Overview of the Circadian Clock in the Hair Follicle Cycle
Abstract
1. Introduction
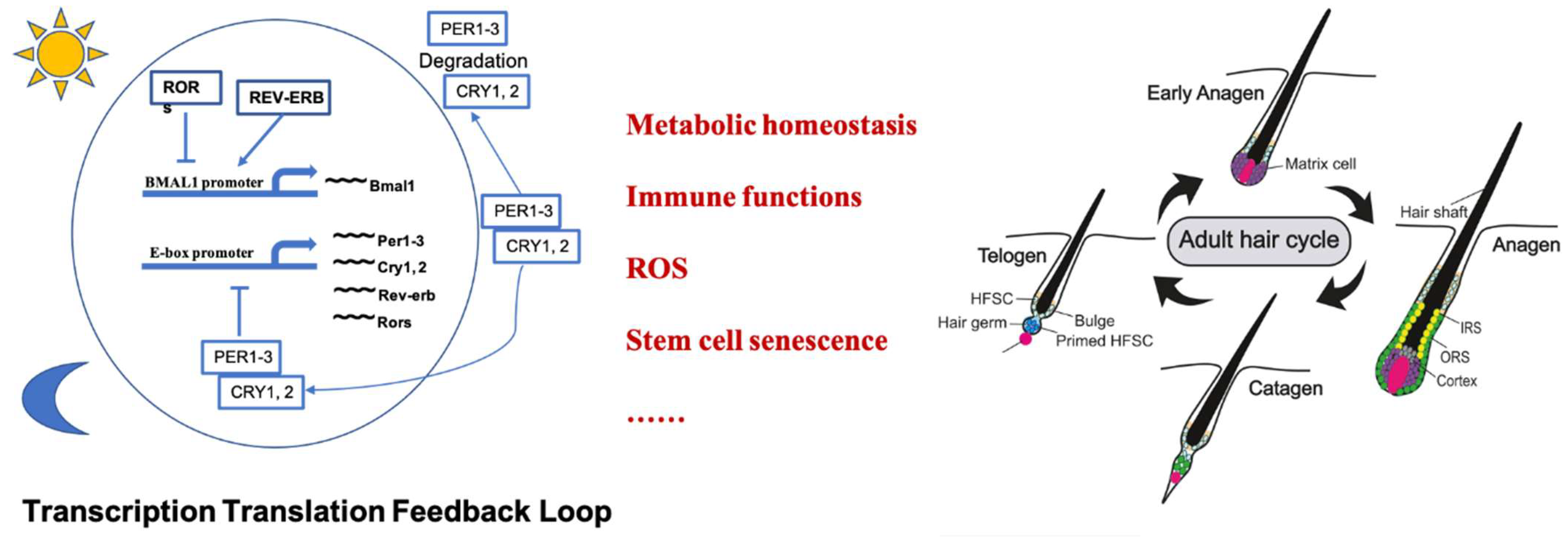
2. Hair Follicle Cycling
3. Molecular Circadian Clock in Hair Follicle Cycling
4. The Circadian Clock Modulates the Hair Follicle Cycle
5. Metabolic Reprogramming in the Hair Follicle Cycle
6. The Circadian Clock Controls Metabolism
6.1. Core Circadian Clock Genes and Metabolism
6.2. Environmental Cues That Regulate Circadian Rhythms and Metabolism
7. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Plikus, M.V.; Van Spyk, E.N.; Pham, K.; Geyfman, M.; Kumar, V.; Takahashi, J.S.; Andersen, B. The Circadian Clock in Skin: Implications for Adult Stem Cells, Tissue Regeneration, Cancer, Aging, and Immunity. J. Biol. Rhythms 2015, 30, 163–182. [Google Scholar] [CrossRef] [PubMed]
- Roenneberg, T.; Merrow, M. The Circadian Clock and Human Health. Curr. Biol. 2016, 26, R432–R443. [Google Scholar] [CrossRef] [PubMed]
- Alonso, L.; Fuchs, E. The Hair Cycle. J. Cell Sci. 2006, 119, 391–393. [Google Scholar] [CrossRef]
- Molecular Mechanisms Regulating Hair Follicle Development|Elsevier Enhanced Reader. Available online: https://reader.elsevier.com/reader/sd/pii/S0022202X15415532?token=DC4D45ADDA7A915A09E1FDA2914EE457433C93A91A0C6723D3E5557F73AFB33D35A8DA156189C32EBAA884878856B8FF&originRegion=us-east-1&originCreation=20220415071753 (accessed on 15 April 2022).
- Lee, S.A.; Li, K.N.; Tumbar, T. Stem Cell-Intrinsic Mechanisms Regulating Adult Hair Follicle Homeostasis. Exp. Dermatol. 2021, 30, 430–447. [Google Scholar] [CrossRef]
- Morita, R.; Sanzen, N.; Sasaki, H.; Hayashi, T.; Umeda, M.; Yoshimura, M.; Yamamoto, T.; Shibata, T.; Abe, T.; Kiyonari, H.; et al. Tracing the Origin of Hair Follicle Stem Cells. Nature 2021, 594, 547–552. [Google Scholar] [CrossRef]
- Kim, C.S.; Ding, X.; Allmeroth, K.; Biggs, L.C.; Kolenc, O.I.; L’Hoest, N.; Chacón-Martínez, C.A.; Edlich-Muth, C.; Giavalisco, P.; Quinn, K.P.; et al. Glutamine Metabolism Controls Stem Cell Fate Reversibility and Long-Term Maintenance in the Hair Follicle. Cell Metab. 2020, 32, 629–642.e8. [Google Scholar] [CrossRef]
- Finger, A.-M.; Kramer, A. Peripheral Clocks Tick Independently of Their Master. Genes Dev. 2021, 35, 304–306. [Google Scholar] [CrossRef]
- Serin, Y.; Acar Tek, N. Effect of Circadian Rhythm on Metabolic Processes and the Regulation of Energy Balance. ANM 2019, 74, 322–330. [Google Scholar] [CrossRef]
- Guan, D.; Lazar, M.A. Interconnections between Circadian Clocks and Metabolism. Available online: https://www.jci.org/articles/view/148278/pdf (accessed on 13 May 2022).
- Curtis, A.M.; Fagundes, C.T.; Yang, G.; Palsson-McDermott, E.M.; Wochal, P.; McGettrick, A.F.; Foley, N.H.; Early, J.O.; Chen, L.; Zhang, H.; et al. Circadian Control of Innate Immunity in Macrophages by MiR-155 Targeting Bmal1. Proc. Natl. Acad. Sci. USA 2015, 112, 7231–7236. [Google Scholar] [CrossRef]
- Akashi, M.; Soma, H.; Yamamoto, T.; Tsugitomi, A.; Yamashita, S.; Yamamoto, T.; Nishida, E.; Yasuda, A.; Liao, J.K.; Node, K. Noninvasive Method for Assessing the Human Circadian Clock Using Hair Follicle Cells. Proc. Natl. Acad. Sci. USA 2010, 107, 15643–15648. [Google Scholar] [CrossRef]
- Liu, L.-P.; Li, M.-H.; Zheng, Y.-W. Hair Follicles as a Critical Model for Monitoring the Circadian Clock. Int. J. Mol. Sci. 2023, 24, 2407. [Google Scholar] [CrossRef]
- Lin, K.K.; Kumar, V.; Geyfman, M.; Chudova, D.; Ihler, A.T.; Smyth, P.; Paus, R.; Takahashi, J.S.; Andersen, B. Circadian Clock Genes Contribute to the Regulation of Hair Follicle Cycling. PLoS Genet 2009, 5, e1000573. [Google Scholar] [CrossRef]
- Panda, S. Circadian Physiology of Metabolism. Science 2016, 354, 1008–1015. [Google Scholar] [CrossRef]
- Greenhill, C. Circadian Protein Affects MTORC1 Activity. Nat. Rev. Endocrinol. 2019, 15, 65. [Google Scholar] [CrossRef]
- Peek, C.B. Metabolic Implications of Circadian–HIF Crosstalk. Trends Endocrinol. Metab. 2020, 31, 459–468. [Google Scholar] [CrossRef]
- Alexander, R.K.; Liou, Y.-H.; Knudsen, N.H.; Starost, K.A.; Xu, C.; Hyde, A.L.; Liu, S.; Jacobi, D.; Liao, N.-S.; Lee, C.-H. Bmal1 Integrates Mitochondrial Metabolism and Macrophage Activation. eLife 2020, 9, e54090. [Google Scholar] [CrossRef]
- Samoilova, E.M.; Belopasov, V.V.; Ekusheva, E.V.; Zhang, C.; Troitskiy, A.V.; Baklaushev, V.P. Epigenetic Clock and Circadian Rhythms in Stem Cell Aging and Rejuvenation. J. Pers. Med. 2021, 11, 1050. [Google Scholar] [CrossRef]
- Deng, Z.; Lei, X.; Zhang, X.; Zhang, H.; Liu, S.; Chen, Q.; Hu, H.; Wang, X.; Ning, L.; Cao, Y.; et al. MTOR Signaling Promotes Stem Cell Activation via Counterbalancing BMP-Mediated Suppression during Hair Regeneration. J. Mol. Cell Biol. 2015, 7, 62–72. [Google Scholar] [CrossRef]
- Rahmani, W.; Sinha, S.; Biernaskie, J. Immune Modulation of Hair Follicle Regeneration. NPJ Regen. Med. 2020, 5, 9. [Google Scholar] [CrossRef]
- Hamanaka, R.B.; Glasauer, A.; Hoover, P.; Yang, S.; Blatt, H.; Mullen, A.R.; Getsios, S.; Gottardi, C.J.; DeBerardinis, R.J.; Lavker, R.M.; et al. Mitochondrial Reactive Oxygen Species Promote Epidermal Differentiation and Hair Follicle Development. Sci. Signal 2013, 6, ra8. [Google Scholar] [CrossRef]
- Morinaga, H.; Mohri, Y.; Grachtchouk, M.; Asakawa, K.; Matsumura, H.; Oshima, M.; Takayama, N.; Kato, T.; Nishimori, Y.; Sorimachi, Y.; et al. Obesity Accelerates Hair Thinning by Stem Cell-Centric Converging Mechanisms. Nature 2021, 595, 266–271. [Google Scholar] [CrossRef]
- Mok, K.-W.; Saxena, N.; Heitman, N.; Grisanti, L.; Srivastava, D.; Muraro, M.J.; Jacob, T.; Sennett, R.; Wang, Z.; Su, Y.; et al. Dermal Condensate Niche Fate Specification Occurs Prior to Formation and Is Placode Progenitor Dependent. Dev. Cell 2019, 48, 32–48.e5. [Google Scholar] [CrossRef]
- Fuchs, E. Scratching the Surface of Skin Development. Nature 2007, 445, 834–842. [Google Scholar] [CrossRef]
- A Comprehensive Guide for the Accurate Classification of Murine Hair Follicles in Distinct Hair Cycle Stages|Elsevier Enhanced Reader. Available online: https://reader.elsevier.com/reader/sd/pii/S0022202X15412825?token=C10FDA112D5DF5382E2C78542930D75919205B0CA280DA382CBA2BC8E65857F450A983385735AD741658F8ABF94E1026&originRegion=us-east-1&originCreation=20220907150920 (accessed on 7 September 2022).
- Koike, N.; Yoo, S.-H.; Huang, H.-C.; Kumar, V.; Lee, C.; Kim, T.-K.; Takahashi, J.S. Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef]
- The Mammalian Circadian Timing System: Organization and Coordination of Central and Peripheral Clocks|Annual Review of Physiology. Available online: https://www.annualreviews.org/doi/pdf/10.1146/annurev-physiol-021909-135821 (accessed on 17 May 2022).
- Kawara, S.; Mydlarski, R.; Mamelak, A.J.; Freed, I.; Wang, B.; Watanabe, H.; Shivji, G.; Tavadia, S.K.; Suzuki, H.; Bjarnason, G.A.; et al. Low-Dose Ultraviolet B Rays Alter the MRNA Expression of the Circadian Clock Genes in Cultured Human Keratinocytes. J. Investig. Dermatol. 2002, 119, 1220–1223. [Google Scholar] [CrossRef]
- Lin, K.K.; Chudova, D.; Hatfield, G.W.; Smyth, P.; Andersen, B. Identification of Hair Cycle-Associated Genes from Time-Course Gene Expression Profile Data by Using Replicate Variance. Proc. Natl. Acad. Sci. USA 2004, 101, 15955–15960. [Google Scholar] [CrossRef]
- Zanello, S.B.; Jackson, D.M.; Holick, M.F. Expression of the Circadian Clock Genes Clock and Period1 in Human Skin. J. Investig. Dermatol. 2000, 115, 757–760. [Google Scholar] [CrossRef]
- Tanioka, M.; Yamada, H.; Doi, M.; Bando, H.; Yamaguchi, Y.; Nishigori, C.; Okamura, H. Molecular Clocks in Mouse Skin. J. Investig. Dermatol. 2009, 129, 1225–1231. [Google Scholar] [CrossRef]
- Al-Nuaimi, Y.; Hardman, J.A.; Bíró, T.; Haslam, I.S.; Philpott, M.P.; Tóth, B.I.; Farjo, N.; Farjo, B.; Baier, G.; Watson, R.E.B.; et al. A Meeting of Two Chronobiological Systems: Circadian Proteins Period1 and BMAL1 Modulate the Human Hair Cycle Clock. J. Investig. Dermatol. 2014, 134, 610–619. [Google Scholar] [CrossRef]
- Brain and Muscle Arnt-like Protein-1 (BMAL1) Controls Circadian Cell Proliferation and Susceptibility to UVB-Induced DNA Damage in the Epidermis. Available online: https://www.pnas.org/doi/10.1073/pnas.1209592109 (accessed on 27 March 2022).
- Gekakis, N.; Staknis, D.; Nguyen, H.B.; Davis, F.C.; Wilsbacher, L.D.; King, D.P.; Takahashi, J.S.; Weitz, C.J. Role of the CLOCK Protein in the Mammalian Circadian Mechanism. Science 1998, 280, 1564–1569. [Google Scholar] [CrossRef]
- Bunger, M.K.; Wilsbacher, L.D.; Moran, S.M.; Clendenin, C.; Radcliffe, L.A.; Hogenesch, J.B.; Simon, M.C.; Takahashi, J.S.; Bradfield, C.A. Mop3 Is an Essential Component of the Master Circadian Pacemaker in Mammals. Cell 2000, 103, 1009–1017. [Google Scholar] [CrossRef]
- Crane, B.R.; Young, M.W. Interactive Features of Proteins Composing Eukaryotic Circadian Clocks. Annu. Rev. Biochem. 2014, 83, 191–219. [Google Scholar] [CrossRef]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The Orphan Nuclear Receptor REV-ERBalpha Controls Circadian Transcription within the Positive Limb of the Mammalian Circadian Oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Duez, H.; Staels, B. The Nuclear Receptors Rev-Erbs and RORs Integrate Circadian Rhythms and Metabolism. Diabetes Vasc. Dis. Res. 2008, 5, 82–88. [Google Scholar] [CrossRef]
- Solt, L.A.; Kojetin, D.J.; Burris, T.P. The REV-ERBs and RORs: Molecular Links between Circadian Rhythms and Lipid Homeostasis. Future Med. Chem. 2011, 3, 623–638. [Google Scholar] [CrossRef]
- Mechanisms Linking Circadian Clocks, Sleep, and Neurodegeneration|Science. Available online: https://www.science.org/doi/10.1126/science.aah4968 (accessed on 3 November 2022).
- Fang, B.; Everett, L.J.; Jager, J.; Briggs, E.; Armour, S.M.; Feng, D.; Roy, A.; Gerhart-Hines, Z.; Sun, Z.; Lazar, M.A. Circadian Enhancers Coordinate Multiple Phases of Rhythmic Gene Transcription In Vivo. Cell 2014, 159, 1140–1152. [Google Scholar] [CrossRef]
- Blue Light Disrupts the Circadian Rhythm and Create Damage in Skin Cells—Dong—2019—International Journal of Cosmetic Science—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/10.1111/ics.12572 (accessed on 31 March 2022).
- Sun, Y.; Wang, P.; Li, H.; Dai, J. BMAL1 and CLOCK Proteins in Regulating UVB-Induced Apoptosis and DNA Damage Responses in Human Keratinocytes. J. Cell. Physiol. 2018, 233, 9563–9574. [Google Scholar] [CrossRef]
- Husse, J.; Leliavski, A.; Tsang, A.H.; Oster, H.; Eichele, G. The Light-dark Cycle Controls Peripheral Rhythmicity in Mice with a Genetically Ablated Suprachiasmatic Nucleus Clock. FASEB J. 2014, 28, 4950–4960. [Google Scholar] [CrossRef]
- Festa, E.; Fretz, J.; Berry, R.; Schmidt, B.; Rodeheffer, M.; Horowitz, M.; Horsley, V. Adipocyte Lineage Cells Contribute to the Skin Stem Cell Niche to Drive Hair Cycling. Cell 2011, 146, 761–771. [Google Scholar] [CrossRef]
- Enshell-Seijffers, D.; Lindon, C.; Kashiwagi, M.; Morgan, B.A. β-Catenin Activity in the Dermal Papilla Regulates Morphogenesis and Regeneration of Hair. Dev. Cell 2010, 18, 633–642. [Google Scholar] [CrossRef]
- Hardman, J.A. The Peripheral Clock Regulates Human Pigmentation. J. Investig. Dermatol. 2015, 135, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Welz, P.-S.; Zinna, V.M.; Symeonidi, A.; Koronowski, K.B.; Kinouchi, K.; Smith, J.G.; Guillén, I.M.; Castellanos, A.; Furrow, S.; Aragón, F.; et al. BMAL1-Driven Tissue Clocks Respond Independently to Light to Maintain Homeostasis. Cell 2019, 177, 1436–1447.e12. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.M.-Y.; Chang, Y.-T.; Chen, C.-L.; Wang, W.-H.; Pan, M.-K.; Chen, W.-P.; Huang, W.-Y.; Xu, Z.; Huang, H.-E.; Chen, T.; et al. External Light Activates Hair Follicle Stem Cells through Eyes via an IpRGC–SCN–Sympathetic Neural Pathway. Proc. Natl. Acad. Sci. USA 2018, 115, E6880–E6889. [Google Scholar] [CrossRef] [PubMed]
- Welz, P.-S.; Benitah, S.A. Molecular Connections Between Circadian Clocks and Aging. J. Mol. Biol. 2020, 432, 3661–3679. [Google Scholar] [CrossRef]
- Yu, Y. A Stress-Induced MiR-31–CLOCK–ERK Pathway Is a Key Driver and Therapeutic Target for Skin Aging. Nat. Aging 2021, 1, 39. [Google Scholar] [CrossRef]
- Kanathur, N.; Harrington, J.; Lee-Chiong, T. Circadian Rhythm Sleep Disorders. Clin. Chest Med. 2010, 31, 319–325. [Google Scholar] [CrossRef]
- Seo, H.-M.; Kim, T.L.; Kim, J.S. The Risk of Alopecia Areata and Other Related Autoimmune Diseases in Patients with Sleep Disorders: A Korean Population-Based Retrospective Cohort Study. Sleep 2018, 41, zsy111. [Google Scholar] [CrossRef]
- Flores, A.; Schell, J.; Krall, A.S.; Jelinek, D.; Miranda, M.; Grigorian, M.; Braas, D.; White, A.C.; Zhou, J.L.; Graham, N.A.; et al. Lactate Dehydrogenase Activity Drives Hair Follicle Stem Cell Activation. Nat. Cell Biol. 2017, 19, 1017–1026. [Google Scholar] [CrossRef]
- Williams, R.; Philpott, M.P.; Kealey, T. Metabolism of Freshly Isolated Human Hair Follicles Capable of Hair Elongation: A Glutaminolytic, Aerobic Glycolytic Tissue. J. Investig. Dermatol. 1993, 100, 834–840. [Google Scholar] [CrossRef]
- Choi, B.Y. Targeting Wnt/β-Catenin Pathway for Developing Therapies for Hair Loss. Int. J. Mol. Sci. 2020, 21, 4915. [Google Scholar] [CrossRef]
- Carrasco, E.; Calvo, M.I.; Blázquez-Castro, A.; Vecchio, D.; Zamarrón, A.; de Almeida, I.J.D.; Stockert, J.C.; Hamblin, M.R.; Juarranz, Á.; Espada, J. Photoactivation of ROS Production in Situ Transiently Activates Cell Proliferation in Mouse Skin and in the Hair Follicle Stem Cell Niche Promoting Hair Growth and Wound Healing. J. Investig. Dermatol. 2015, 135, 2611–2622. [Google Scholar] [CrossRef] [PubMed]
- Liu, M. Intrinsic ROS Drive Hair Follicle Cycle Progression by Modulating DNA Damage and Repair, and Subsequently, Hair Follicle Apoptosis and Macrophage Polarization. Oxidative Med. Cell. Longev. 2022, 2022, 8279269. [Google Scholar] [CrossRef] [PubMed]
- Gharibi, B.; Farzadi, S.; Ghuman, M.; Hughes, F.J. Inhibition of Akt/MTOR Attenuates Age-Related Changes in Mesenchymal Stem Cells. Stem Cells 2014, 32, 2256–2266. [Google Scholar] [CrossRef]
- Harshuk-Shabso, S.; Dressler, H.; Niehrs, C.; Aamar, E.; Enshell-Seijffers, D. Fgf and Wnt Signaling Interaction in the Mesenchymal Niche Regulates the Murine Hair Cycle Clock. Nat. Commun. 2020, 11, 5114. [Google Scholar] [CrossRef]
- Yu, Z.; Jiang, K.; Xu, Z.; Huang, H.; Qian, N.; Lu, Z.; Chen, D.; Di, R.; Yuan, T.; Du, Z.; et al. Hoxc-Dependent Mesenchymal Niche Heterogeneity Drives Regional Hair Follicle Regeneration. Cell Stem Cell 2018, 23, 487–500.e6. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Quondamatteo, F.; Lefever, T.; Czuchra, A.; Meyer, H.; Chrostek, A.; Paus, R.; Langbein, L.; Brakebusch, C. Cdc42 Controls Progenitor Cell Differentiation and β-Catenin Turnover in Skin. Genes Dev. 2006, 20, 571–585. [Google Scholar] [CrossRef]
- Wang, G.; Sweren, E.; Andrews, W.; Li, Y.; Chen, J.; Xue, Y.; Wier, E.; Alphonse, M.P.; Luo, L.; Miao, Y.; et al. Commensal Microbiome Promotes Hair Follicle Regeneration by Inducing Keratinocyte HIF-1α Signaling and Glutamine Metabolism. Sci. Adv. 2023, 9, eabo7555. [Google Scholar] [CrossRef]
- Zhang, E.E.; Kay, S.A. Clocks Not Winding down: Unravelling Circadian Networks. Nat. Rev. Mol. Cell Biol. 2010, 11, 764–776. [Google Scholar] [CrossRef]
- Koronowski, K.B.; Sassone-Corsi, P. Communicating Clocks Shape Circadian Homeostasis. Science 2021, 371, eabd0951. [Google Scholar] [CrossRef]
- Peng, F.; Li, X.; Xiao, F.; Zhao, R.; Sun, Z. Circadian Clock, Diurnal Glucose Metabolic Rhythm, and Dawn Phenomenon. Trends Neurosci. 2022, 45, 471–482. [Google Scholar] [CrossRef]
- Krishnaiah, S.Y.; Wu, G.; Altman, B.J.; Growe, J.; Rhoades, S.D.; Coldren, F.; Venkataraman, A.; Olarerin-George, A.O.; Francey, L.J.; Mukherjee, S.; et al. Clock Regulation of Metabolites Reveals Coupling between Transcription and Metabolism. Cell Metab. 2017, 25, 961–974.e4. [Google Scholar] [CrossRef]
- Rudic, R.D.; McNamara, P.; Curtis, A.-M.; Boston, R.C.; Panda, S.; Hogenesch, J.B.; FitzGerald, G.A. BMAL1 and CLOCK, Two Essential Components of the Circadian Clock, Are Involved in Glucose Homeostasis. PLoS Biol. 2004, 2, e377. [Google Scholar] [CrossRef] [PubMed]
- Dissecting the Functions of the Mammalian Clock Protein BMAL1 by Tissue-Specific Rescue in Mice. Available online: https://www.science.org/doi/10.1126/science.1132430 (accessed on 19 July 2022).
- Harfmann, B.D.; Schroder, E.A.; Kachman, M.T.; Hodge, B.A.; Zhang, X.; Esser, K.A. Muscle-Specific Loss of Bmal1 Leads to Disrupted Tissue Glucose Metabolism and Systemic Glucose Homeostasis. Skelet. Muscle 2016, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Obesity in Mice with Adipocyte-Specific Deletion of Clock Component Arntl|Nature Medicine. Available online: https://www.nature.com/articles/nm.2979 (accessed on 26 July 2022).
- Pan, X.; Zhang, Y.; Wang, L.; Hussain, M.M. Diurnal Regulation of MTP and Plasma Triglyceride by CLOCK Is Mediated by SHP. Cell Metab. 2010, 12, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Obesity and Metabolic Syndrome in Circadian Clock Mutant Mice|Science. Available online: https://www.science.org/doi/10.1126/science.1108750 (accessed on 3 August 2022).
- Attenuating Effect of Clock Mutation on Triglyceride Contents in the ICR Mouse Liver under a High-Fat Diet—Takashi Kudo, Toru Tamagawa, Mihoko Kawashima, Natsuko Mito, Shigenobu Shibata. 2007. Available online: https://journals.sagepub.com/doi/10.1177/0748730407302625 (accessed on 3 August 2022).
- Ameneiro, C.; Moreira, T.; Fuentes-Iglesias, A.; Coego, A.; Garcia-Outeiral, V.; Escudero, A.; Torrecilla, D.; Mulero-Navarro, S.; Carvajal-Gonzalez, J.M.; Guallar, D.; et al. BMAL1 Coordinates Energy Metabolism and Differentiation of Pluripotent Stem Cells. Life Sci. Alliance 2020, 3, e201900534. [Google Scholar] [CrossRef]
- Wu, Y.; Tang, D.; Liu, N.; Xiong, W.; Huang, H.; Li, Y.; Ma, Z.; Zhao, H.; Chen, P.; Qi, X.; et al. Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab. 2017, 25, 73–85. [Google Scholar] [CrossRef]
- Peek, C.B.; Levine, D.C.; Cedernaes, J.; Taguchi, A.; Kobayashi, Y.; Tsai, S.J.; Bonar, N.A.; McNulty, M.R.; Ramsey, K.M.; Bass, J. Circadian Clock Interaction with HIF1α Mediates Oxygenic Metabolism and Anaerobic Glycolysis in Skeletal Muscle. Cell Metab. 2017, 25, 86–92. [Google Scholar] [CrossRef]
- Adamovich, Y.; Ladeuix, B.; Golik, M.; Koeners, M.P.; Asher, G. Rhythmic Oxygen Levels Reset Circadian Clocks through HIF1α. Cell Metab. 2017, 25, 93–101. [Google Scholar] [CrossRef]
- Early, J.O.; Menon, D.; Wyse, C.A.; Cervantes-Silva, M.P.; Zaslona, Z.; Carroll, R.G.; Palsson-McDermott, E.M.; Angiari, S.; Ryan, D.G.; Corcoran, S.E.; et al. Circadian Clock Protein BMAL1 Regulates IL-1β in Macrophages via NRF2. Proc. Natl. Acad. Sci. USA 2018, 115, E8460–E8468. [Google Scholar] [CrossRef]
- Wang, X.-L.; Li, L. Circadian Clock Regulates Inflammation and the Development of Neurodegeneration. Front. Cell. Infect. Microbiol. 2021, 11, 696554. [Google Scholar] [CrossRef]
- Garten, A.; Schuster, S.; Penke, M.; Gorski, T.; de Giorgis, T.; Kiess, W. Physiological and Pathophysiological Roles of NAMPT and NAD Metabolism. Nat. Rev. Endocrinol. 2015, 11, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, Y.; Sahar, S.; Astarita, G.; Kaluzova, M.; Sassone-Corsi, P. Circadian Control of the NAD+ Salvage Pathway by CLOCK-SIRT1. Science 2009, 324, 654–657. [Google Scholar] [CrossRef]
- Ramsey, K.M.; Yoshino, J.; Brace, C.S.; Abrassart, D.; Kobayashi, Y.; Marcheva, B.; Hong, H.-K.; Chong, J.L.; Buhr, E.D.; Lee, C.; et al. Circadian Clock Feedback Cycle Through NAMPT-Mediated NAD+ Biosynthesis. Science 2009, 324, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Asher, G.; Gatfield, D.; Stratmann, M.; Reinke, H.; Dibner, C.; Kreppel, F.; Mostoslavsky, R.; Alt, F.W.; Schibler, U. SIRT1 Regulates Circadian Clock Gene Expression through PER2 Deacetylation. Cell 2008, 134, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Asher, G.; Reinke, H.; Altmeyer, M.; Gutierrez-Arcelus, M.; Hottiger, M.O.; Schibler, U. Poly(ADP-Ribose) Polymerase 1 Participates in the Phase Entrainment of Circadian Clocks to Feeding. Cell 2010, 142, 943–953. [Google Scholar] [CrossRef]
- Wu, R.; Dang, F.; Li, P.; Wang, P.; Xu, Q.; Liu, Z.; Li, Y.; Wu, Y.; Chen, Y.; Liu, Y. The Circadian Protein Period2 Suppresses MTORC1 Activity via Recruiting Tsc1 to MTORC1 Complex. Cell Metab. 2019, 29, 653–667.e6. [Google Scholar] [CrossRef]
- Khapre, R.V.; Kondratova, A.A.; Patel, S.; Dubrovsky, Y.; Wrobel, M.; Antoch, M.P.; Kondratov, R.V. BMAL1-Dependent Regulation of the MTOR Signaling Pathway Delays Aging. Aging 2014, 6, 48–57. [Google Scholar] [CrossRef]
- Fukuda, Y.; Morita, T. Effects of the Light–Dark Cycle on Diurnal Rhythms of Diet-Induced Thermogenesis in Humans. Chronobiol. Int. 2017, 34, 1465–1472. [Google Scholar] [CrossRef]
- Lund, J.; Arendt, J.; Hampton, S.M.; English, J.; Morgan, L.M. Postprandial Hormone and Metabolic Responses amongst Shift Workers in Antarctica. J. Endocrinol. 2001, 171, 557–564. Available online: https://joe.bioscientifica.com/view/journals/joe/171/3/557.xml (accessed on 8 July 2022).
- Coomans, C.P.; van den Berg, S.A.A.; Houben, T.; van Klinken, J.-B.; van den Berg, R.; Pronk, A.C.M.; Havekes, L.M.; Romijn, J.A.; van Dijk, K.W.; Biermasz, N.R.; et al. Detrimental Effects of Constant Light Exposure and High-Fat Diet on Circadian Energy Metabolism and Insulin Sensitivity. FASEB J. 2013, 27, 1721–1732. [Google Scholar] [CrossRef]
- Borck, P.C.; Batista, T.M.; Vettorazzi, J.F.; Soares, G.M.; Lubaczeuski, C.; Guan, D.; Boschero, A.C.; Vieira, E.; Lazar, M.A.; Carneiro, E.M. Nighttime Light Exposure Enhances Rev-Erbα-Targeting MicroRNAs and Contributes to Hepatic Steatosis. Metabolism 2018, 85, 250–258. [Google Scholar] [CrossRef]
- Light at Night Increases Body Mass by Shifting the Time of Food Intake. Available online: https://www.pnas.org/doi/10.1073/pnas.1008734107 (accessed on 11 September 2022).
- Alaasam, V.J.; Liu, X.; Niu, Y.; Habibian, J.S.; Pieraut, S.; Ferguson, B.S.; Zhang, Y.; Ouyang, J.Q. Effects of Dim Artificial Light at Night on Locomotor Activity, Cardiovascular Physiology, and Circadian Clock Genes in a Diurnal Songbird. Environ. Pollut. 2021, 282, 117036. [Google Scholar] [CrossRef] [PubMed]
- Mezhnina, V.; Ebeigbe, O.P.; Poe, A.; Kondratov, R.V. Circadian Control of Mitochondria in Reactive Oxygen Species Homeostasis. Antioxid. Redox Signal. 2022, 37, 647–663. [Google Scholar] [CrossRef] [PubMed]
- Fanjul-Moles, M.L.; López-Riquelme, G.O. Relationship between Oxidative Stress, Circadian Rhythms, and AMD. Oxid. Med. Cell. Longev. 2016, 2016, 7420637. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.; Li, H.; Gao, L.; Guo, W.; Xu, S.; Li, L. PER2 Regulates Reactive Oxygen Species Production in the Circadian Susceptibility to Ischemia/Reperfusion Injury in the Heart. Oxid. Med. Cell. Longev. 2021, 2021, 6256399. [Google Scholar] [CrossRef] [PubMed]
- Son, M.J.; Jeong, J.K.; Kwon, Y.; Ryu, J.-S.; Mun, S.J.; Kim, H.J.; Kim, S.-W.; Yoo, S.; Kook, J.; Lee, H.; et al. A Novel and Safe Small Molecule Enhances Hair Follicle Regeneration by Facilitating Metabolic Reprogramming. Exp. Mol. Med. 2018, 50, 1–15. [Google Scholar] [CrossRef]
- Morgan, B.A. The Dermal Papilla: An Instructive Niche for Epithelial Stem and Progenitor Cells in Development and Regeneration of the Hair Follicle. Cold Spring Harb. Perspect. Med. 2014, 4, a015180. [Google Scholar] [CrossRef]
- Avigad Laron, E.; Aamar, E.; Enshell-Seijffers, D. The Mesenchymal Niche of the Hair Follicle Induces Regeneration by Releasing Primed Progenitors from Inhibitory Effects of Quiescent Stem Cells. Cell Rep. 2018, 24, 909–921. [Google Scholar] [CrossRef]
- Legrand, J.M.D.; Roy, E.; Ellis, J.J.; Francois, M.; Brooks, A.J.; Khosrotehrani, K. STAT5 Activation in the Dermal Papilla is Important for Hair Follicle Growth Phase Induction. J. Investig. Dermatol. 2016, 136, 1781–1791. [Google Scholar] [CrossRef]
- Wang, B.; Liu, X.-M.; Liu, Z.-N.; Wang, Y.; Han, X.; Lian, A.-B.; Mu, Y.; Jin, M.-H.; Liu, J.-Y. Human Hair Follicle-Derived Mesenchymal Stem Cells: Isolation, Expansion, and Differentiation. World J. Stem Cells 2020, 12, 462–470. [Google Scholar] [CrossRef]
- Timmons, G.A.; Carroll, R.G.; O’Siorain, J.R.; Cervantes-Silva, M.P.; Fagan, L.E.; Cox, S.L.; Palsson-McDermott, E.; Finlay, D.K.; Vincent, E.E.; Jones, N.; et al. The Circadian Clock Protein BMAL1 Acts as a Metabolic Sensor In Macrophages to Control the Production of Pro IL-1β. Front. Immunol. 2021, 12, 700431. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, Y.; Wang, Y.; Chen, H.; Liu, X.; Liu, J. Overview of the Circadian Clock in the Hair Follicle Cycle. Biomolecules 2023, 13, 1068. https://doi.org/10.3390/biom13071068
Niu Y, Wang Y, Chen H, Liu X, Liu J. Overview of the Circadian Clock in the Hair Follicle Cycle. Biomolecules. 2023; 13(7):1068. https://doi.org/10.3390/biom13071068
Chicago/Turabian StyleNiu, Ye, Yujie Wang, Hao Chen, Xiaomei Liu, and Jinyu Liu. 2023. "Overview of the Circadian Clock in the Hair Follicle Cycle" Biomolecules 13, no. 7: 1068. https://doi.org/10.3390/biom13071068
APA StyleNiu, Y., Wang, Y., Chen, H., Liu, X., & Liu, J. (2023). Overview of the Circadian Clock in the Hair Follicle Cycle. Biomolecules, 13(7), 1068. https://doi.org/10.3390/biom13071068




