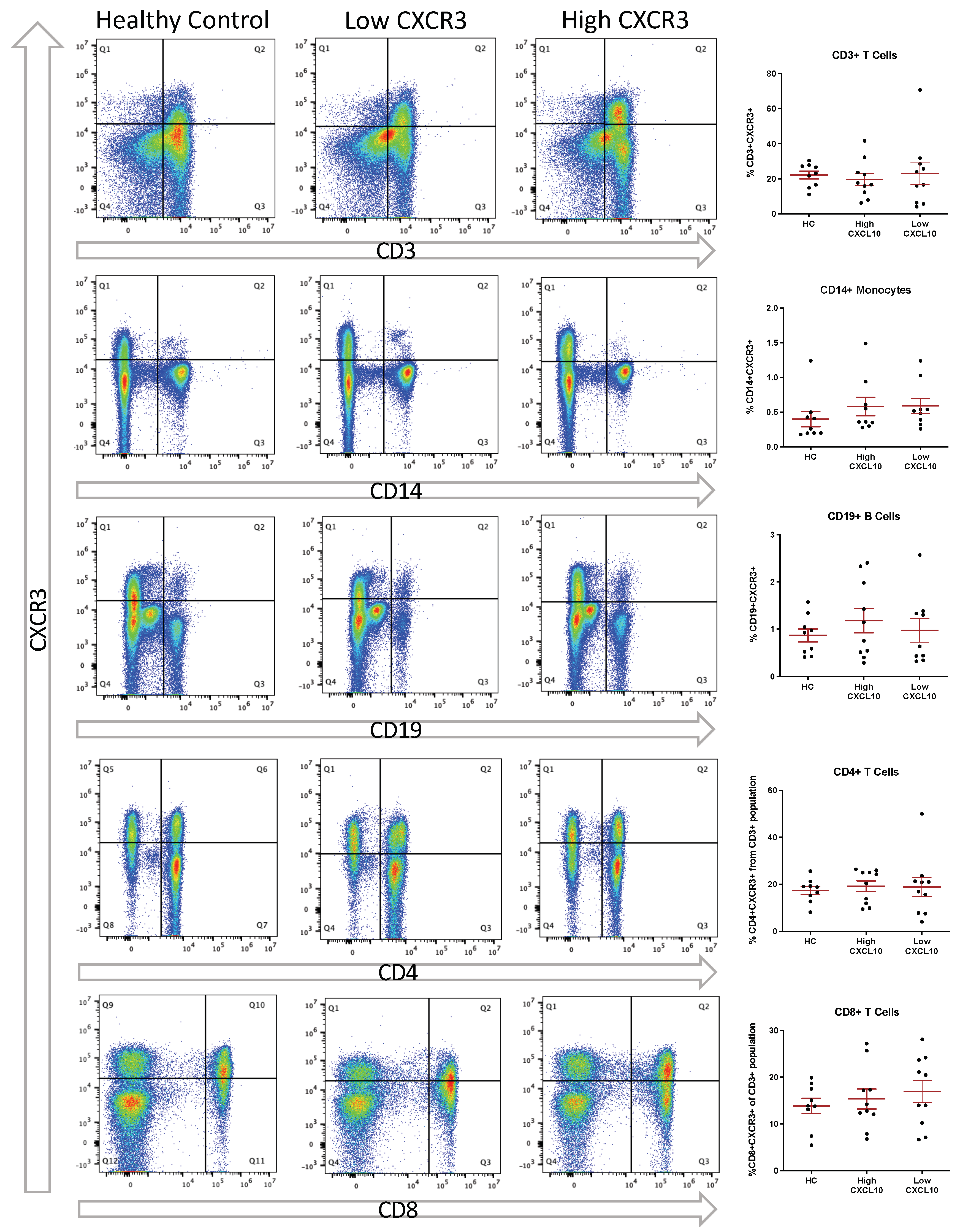
3.4. Elevated CSF CXCL10 Is Associated with Increased T Cell Presence within the CSF
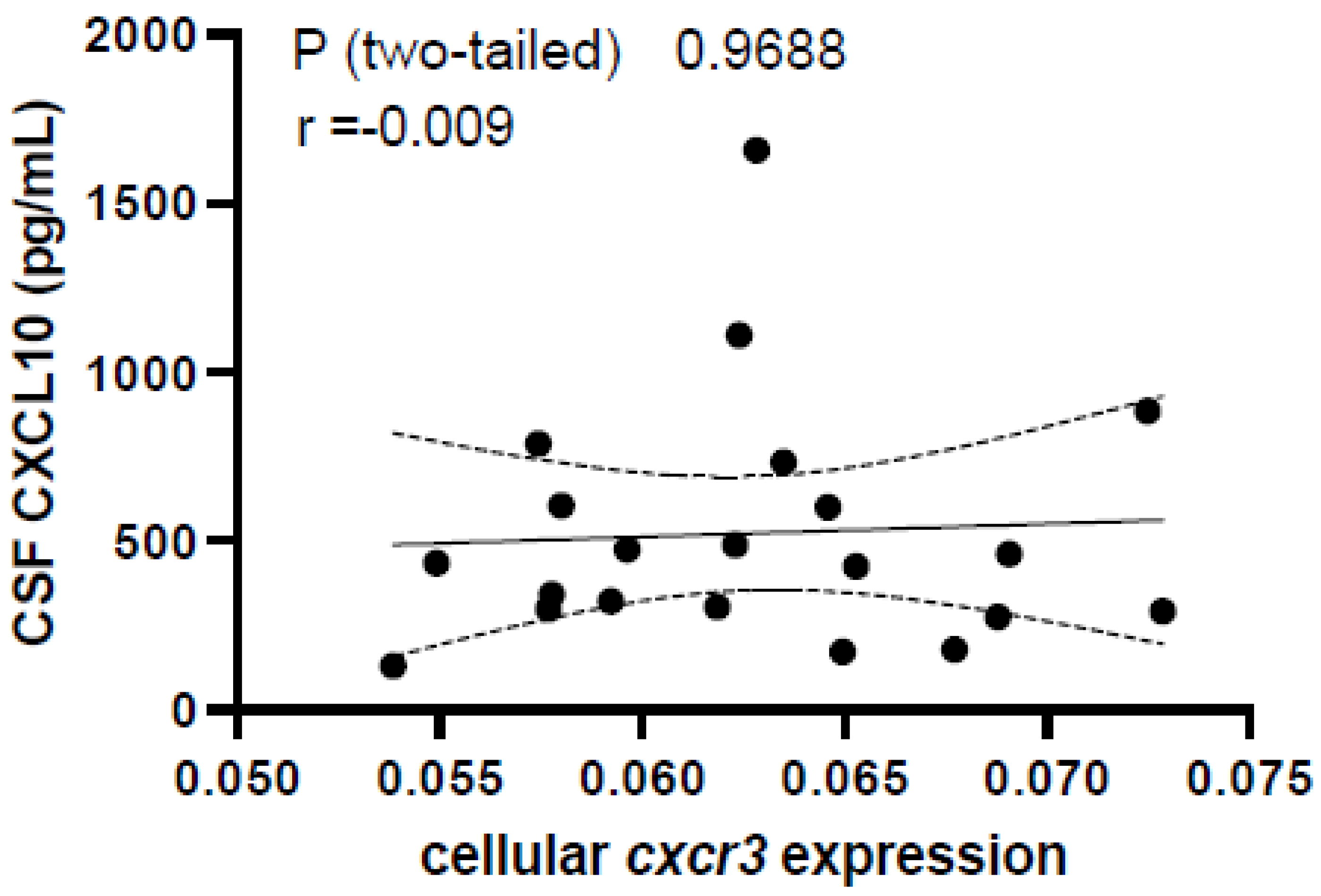
3.5. Peripheral Expression of CXCR3 Does Not Differ between HC and RRMS and Is Not Associated with CSF CXCL10 Levels
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Croese, T.; Castellani, G.; Schwartz, M. Immune cell compartmentalization for brain surveillance and protection. Nat. Immunol. 2021, 22, 1083–1092. [Google Scholar] [CrossRef]
- de Graaf, M.T.; Smitt, P.A.; Luitwieler, R.L.; van Velzen, C.; van den Broek, P.D.; Kraan, J.; Gratama, J.W. Central memory CD4+ T cells dominate the normal cerebrospinal fluid. Cytom. B Clin. Cytom. 2011, 80, 43–50. [Google Scholar] [CrossRef]
- Han, S.; Lin, Y.C.; Wu, T.; Salgado, A.D.; Mexhitaj, I.; Wuest, S.C.; Romm, E.; Ohayon, J.; Goldbach-Mansky, R.; Vanderver, A.; et al. Comprehensive immunophenotyping of cerebrospinal fluid cells in patients with neuroimmunological diseases. J. Immunol. 2014, 192, 2551–2563. [Google Scholar] [CrossRef]
- Schafflick, D.; Xu, C.A.; Hartlehnert, M.; Cole, M.; Schulte-Mecklenbeck, A.; Lautwein, T.; Wolbert, J.; Heming, M.; Meuth, S.G.; Kuhlmann, T.; et al. Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis. Nat. Commun. 2020, 11, 247. [Google Scholar] [CrossRef]
- Esaulova, E.; Cantoni, C.; Shchukina, I.; Zaitsev, K.; Bucelli, R.C.; Wu, G.F.; Artyomov, M.N.; Cross, A.H.; Edelson, B.T. Single-cell RNA-seq analysis of human CSF microglia and myeloid cells in neuroinflammation. Neurol. Neuroimmunol. Neuroinflamm. 2020, 7. [Google Scholar] [CrossRef]
- Ramesh, A.; Schubert, R.D.; Greenfield, A.L.; Dandekar, R.; Loudermilk, R.; Sabatino, J.J., Jr.; Koelzer, M.T.; Tran, E.B.; Koshal, K.; Kim, K.; et al. A pathogenic and clonally expanded B cell transcriptome in active multiple sclerosis. Proc. Natl. Acad. Sci. USA 2020, 117, 22932–22943. [Google Scholar] [CrossRef]
- Khaibullin, T.; Ivanova, V.; Martynova, E.; Cherepnev, G.; Khabirov, F.; Granatov, E.; Rizvanov, A.; Khaiboullina, S. Elevated Levels of Proinflammatory Cytokines in Cerebrospinal Fluid of Multiple Sclerosis Patients. Front. Immunol. 2017, 8, 531. [Google Scholar] [CrossRef]
- Maimone, D.; Gregory, S.; Arnason, B.G.; Reder, A.T. Cytokine levels in the cerebrospinal fluid and serum of patients with multiple sclerosis. J. Neuroimmunol. 1991, 32, 67–74. [Google Scholar] [CrossRef]
- Rodríguez-Sáinz Mdel, C.; Sánchez-Ramón, S.; de Andrés, C.; Rodríguez-Mahou, M.; Muñoz-Fernández, M.A. Th1/Th2 cytokine balance and nitric oxide in cerebrospinal fluid and serum from patients with multiple sclerosis. Eur. Cytokine Netw. 2002, 13, 110–114. [Google Scholar]
- Hauser, S.L.; Doolittle, T.H.; Lincoln, R.; Brown, R.H.; Dinarello, C.A. Cytokine accumulations in CSF of multiple sclerosis patients: Frequent detection of interleukin-1 and tumor necrosis factor but not interleukin-6. Neurology 1990, 40, 1735–1739. [Google Scholar] [CrossRef]
- Burman, J.; Svensson, E.; Fransson, M.; Loskog, A.S.; Zetterberg, H.; Raininko, R.; Svenningsson, A.; Fagius, J.; Mangsbo, S.M. The cerebrospinal fluid cytokine signature of multiple sclerosis: A homogenous response that does not conform to the Th1/Th2/Th17 convention. J. Neuroimmunol. 2014, 277, 153–159. [Google Scholar] [CrossRef]
- Kothur, K.; Wienholt, L.; Brilot, F.; Dale, R.C. CSF cytokines/chemokines as biomarkers in neuroinflammatory CNS disorders: A systematic review. Cytokine 2016, 77, 227–237. [Google Scholar] [CrossRef]
- Vazirinejad, R.; Ahmadi, Z.; Kazemi Arababadi, M.; Hassanshahi, G.; Kennedy, D. The Biological Functions, Structure and Sources of CXCL10 and Its Outstanding Part in the Pathophysiology of Multiple Sclerosis. Neuroimmunomodulation 2014, 21, 322–330. [Google Scholar] [CrossRef]
- Balashov, K.E.; Rottman, J.B.; Weiner, H.L.; Hancock, W.W. CCR5(+) and CXCR3(+) T cells are increased in multiple sclerosis and their ligands MIP-1alpha and IP-10 are expressed in demyelinating brain lesions. Proc. Natl. Acad. Sci. USA 1999, 96, 6873–6878. [Google Scholar] [CrossRef]
- Mahad, D.J.; Howell, S.J.L.; Woodroofe, M.N. Expression of chemokines in the CSF and correlation with clinical disease activity in patients with multiple sclerosis. J. Neurol. Neurosurg. Psychiatry 2002, 72, 498. [Google Scholar] [CrossRef]
- Roberts, W.K.; Blachère, N.E.; Frank, M.O.; Dousmanis, A.; Ransohoff, R.M.; Darnell, R.B. A destructive feedback loop mediated by CXCL 10 in central nervous system inflammatory disease. Ann. Neurol. 2015, 78, 619–629. [Google Scholar] [CrossRef]
- Scarpini, E.; Galimberti, D.; Baron, P.; Clerici, R.; Ronzoni, M.; Conti, G.; Scarlato, G. IP-10 and MCP-1 levels in CSF and serum from multiple sclerosis patients with different clinical subtypes of the disease. J. Neurol. Sci. 2002, 195, 41–46. [Google Scholar] [CrossRef]
- Simpson, J.E.; Newcombe, J.; Cuzner, M.L.; Woodroofe, M.N. Expression of the interferon-gamma-inducible chemokines IP-10 and Mig and their receptor, CXCR3, in multiple sclerosis lesions. Neuropathol. Appl. Neurobiol. 2000, 26, 133–142. [Google Scholar] [CrossRef]
- Sorensen, T.L.; Sellebjerg, F.; Jensen, C.V.; Strieter, R.M.; Ransohoff, R.M. Chemokines CXCL10 and CCL2: Differential involvement in intrathecal inflammation in multiple sclerosis. Eur. J. Neurol. 2001, 8, 665–672. [Google Scholar] [CrossRef]
- Moore, C.S.; Rao, V.T.; Durafourt, B.A.; Bedell, B.J.; Ludwin, S.K.; Bar-Or, A.; Antel, J.P. miR-155 as a multiple sclerosis-relevant regulator of myeloid cell polarization. Ann. Neurol. 2013, 74, 709–720. [Google Scholar] [CrossRef]
- Iwanowski, P.; Losy, J.; Kramer, L.; Wójcicka, M.; Kaufman, E. CXCL10 and CXCL13 chemokines in patients with relapsing remitting and primary progressive multiple sclerosis. J. Neurol. Sci. 2017, 380, 22–26. [Google Scholar] [CrossRef]
- Franciotta, D.; Martino, G.; Zardini, E.; Furlan, R.; Bergamaschi, R.; Andreoni, L.; Cosi, V. Serum and CSF levels of MCP-1 and IP-10 in multiple sclerosis patients with acute and stable disease and undergoing immunomodulatory therapies. J. Neuroimmunol. 2001, 115, 192–198. [Google Scholar] [CrossRef]
- Teleshova, N.; Pashenkov, M.; Huang, Y.M.; Söderström, M.; Kivisäkk, P.; Kostulas, V.; Haglund, M.; Link, H. Multiple sclerosis and optic neuritis: CCR5 and CXCR3 expressing T cells are augmented in blood and cerebrospinal fluid. J. Neurol. 2002, 249, 723–729. [Google Scholar] [CrossRef]
- Sørensen, T.L.; Trebst, C.; Kivisäkk, P.; Klaege, K.L.; Majmudar, A.; Ravid, R.; Lassmann, H.; Olsen, D.B.; Strieter, R.M.; Ransohoff, R.M.; et al. Multiple sclerosis: A study of CXCL10 and CXCR3 co-localization in the inflamed central nervous system. J. Neuroimmunol. 2002, 127, 59–68. [Google Scholar] [CrossRef]
- Matsui, M.; Araya, S.; Wang, H.Y.; Matsushima, K.; Saida, T. Differences in systemic and central nervous system cellular immunity relevant to relapsing-remitting multiple sclerosis. J. Neurol. 2005, 252, 908–915. [Google Scholar] [CrossRef]
- Takeshita, Y.; Ransohoff, R.M. Inflammatory cell trafficking across the blood-brain barrier: Chemokine regulation and in vitro models. Immunol. Rev. 2012, 248, 228–239. [Google Scholar] [CrossRef]
- Metzemaekers, M.; Vanheule, V.; Janssens, R.; Struyf, S.; Proost, P. Overview of the Mechanisms that May Contribute to the Non-Redundant Activities of Interferon-Inducible CXC Chemokine Receptor 3 Ligands. Front. Immunol. 2018, 8, 1970. [Google Scholar] [CrossRef]
- Pezzini, F.; Pisani, A.; Mazziotti, V.; Marastoni, D.; Tamanti, A.; Borroni, E.; Magon, S.; Zinnhardt, B.; Magliozzi, R.; Calabrese, M. Intrathecal versus Peripheral Inflammatory Protein Profile in MS Patients at Diagnosis: A Comprehensive Investigation on Serum and CSF. Int. J. Mol. Sci. 2023, 24, 3768. [Google Scholar] [CrossRef]
- Huang, J.; Khademi, M.; Fugger, L.; Lindhe, O.; Novakova, L.; Axelsson, M.; Malmestrom, C.; Constantinescu, C.; Lycke, J.; Piehl, F.; et al. Inflammation-related plasma and CSF biomarkers for multiple sclerosis. Proc. Natl. Acad. Sci. USA 2020, 117, 12952–12960. [Google Scholar] [CrossRef]



| RRMS (n = 37) | NIND (n = 19) | |
|---|---|---|
| Age (years; mean ± SD) | 39.3 ± 13.7 | 50.3 ± 17.4 |
| F:M ratio | 3.2:1 | 2.2:1 |
| Most recent relapse (months; mean ± SD) | 13.2 ± 18.7 | N/A |
| RRMS (n = 61) | NIND (n = 21) | |
|---|---|---|
| Age (years; mean ± SD) | 40.9 ± 13.6 | 46.0 ± 14.7 |
| F:M ratio | 2.6:1 | 4.2:1 |
| Most recent relapse (months; mean ± SD) | 18.2 ± 30.2 | N/A |
| RRMS (n = 116) | |
|---|---|
| Age (years; mean ± SD) | 44.1 ± 10.2 |
| F:M ratio | 2.9:1 |
| Most recent relapse (months; mean ± SD) | 60.0 ± 58.9 |
| Disease duration (years; mean ± SD) | 11.6 ± 7.36 |
| Disease modifying therapy use | |
| None | 16.4% |
| GA | 17.2% |
| Natalizumab | 3.45% |
| Fingolimod | 11.2% |
| DMF | 29.3% |
| Teriflunomide | 12.9% |
| IFNβ-1a | 9.48% |
| Cytokine | % Detected | % Extrapolated | t Ratio | df | p-Value | ||
|---|---|---|---|---|---|---|---|
| IL-1β | RRMS | 100% | RRMS | 55.3% | 1.454 | 20.51 | 0.970268 |
| NIND | 100% | NIND | 21.1% | ||||
| IL-1RA | RRMS | 100% | RRMS | 0% | 0.6334 | 21.45 | 0.995249 |
| NIND | 100% | NIND | 0% | ||||
| IL-2 | RRMS | 100% | RRMS | 78.9% | 0.8841 | 24.84 | 0.995249 |
| NIND | 100% | NIND | 25.6% | ||||
| IL-4 | RRMS | 100% | RRMS | 0% | 1.099 | 24.20 | 0.992497 |
| NIND | 100% | NIND | 5.26% | ||||
| IL-5 | RRMS | 84.2% | RRMS | 59.4% | 0.5702 | 21.61 | 0.995249 |
| NIND | 84.2% | NIND | 50.0% | ||||
| IL-6 | RRMS | 100% | RRMS | 13.2% | 1.253 | 25.75 | 0.98893 |
| NIND | 100% | NIND | 0% | ||||
| IL-7 | RRMS | 100% | RRMS | 2.63% | 1.126 | 31.14 | 0.992497 |
| NIND | 100% | NIND | 5.26% | ||||
| IL-8 | RRMS | 100% | RRMS | 0% | 0.7736 | 22.40 | 0.995249 |
| NIND | 100% | NIND | 0% | ||||
| IL-9 | RRMS | 100% | RRMS | 5.26% | 0.8327 | 24.40 | 0.995249 |
| NIND | 100% | NIND | 5.26% | ||||
| IL-10 | RRMS | 92.1% | RRMS | 20.0% | 1.008 | 21.56 | 0.992497 |
| NIND | 94.7% | NIND | 16.7% | ||||
| IL-12p70 | RRMS | 97.4% | RRMS | 32.4% | 0.6928 | 23.25 | 0.995249 |
| NIND | 100% | NIND | 36.8% | ||||
| IL-13 | RRMS | 100% | RRMS | 0% | 0.06743 | 28.31 | 0.997161 |
| NIND | 100% | NIND | 0% | ||||
| IL-15 | RRMS | 47.4% | RRMS | 0% | n/a | n/a | n/a |
| NIND | 42.1% | NIND | 25.0% | ||||
| IL-17 | RRMS | 100% | RRMS | 63.2% | 1.145 | 24.37 | 0.992497 |
| NIND | 100% | NIND | 47.4% | ||||
| CXCL10 | RRMS | 100% | RRMS | 0% | 3.252 | 44.00 | 0.04945* |
| NIND | 100% | NIND | 0% | ||||
| Eotaxin | RRMS | 100% | RRMS | 0% | 0.3407 | 24.91 | 0.995249 |
| NIND | 100% | NIND | 0% | ||||
| FGF Basic | RRMS | 97.4% | RRMS | 100% | 1.108 | 29.20 | 0.992497 |
| NIND | 94.7% | NIND | 88.9% | ||||
| G-CSF | RRMS | 100% | RRMS | 0% | 1.359 | 22.04 | 0.980866 |
| NIND | 100% | NIND | 0% | ||||
| GM-CSF | RRMS | 89.4% | RRMS | 20.6% | 0.3552 | 28.87 | 0.995249 |
| NIND | 89.4% | NIND | 11.8% | ||||
| IFN-γ | RRMS | 73.7% | RRMS | 10.7% | n/a | n/a | n/a |
| NIND | 78.9% | NIND | 20.0% | ||||
| MCP-1 | RRMS | 100% | RRMS | 0% | 1.603 | 18.75 | 0.940469 |
| NIND | 100% | NIND | 0% | ||||
| MIP-1a | RRMS | 100% | RRMS | 0% | 2.158 | 50.76 | 0.550753 |
| NIND | 94.7% | NIND | 0% | ||||
| PDGF-bb | RRMS | 42.1% | RRMS | 50.0% | n/a | n/a | n/a |
| NIND | 47.4% | NIND | 22.2% | ||||
| MIP-1b | RRMS | 100% | RRMS | 0% | 1.193 | 20.56 | 0.991874 |
| NIND | 100% | NIND | 0% | ||||
| RANTES | RRMS | 100% | RRMS | 26.3% | 0.01761 | 34.10 | 0.997161 |
| NIND | 100% | NIND | 26.3% | ||||
| TNFα | RRMS | 100% | RRMS | 34.2% | 0.736 | 33.74 | 0.995249 |
| NIND | 100% | NIND | 15.8% | ||||
| VEGF | RRMS | 76.3% | RRMS | 3.44% | n/a | n/a | n/a |
| NIND | 84.2% | NIND | 6.25% | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blandford, S.N.; Fudge, N.J.; Moore, C.S. CXCL10 Is Associated with Increased Cerebrospinal Fluid Immune Cell Infiltration and Disease Duration in Multiple Sclerosis. Biomolecules 2023, 13, 1204. https://doi.org/10.3390/biom13081204
Blandford SN, Fudge NJ, Moore CS. CXCL10 Is Associated with Increased Cerebrospinal Fluid Immune Cell Infiltration and Disease Duration in Multiple Sclerosis. Biomolecules. 2023; 13(8):1204. https://doi.org/10.3390/biom13081204
Chicago/Turabian StyleBlandford, Stephanie N., Neva J. Fudge, and Craig S. Moore. 2023. "CXCL10 Is Associated with Increased Cerebrospinal Fluid Immune Cell Infiltration and Disease Duration in Multiple Sclerosis" Biomolecules 13, no. 8: 1204. https://doi.org/10.3390/biom13081204
APA StyleBlandford, S. N., Fudge, N. J., & Moore, C. S. (2023). CXCL10 Is Associated with Increased Cerebrospinal Fluid Immune Cell Infiltration and Disease Duration in Multiple Sclerosis. Biomolecules, 13(8), 1204. https://doi.org/10.3390/biom13081204






