Endogenous Caulimovirids: Fossils, Zombies, and Living in Plant Genomes
Abstract
1. Introduction
2. ECVs as Molecular Fossils of Caulimoviridae
2.1. Classification of ECVs
2.2. Caulimovirids Have Colonized Almost All Plant Families
2.3. ECVs Provide Insights into Caulimovirid Genome Evolution
2.4. Dating the Integration of ECVs
2.5. Using ECVs to Infer the Long-Term Evolution of the Caulimoviridae
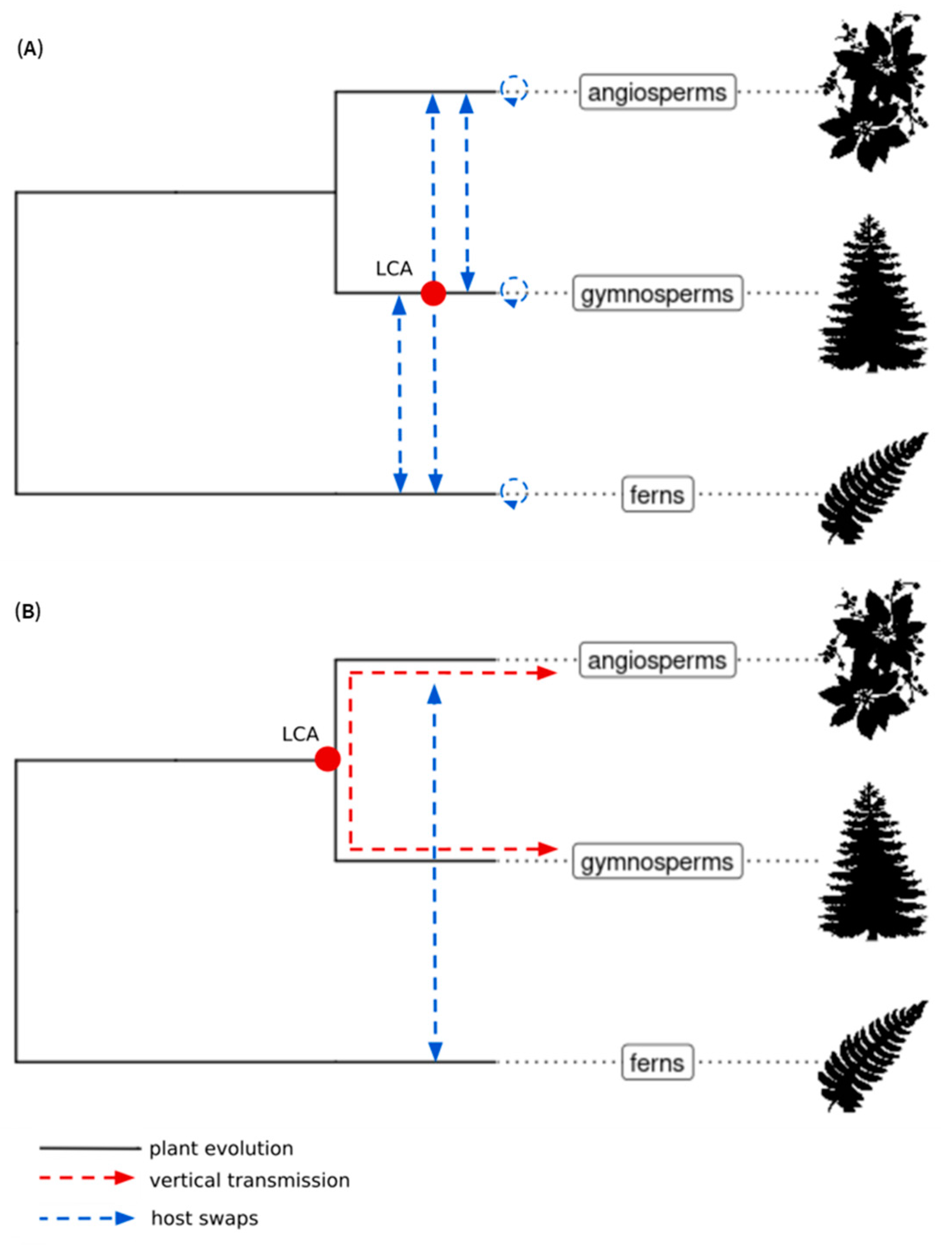

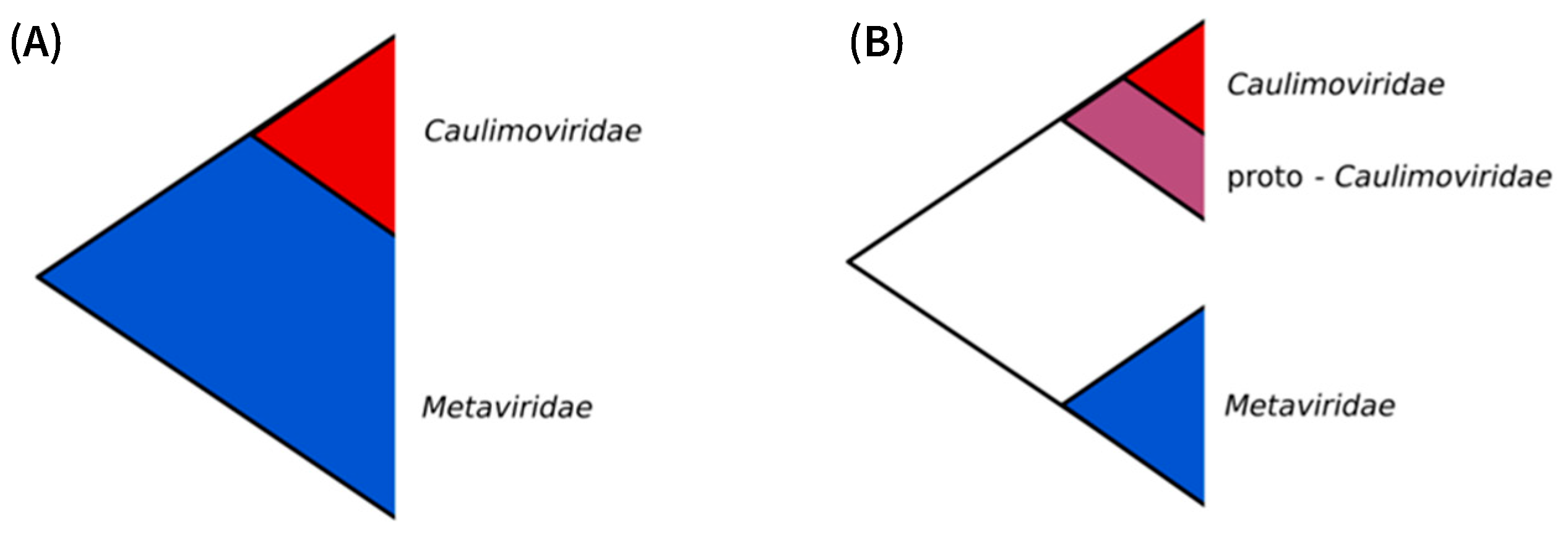
2.6. Origin of the Caulimoviridae
3. Integration, Genomic Distribution, and Expression of ECVs
3.1. Mechanisms of Integration
3.2. How Caulimovirid EVEs Have Invaded Plant Genomes
3.3. Distribution of Caulimovirid EVEs in Plant Genomes
3.4. Replication-Competent ECVs
3.5. Epigenetics and Silencing
4. Impact of ECVs on the Structure and Functions of Plant Genomes
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Weiss, R.A. The discovery of endogenous retroviruses. Retrovirology 2006, 3, 67. [Google Scholar] [CrossRef] [PubMed]
- Lwoff, A. The Concept of Virus. Microbiology 1957, 17, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yu, X.-F.; Chen, T. Human endogenous retroviruses in cancer: Expression, regulation and function (Review). Oncol. Lett. 2021, 21, 121. [Google Scholar] [CrossRef]
- Feschotte, C.; Gilbert, C. Endogenous viruses: Insights into viral evolution and impact on host biology. Nat. Rev. Genet 2012, 13, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Aiewsakun, P.; Katzourakis, A. Endogenous viruses: Connecting recent and ancient viral evolution. Virology 2015, 479–480, 26–37. [Google Scholar] [CrossRef]
- Bejarano, E.R.; Khashoggi, A.; Witty, M. Integration of multiple repeats of geminiviral DNA into the nuclear genome of tobacco during evolution. Proc. Natl. Acad. Sci. USA 1996, 93, 759–764. [Google Scholar] [CrossRef]
- Sharma, V.; Lefeuvre, P.; Roumagnac, P.; Filloux, D.; Teycheney, P.Y.; Martin, D.P.; Maumus, F. Large-scale survey reveals pervasiveness and potential function of endogenous geminiviral sequences in plants. Virus Evol. 2020, 6, veaa071. [Google Scholar] [CrossRef]
- Ndowora, T.; Dahal, G.; LaFleur, D.; Harper, G.; Hull, R.; Olszewski, N.E.; Lockhart, B. Evidence that badnavirus infection in Musa can originate from integrated pararetroviral sequences. Virology 1999, 255, 214–220. [Google Scholar] [CrossRef]
- Harper, G.; Osuji, J.O.; Heslop-Harrison, J.S.; Hull, R. Integration of banana streak badnavirus into the Musa genome: Molecular and cytogenetic evidence. Virology 1999, 255, 207–213. [Google Scholar] [CrossRef]
- Jakowitsch, J.; Mette, M.F.; van Der Winden, J.; Matzke, M.A.; Matzke, A.J.M. Integrated pararetroviral sequences define a unique class of dispersed repetitive DNA in plants. Proc. Natl. Acad. Sci. USA 1999, 96, 13241–13246. [Google Scholar] [CrossRef]
- Diop, S.I.; Geering, A.D.W.; Alfama-Depauw, F.; Loaec, M.; Teycheney, P.-Y.; Maumus, F. Tracheophyte genomes keep track of the deep evolution of the Caulimoviridae. Sci. Rep. 2018, 8, 572. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Blomberg, J.; Coffin, J.M.; Dasgupta, I.; Fan, H.; Geering, A.D.; Gifford, R.; Harrach, B.; Hull, R.; Johnson, W.; et al. Ortervirales: New virus order unifying five families of reverse-transcribing viruses. J. Virol. 2018, 92, e00515-18. [Google Scholar] [CrossRef]
- Teycheney, P.-Y.; Geering, A.D.W.; Dasgupta, I.; Hull, R.; Kreuze, J.F.; Lockhart, B.; Muller, E.; Olszewski, N.; Pappu, H.; Pooggin, M.M.; et al. ICTV Virus Taxonomy Profile: Caulimoviridae. J. Gen. Virol. 2020, 101, 1025–1026. [Google Scholar] [CrossRef] [PubMed]
- Medberry, S.L.; Lockhart, B.E.; Olszewski, N.E. Properties of Commelina yellow mottle virus’s complete DNA sequence, genomic discontinuities and transcript suggest that it is a pararetrovirus. Nucleic Acids Res. 1990, 18, 5505–5513. [Google Scholar] [CrossRef] [PubMed]
- Stavolone, L.; Herzog, E.; Leclerc, D.; Hohn, T. Tetramerization Is a Conserved Feature of the Virion-Associated Protein in Plant Pararetroviruses. J. Virol. 2001, 75, 7739–7743. [Google Scholar] [CrossRef] [PubMed]
- de Tomás, C.; Vicient, C.M. Genome-wide identification of reverse transcriptase domains of recently inserted endogenous plant pararetrovirus (Caulimoviridae). Front. Plant Sci. 2022, 13, 1011565. [Google Scholar] [CrossRef]
- Geering, A.D.W.; Maumus, F.; Copetti, D.; Choisne, N.; Zwickl, D.J.; Zytnicki, M.; McTaggart, A.R.; Scalabrin, S.; Vezzulli, S.; Wing, R.A.; et al. Endogenous florendoviruses are major components of plant genomes and hallmarks of virus evolution. Nat. Commun. 2014, 5, 5269. [Google Scholar] [CrossRef]
- Pooggin, M.M.; Ryabova, L.A. Ribosome Shunting, Polycistronic Translation, and Evasion of Antiviral Defenses in Plant Pararetroviruses and Beyond. Front. Microbiol. 2018, 9, 644. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.-D.; Chen, X.; Tian, J.-H.; Chen, L.-J.; Li, K.; Wang, W.; Eden, J.-S.; Shen, J.-J.; Liu, L.; et al. The evolutionary history of vertebrate RNA viruses. Nature 2018, 556, 197–202. [Google Scholar] [CrossRef]
- Dutilh, B.E.; Varsani, A.; Tong, Y.; Simmonds, P.; Sabanadzovic, S.; Rubino, L.; Roux, S.; Muñoz, A.R.; Lood, C.; Lefkowitz, E.J.; et al. Perspective on taxonomic classification of uncultivated viruses. Curr. Opin. Virol. 2021, 51, 207–215. [Google Scholar] [CrossRef]
- Simmonds, P.; Adriaenssens, E.M.; Zerbini, F.M.; Abrescia, N.G.A.; Aiewsakun, P.; Alfenas-Zerbini, P.; Bao, Y.; Barylski, J.; Drosten, C.; Duffy, S.; et al. Four principles to establish a universal virus taxonomy. PLoS Biol. 2023, 21, e3001922. [Google Scholar] [CrossRef]
- Geering, A.D.; Scharaschkin, T.; Teycheney, P.-Y. The classification and nomenclature of endogenous viruses of the family Caulimoviridae. Arch. Virol. 2010, 155, 123–131. [Google Scholar] [CrossRef]
- Kunii, M.; Kanda, M.; Nagano, H.; Uyeda, I.; Kishima, Y.; Sano, Y. Reconstruction of putative DNA virus from endogenous rice tungro bacilliform virus-like sequences in the rice genome: Implications for integration and evolution. BMC Genom. 2004, 5, 80. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Han, G.-Z. Euphyllophyte Paleoviruses illuminate hidden diversity and macroevolutionary mode of Caulimoviridae. J. Virol. 2018, 92, e02043-17. [Google Scholar] [CrossRef]
- Diouf, M.B.; Guyader, S.; Gaspard, O.; Francius, E.; Teycheney, P.-Y.; Umber, M. Epidemiology of yam viruses in Guadeloupe: Role of cropping practices and seed-tuber supply. Viruses 2022, 14, 2366. [Google Scholar] [CrossRef]
- Bouhida, M.; Lockhart, B.E.L.; Olszewski, N.E. An analysis of the complete sequence of a sugarcane bacilliform virus genome infectious to banana and rice. J. Gen. Virol. 1993, 74, 15–22. [Google Scholar] [CrossRef]
- Lockhart, B.E.; Menke, J.; Dahal, G.; Olszewski, N.E. Characterization and genomic analysis of tobacco vein clearing virus, a plant pararetrovirus that is transmitted vertically and related to sequences integrated in the host genome. J. Gen. Virol. 2000, 81, 1579–1585. [Google Scholar] [CrossRef] [PubMed]
- Mushegian, A.R.; Elena, S.F. Evolution of plant virus movement proteins from the 30K superfamily and of their homologs integrated in plant genomes. Virology 2015, 476, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Bloemendal, S.; Kück, U. Cell-to-cell communication in plants, animals, and fungi: A comparative review. Naturwissenschaften 2013, 100, 3–19. [Google Scholar] [CrossRef]
- Rodriguez, A.; Angel, C.A.; Lutz, L.; Leisner, S.M.; Nelson, R.S.; Schoelz, J.E. Association of the P6 protein of Cauliflower mosaic virus with plasmodesmata and plasmodesmal proteins. Plant Physiol. 2014, 166, 1345–1358. [Google Scholar] [CrossRef]
- Schmidt, N.; Seibt, K.M.; Weber, B.; Schwarzacher, T.; Schmidt, T.; Heitkam, T. Broken, silent, and in hiding: Tamed endogenous pararetroviruses escape elimination from the genome of sugar beet (Beta vulgaris). Ann. Bot. 2021, 128, 281–299. [Google Scholar] [CrossRef]
- Chao, L. Evolution of sex in RNA viruses. J. Theor. Biol. 1988, 133, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Nee, S. The evolution of multicompartmental genomes in viruses. J. Mol. Evol. 1987, 25, 277–281. [Google Scholar] [CrossRef]
- Gayral, P.; Noa-Carrazana, J.-C.; Lescot, M.; Lheureux, F.; Lockhart, B.E.L.; Matsumoto, T.; Piffanelli, P.; Iskra-Caruana, M.-L. A Single Banana Streak Virus Integration Event in the Banana Genome as the Origin of Infectious Endogenous Pararetrovirus. J. Virol. 2008, 82, 6697–6710. [Google Scholar] [CrossRef] [PubMed]
- Gayral, P.; Blondin, L.; Guidolin, O.; Carreel, F.; Hippolyte, I.; Perrier, X.; Iskra-Caruana, M.-L. Evolution of endogenous sequences of banana streak virus: What can we learn from banana (Musa sp.) evolution? J. Virol. 2010, 84, 7346–7359. [Google Scholar] [CrossRef]
- Kijima, T.E.; Innan, H. On the Estimation of the Insertion Time of LTR Retrotransposable Elements. Mol. Biol. Evol. 2010, 27, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Nolan, A.; Watson, J.; Tristem, M. Identification of an ancient endogenous retrovirus, predating the divergence of the placental mammals. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120503. [Google Scholar] [CrossRef]
- Katzourakis, A.; Gifford, R.J.; Tristem, M.; Gilbert, M.T.P.; Pybus, O.G. Macroevolution of complex retroviruses. Science 2009, 325, 1512. [Google Scholar] [CrossRef]
- Aiewsakun, P.; Katzourakis, A. Marine origin of retroviruses in the early Palaeozoic Era. Nat. Commun. 2017, 8, 13954. [Google Scholar] [CrossRef]
- Xiong, Y.; Eickbush, T.H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990, 9, 3353–3362. [Google Scholar] [CrossRef]
- Menéndez-Arias, L.; Sebastián-Martín, A.; Álvarez, M. Viral reverse transcriptases. Virus Res. 2017, 234, 153–176. [Google Scholar] [CrossRef] [PubMed]
- Springer, M.S.; Britten, R.J. Phylogenetic relationships of reverse transcriptase and RNase H sequences and aspects of genome structure in the gypsy group of retrotransposons. Mol. Biol. Evol. 1993, 10, 1370–1379. [Google Scholar] [PubMed][Green Version]
- Chang, G.S.; Hong, Y.; Ko, K.D.; Bhardwaj, G.; Holmes, E.C.; Patterson, R.L.; van Rossum, D.B. Phylogenetic profiles reveal evolutionary relationships within the “twilight zone” of sequence similarity. Proc. Natl. Acad. Sci. USA 2008, 105, 13474–13479. [Google Scholar] [CrossRef]
- Sperber, G.O.; Airola, T.; Jern, P.; Blomberg, J. Automated recognition of retroviral sequences in genomic data—RetroTector©. Nucleic Acids Res. 2007, 35, 4964–4976. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.; Cornwallis, C.K.; Jern, P. Pan-vertebrate comparative genomics unmasks retrovirus macroevolution. Proc. Natl. Acad Sci 2015, 112, 464–469. [Google Scholar] [CrossRef]
- Llorens, C.; Muñoz-Pomer, A.; Bernad, L.; Botella, H.; Moya, A. Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol. Direct 2009, 4, 41. [Google Scholar] [CrossRef]
- Butterfield, N.J. Early evolution of the Eukaryota. Palaeontology 2015, 58, 5–17. [Google Scholar] [CrossRef]
- Koonin, E.V.; Mushegian, A.R.; Ryabov, E.V.; Dolja, V.V. Diverse groups of plant RNA and DNA viruses share related movement proteins that may possess chaperone-like activity. J. Gen. Virol. 1991, 72, 2895–2903. [Google Scholar] [CrossRef]
- Harrison, C.J.; Morris, J.L. The origin and early evolution of vascular plant shoots and leaves. Philos. Trans. R. Soc. B Biol. Sci. 2017, 373, 20160496. [Google Scholar] [CrossRef]
- Donoghue, P.C.J.; Harrison, C.J.; Paps, J.; Schneider, H. The evolutionary emergence of land plants. Curr. Biol. 2021, 31, R1281–R1298. [Google Scholar] [CrossRef]
- Liu, R.; Koyanagi, K.O.; Chen, S.; Kishima, Y. Evolutionary force of AT-rich repeats to trap genomic and episomal DNAs into the rice genome: Lessons from endogenous pararetrovirus. Plant J. 2012, 72, 817–828. [Google Scholar] [CrossRef]
- Yu, H.; Wang, X.; Lu, Z.; Xu, Y.; Deng, X.; Xu, Q. Endogenous pararetrovirus sequences are widely present in Citrinae genomes. Virus Res. 2019, 262, 48–53. [Google Scholar] [CrossRef]
- Brown, R.E.; Freudenreich, C.H. Structure-forming repeats and their impact on genome stability. Curr. Opin. Genet. Dev. 2021, 67, 41–51. [Google Scholar] [CrossRef]
- Vítor, A.C.; Huertas, P.; Legube, G.; de Almeida, S.F. Studying DNA Double-Strand Break Repair: An Ever-Growing Toolbox. Front. Mol. Biosci. 2020, 7, 24. [Google Scholar] [CrossRef]
- Richert-Pöggeler, K.R.; Noreen, F.; Schwarzacher, T.; Harper, G.; Hohn, T. Induction of infectious petunia vein clearing (pararetro) virus from endogenous provirus in petunia. EMBO J. 2003, 22, 4836–4845. [Google Scholar] [CrossRef]
- Umber, M.; Filloux, D.; Muller, E.; Laboureau, N.; Galzi, S.; Roumagnac, P.; Iskra-Caruana, M.-L.; Pavis, C.; Teycheney, P.-Y.; Seal, S.E. The genome of African yam (Dioscorea cayenensis-rotundata complex) hosts endogenous sequences from four distinct Badnavirus species. Mol. Plant Pathol. 2014, 15, 790–801. [Google Scholar] [CrossRef]
- Chabannes, M.; Baurens, F.-C.; Duroy, P.-O.; Bocs, S.; Vernerey, M.-S.; Rodier-Goud, M.; Barbe, V.; Gayral, P.; Iskra-Caruana, M.-L. Three infectious viral species lying in wait in the banana genome. J. Virol. 2013, 87, 8624–8637. [Google Scholar] [CrossRef]
- Li, F.; Lee, M.; Esnault, C.; Wendover, K.; Guo, Y.; Atkins, P.; Zaratiegui, M.; Levin, H.L. Identification of an integrase-independent pathway of retrotransposition. Sci. Adv. 2022, 8, eabm9390. [Google Scholar] [CrossRef] [PubMed]
- Samach, A.; Melamed-Bessudo, C.; Avivi-Ragolski, N.; Pietrokovski, S.; Levy, A.A. Identification of plant RAD52 homologs and characterization of the Arabidopsis thaliana RAD52-like genes. Plant Cell 2011, 23, 4266–4279. [Google Scholar] [CrossRef] [PubMed]
- Horie, M.; Honda, T.; Suzuki, Y.; Kobayashi, Y.; Daito, T.; Oshida, T.; Ikuta, K.; Jern, P.; Gojobori, T.; Coffin, J.M.; et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 2010, 463, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Houé, V.; Bonizzoni, M.; Failloux, A.-B. Endogenous non-retroviral elements in genomes of Aedes mosquitoes and vector competence. Emerg. Microbes Infect. 2019, 8, 542–555. [Google Scholar] [CrossRef]
- Hancks, D.C.; Kazazian, H.H. Roles for retrotransposon insertions in human disease. Mob. DNA 2016, 7, 9. [Google Scholar] [CrossRef]
- Takahashi, H.; Fukuhara, T.; Kitazawa, H.; Kormelink, R. Virus Latency and the Impact on Plants. Front. Microbiol. 2019, 10, 2764. [Google Scholar] [CrossRef]
- Belshaw, R.; Pereira, V.; Katzourakis, A.; Talbot, G.; Pačes, J.; Burt, A.; Tristem, M. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 2004, 101, 4894–4899. [Google Scholar] [CrossRef]
- Kim, S.; Park, M.; Yeom, S.-I.; Kim, Y.-M.; Lee, J.M.; Lee, H.-A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.-T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef]
- Serfraz, S.; Sharma, V.; Maumus, F.; Aubriot, X.; Geering, A.D.W.; Teycheney, P.-Y. Insertion of badnaviral DNA in the late blight resistance gene (R1a) of Brinjal Eggplant (Solanum melongena). Front. Plant Sci. 2021, 12, 683681. [Google Scholar] [CrossRef]
- Boutanaev, A.M.; Nemchinov, L.G. Genome-wide identification of endogenous viral sequences in alfalfa (Medicago sativa L.). Virol. J. 2021, 18, 185. [Google Scholar] [CrossRef]
- Staginnus, C.; Gregor, W.; Mette, M.F.; Teo, C.H.; Borroto-Fernández, E.G.; Machado, M.L.d.C.; Matzke, M.; Schwarzacher, T. Endogenous pararetroviral sequences in tomato (Solanum lycopersicum) and related species. BMC Plant Biol. 2007, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Becher, H.; Ma, L.; Kelly, L.J.; Kovarik, A.; Leitch, I.J.; Leitch, A.R. Endogenous pararetrovirus sequences associated with 24 nt small RNAs at the centromeres of Fritillaria imperialis L. (Liliaceae), a species with a giant genome. Plant J. 2014, 80, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.I.; Mohandas, A.; Sreenayana, B.; Archana, T.S.; Jasna, K. Piper DNA virus 1 and 2 are endogenous pararetroviruses integrated into chromosomes of black pepper (Piper nigrum L). Virusdisease 2022, 33, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.-M.; Miao, L.-K.; Xie, K.-D.; Yin, Z.-P.; Wu, X.-M.; Chen, C.-L.; Grosser, J.W.; Guo, W.-W. Localization and characterization of Citrus centromeres by combining half-tetrad analysis and CenH3-associated sequence profiling. Plant Cell Rep. 2020, 39, 1609–1622. [Google Scholar] [CrossRef]
- Martin, S.L.; Garfinkel, D.J. Survival strategies for transposons and genomes. Genome Biol. 2003, 4, 313. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gao, X.; Hou, Y.; Ebina, H.; Levin, H.L.; Voytas, D.F. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 2008, 18, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Dallot, S.; Acuña, P.; Rivera, C.; Ramírez, P.; Côte, F.; Lockhart, B.E.; Caruana, M.L. Evidence that the proliferation stage of micropropagation procedure is determinant in the expression of banana streak virus integrated into the genome of the FHIA 21 hybrid (Musa AAAB). Arch. Virol. 2001, 146, 2179–2190. [Google Scholar] [CrossRef] [PubMed]
- Côte, F.X.; Galzi, S.; Folliot, M.; Lamagnère, Y.; Teycheney, P.-Y.; Iskra-Caruana, M.-L. Micropropagation by tissue culture triggers differential expression of infectious endogenous Banana streak virus sequences (eBSV) present in the B genome of natural and synthetic interspecific banana plantains. Mol. Plant Pathol. 2010, 11, 137–144. [Google Scholar] [CrossRef]
- Umber, M.; Pressat, G.; Fort, G.; Plaisir Pineau, K.; Guiougiou, C.; Lambert, F.; Farinas, B.; Pichaut, J.-P.; Janzac, B.; Delos, J.-M.; et al. Risk Assessment of Infectious Endogenous Banana Streak Viruses in Guadeloupe. Front. Plant Sci. 2022, 13, 951285. [Google Scholar] [CrossRef] [PubMed]
- Noreen, F.; Akbergenov, R.; Hohn, T.; Richert-Pöggeler, K.R. Distinct expression of endogenous Petunia vein clearing virus and the DNA transposon dTph1 in two Petunia hybrida lines is correlated with differences in histone modification and siRNA production. Plant J. 2007, 50, 219–229. [Google Scholar] [CrossRef]
- Kuriyama, K.; Tabara, M.; Moriyama, H.; Kanazawa, A.; Koiwa, H.; Takahashi, H.; Fukuhara, T. Disturbance of floral colour pattern by activation of an endogenous pararetrovirus, petunia vein clearing virus, in aged petunia plants. Plant J. 2020, 103, 497–511. [Google Scholar] [CrossRef]
- Sequeira-Mendes, J.; Aragüez, I.; Peiró, R.; Mendez-Giraldez, R.; Zhang, X.; Jacobsen, S.E.; Bastolla, U.; Gutierrez, C. The Functional Topography of the Arabidopsis Genome Is Organized in a Reduced Number of Linear Motifs of Chromatin States. Plant Cell 2014, 26, 2351–2366. [Google Scholar] [CrossRef]
- Richert-Pöggeler, K.R.; Vijverberg, K.; Alisawi, O.; Chofong, G.N.; Heslop-Harrison, J.S.; Schwarzacher, T. Participation of Multifunctional RNA in Replication, Recombination and Regulation of Endogenous Plant Pararetroviruses (EPRVs). Front. Plant Sci. 2021, 12, 689307. [Google Scholar] [CrossRef]
- Erdmann, R.M.; Picard, C.L. RNA-directed DNA Methylation. PLoS Genet. 2020, 16, e1009034. [Google Scholar] [CrossRef] [PubMed]
- Cuerda-Gil, D.; Slotkin, R.K. Non-canonical RNA-directed DNA methylation. Nat. Plants 2016, 2, 16163. [Google Scholar] [CrossRef] [PubMed]
- Pooggin, M.M. How Can Plant DNA Viruses Evade siRNA-Directed DNA Methylation and Silencing? Int. J. Mol. Sci. 2013, 14, 15233–15259. [Google Scholar] [CrossRef] [PubMed]
- Pooggin, M.M. Role of Small RNAs in Virus-Host Interaction. In Plant-Virus Interactions: Molecular Biology, Intra- and Intercellular Transport; Kleinow, T., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 161–189. [Google Scholar]
- Mette, M.F.; Kanno, T.; Aufsatz, W.; Jakowitsch, J.; van der Winden, J.; Matzke, M.A.; Matzke, A.J. Endogenous viral sequences and their potential contribution to heritable virus resistance in plants. EMBO J. 2002, 21, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Baidaliuk, A.; Miesen, P.; Frangeul, L.; Crist, A.B.; Merkling, S.H.; Fontaine, A.; Lequime, S.; Moltini-Conclois, I.; Blanc, H.; et al. Non-retroviral Endogenous Viral Element Limits Cognate Virus Replication in Aedes aegypti Ovaries. Curr. Biol. 2020, 30, 3495–3506.e6. [Google Scholar] [CrossRef]
- Valli, A.A.; Gonzalo-Magro, I.; Sanchez, D.H. Rearranged Endogenized Plant Pararetroviruses as Evidence of Heritable RNA-based Immunity. Mol. Biol. Evol. 2023, 40, msac240. [Google Scholar] [CrossRef]
- da Fonseca, G.C.; de Oliveira, L.F.V.; de Morais, G.L.; Abdelnor, R.V.; Nepomuceno, A.L.; Waterhouse, P.M.; Farinelli, L.; Margis, R. Unusual RNA plant virus integration in the soybean genome leads to the production of small RNAs. Plant Sci. 2016, 246, 62–69. [Google Scholar] [CrossRef]
- Jia, J.; Ji, R.; Li, Z.; Yu, Y.; Nakano, M.; Long, Y.; Feng, L.; Qin, C.; Lu, D.; Zhan, J.; et al. Soybean DICER-LIKE2 Regulates Seed Coat Color via Production of Primary 22-Nucleotide Small Interfering RNAs from Long Inverted Repeats. Plant Cell 2020, 32, 3662–3673. [Google Scholar] [CrossRef]
- Lopez-Gomollon, S.; Müller, S.Y.; Baulcombe, D.C. Interspecific hybridization in tomato influences endogenous viral sRNAs and alters gene expression. Genome Biol. 2022, 23, 120. [Google Scholar] [CrossRef]
- Pooggin, M.M. Small RNA-Omics for Plant Virus Identification, Virome Reconstruction, and Antiviral Defense Characterization. Front. Microbiol. 2018, 9, 2779. [Google Scholar] [CrossRef]
- Cho, B.C.; Shaughnessy, J.D.; Largaespada, D.A.; Bedigian, H.G.; Buchberg, A.M.; Jenkins, N.A.; Copeland, N.G. Frequent disruption of the Nf1 gene by a novel murine AIDS virus-related provirus in BXH-2 murine myeloid lymphomas. J. Virol. 1995, 69, 7138–7146. [Google Scholar] [CrossRef]
- Kurth, R.; Bannert, N. Beneficial and detrimental effects of human endogenous retroviruses. Int. J. Cancer 2010, 126, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Meyer, T.J.; Rosenkrantz, J.L.; Carbone, L.; Chavez, S.L. Endogenous Retroviruses: With Us and against Us. Front. Chem. 2017, 5, 23. [Google Scholar] [CrossRef]
- Beyer, U.; Moll-Rocek, J.; Moll, U.M.; Dobbelstein, M. Endogenous retrovirus drives hitherto unknown proapoptotic p63 isoforms in the male germ line of humans and great apes. Proc. Natl. Acad. Sci. USA 2011, 108, 3624–3629. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, G.J.; Kimura, Y.; Daub, C.O.; Wani, S.; Plessy, C.; Irvine, K.M.; Schroder, K.; Cloonan, N.; Steptoe, A.L.; Lassmann, T.; et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009, 41, 563–571. [Google Scholar] [CrossRef]
- Carrasco, J.L.; Sánchez-Navarro, J.A.; Elena, S.F. Exploring the role of cellular homologous of the 30K-superfamily of plant virus movement proteins. Virus Res. 2019, 262, 54–61. [Google Scholar] [CrossRef]
- Barchi, L.; Pietrella, M.; Venturini, L.; Minio, A.; Toppino, L.; Acquadro, A.; Andolfo, G.; Aprea, G.; Avanzato, C.; Bassolino, L.; et al. A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 2019, 9, 11769. [Google Scholar] [CrossRef] [PubMed]
- Vassilieff, H.; Haddad, S.; Jamilloux, V.; Choisne, N.; Sharma, V.; Giraud, D.; Wan, M.; Serfraz, S.; Geering, A.D.W.; Teycheney, P.-Y.; et al. CAULIFINDER: A pipeline for the automated detection and annotation of caulimovirid endogenous viral elements in plant genomes. Mob. DNA 2022, 13, 31. [Google Scholar] [CrossRef]

| Gong & Han [24] | α-GECV PtaeV_2 | β-GECV PglaV_2 | δ-GECV PglaV_1 | ε-GECV PtaeV_1 | Ɣ-GECV GbilV | |
|---|---|---|---|---|---|---|
| Diop et al. [11] | ||||||
| Gymnendovirus 1 Gymnendovirus_1_Pabies | 44.1% | 73.0% | 50.2% | 46.3 | 50.6% | |
| Gymnendovirus 2 Gymnendovirus_2_Gbilo | 51.0% | 52.0% | 47.1% | 48.0% | 56% | |
| Gymnendovirus 3 Gymnendovirus_3_Pabies | 43.7% | 50.6% | 76.1% | 58.6% | 48.2% | |
| Gymnendovirus 4 Gymnendovirus_4_Pabies | 46.2% | 53.7% | 57.7% | 60.7% | 45.1% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vassilieff, H.; Geering, A.D.W.; Choisne, N.; Teycheney, P.-Y.; Maumus, F. Endogenous Caulimovirids: Fossils, Zombies, and Living in Plant Genomes. Biomolecules 2023, 13, 1069. https://doi.org/10.3390/biom13071069
Vassilieff H, Geering ADW, Choisne N, Teycheney P-Y, Maumus F. Endogenous Caulimovirids: Fossils, Zombies, and Living in Plant Genomes. Biomolecules. 2023; 13(7):1069. https://doi.org/10.3390/biom13071069
Chicago/Turabian StyleVassilieff, Héléna, Andrew D. W. Geering, Nathalie Choisne, Pierre-Yves Teycheney, and Florian Maumus. 2023. "Endogenous Caulimovirids: Fossils, Zombies, and Living in Plant Genomes" Biomolecules 13, no. 7: 1069. https://doi.org/10.3390/biom13071069
APA StyleVassilieff, H., Geering, A. D. W., Choisne, N., Teycheney, P.-Y., & Maumus, F. (2023). Endogenous Caulimovirids: Fossils, Zombies, and Living in Plant Genomes. Biomolecules, 13(7), 1069. https://doi.org/10.3390/biom13071069






