Nuclear Phosphoinositides as Key Determinants of Nuclear Functions
Abstract
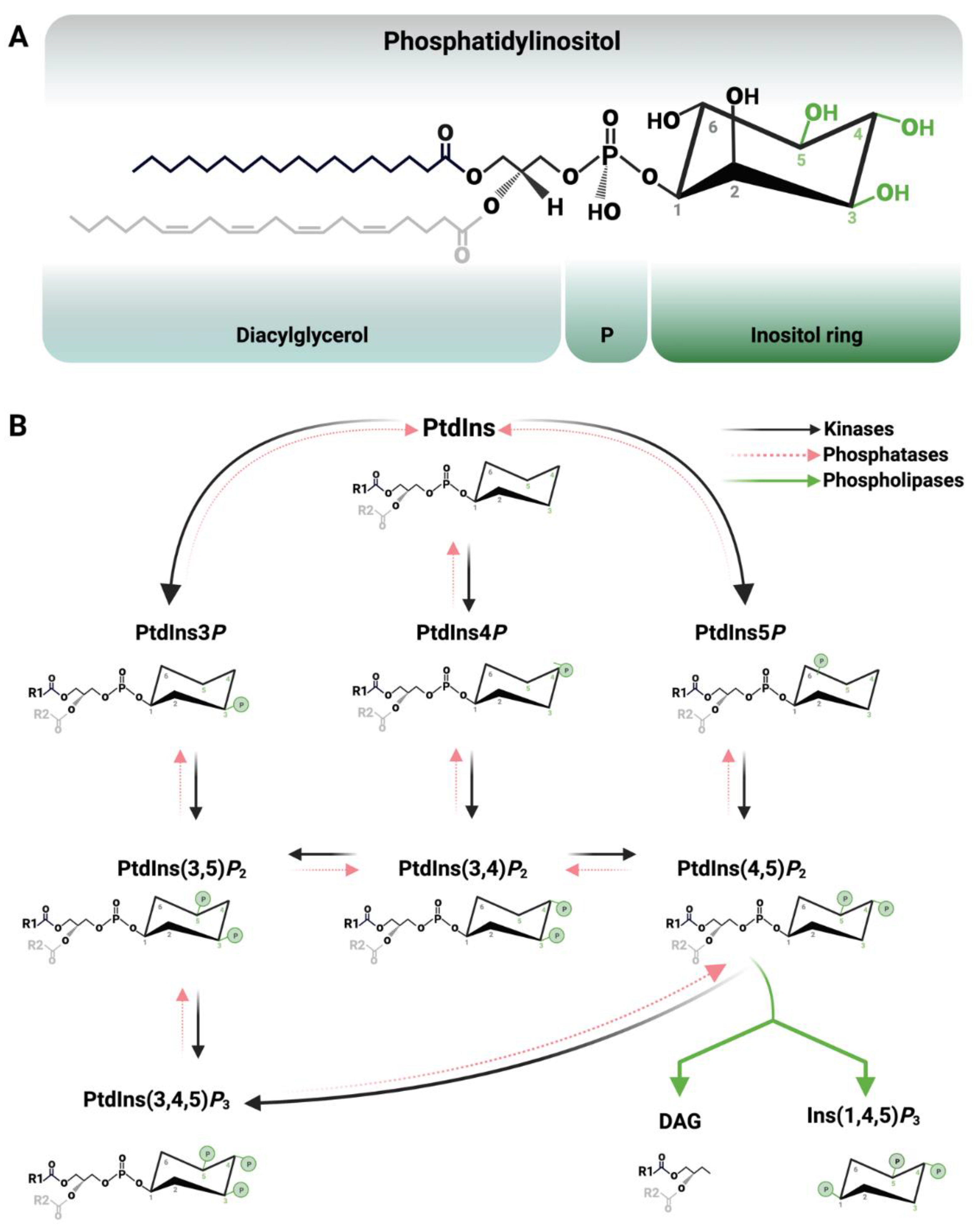
1. Introduction
| Protein Domain | Phosphoinositide(s) Bound | References |
|---|---|---|
| Pleckstrin homology (PH) domain | PtdIns3P, PtdIns4P, PtdIns(4,5)P2, PtdIns(3,4)P2 PtdIns(3,4,5)P3 | [10,11,12,13,14,33,34,35,36] |
| Phox homology (PX) domain | PtdIns3P, PtdIns(3,4)P2, PtdIns(4,5)P2 | [37,38,39] |
| Plant homeodomain (PHD) | PtdIns3P, PtdIns5P | [40,41] |
| FYVE domain | PtdIns3P | [42] |
| ENTH domain | PtdIns(4,5)P2 | [43] |
| ANTH domain | PtdIns(4,5)P2 | [44] |
| Polybasic domains | PtdIns(4,5)P2 | [45,46,47] |
| Tubby | PtdIns(4,5)P2 | [48,49] |
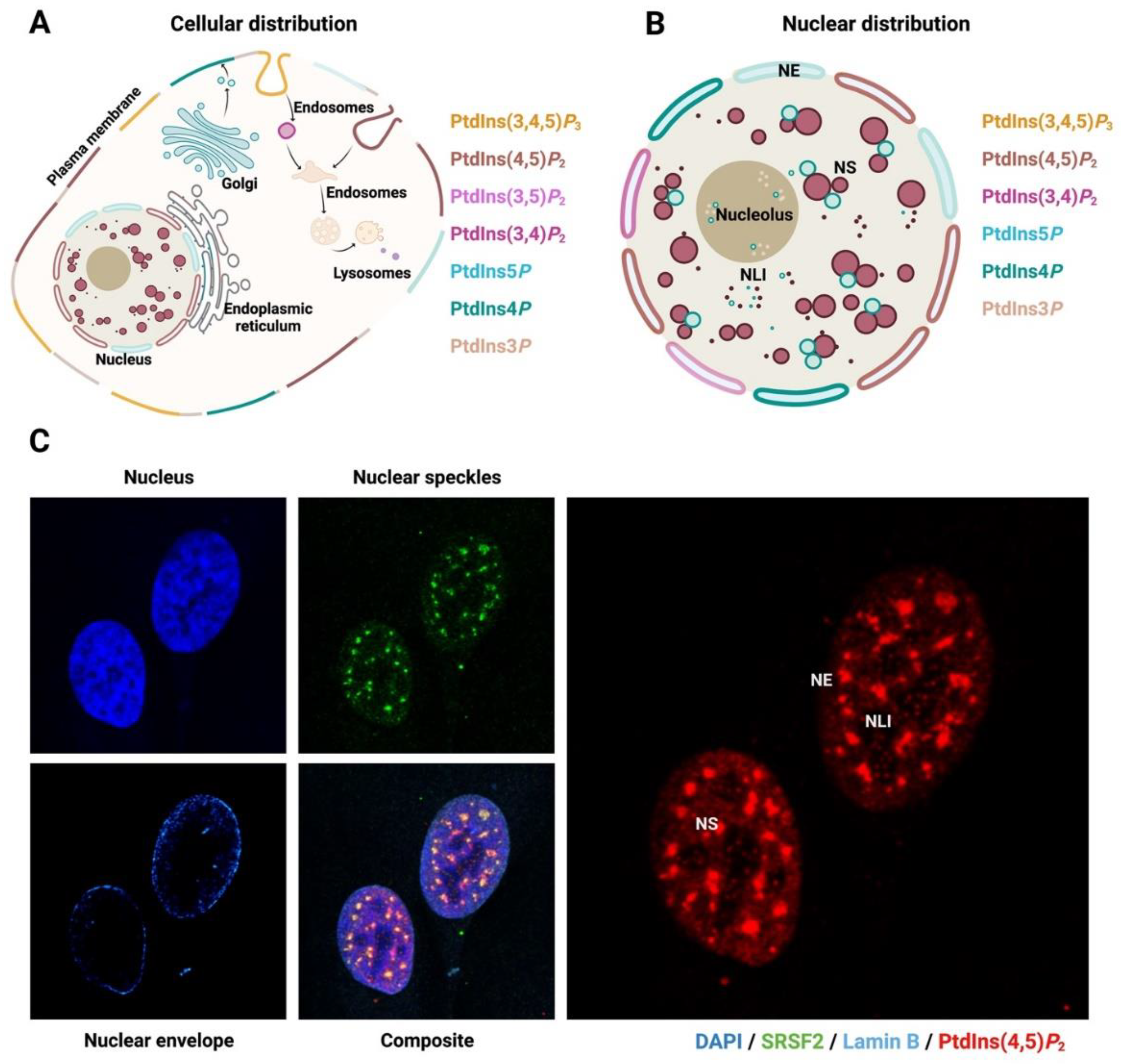
2. The Nucleus and Nuclear PPIn Transport
3. Biogenesis of Nuclear Phosphoinositides and Their Metabolising Enzymes
3.1. PtdIns3P
3.2. PtdIns4P
3.3. PtdIns5P
3.4. PtdIns(3,4)P2
3.5. PtdIns(4,5)P2
3.6. PtdIns(3,4,5)P3
4. Physiological Functions of Nuclear Phosphoinositide
4.1. Nuclear PPIns as Regulators of Histone Modifications
4.2. Nuclear PPIns and Their Role in Defining How Histone Modifications Drive Downstream Signalling Outputs
4.3. Nuclear PPIns and Regulation of Transcription Factors
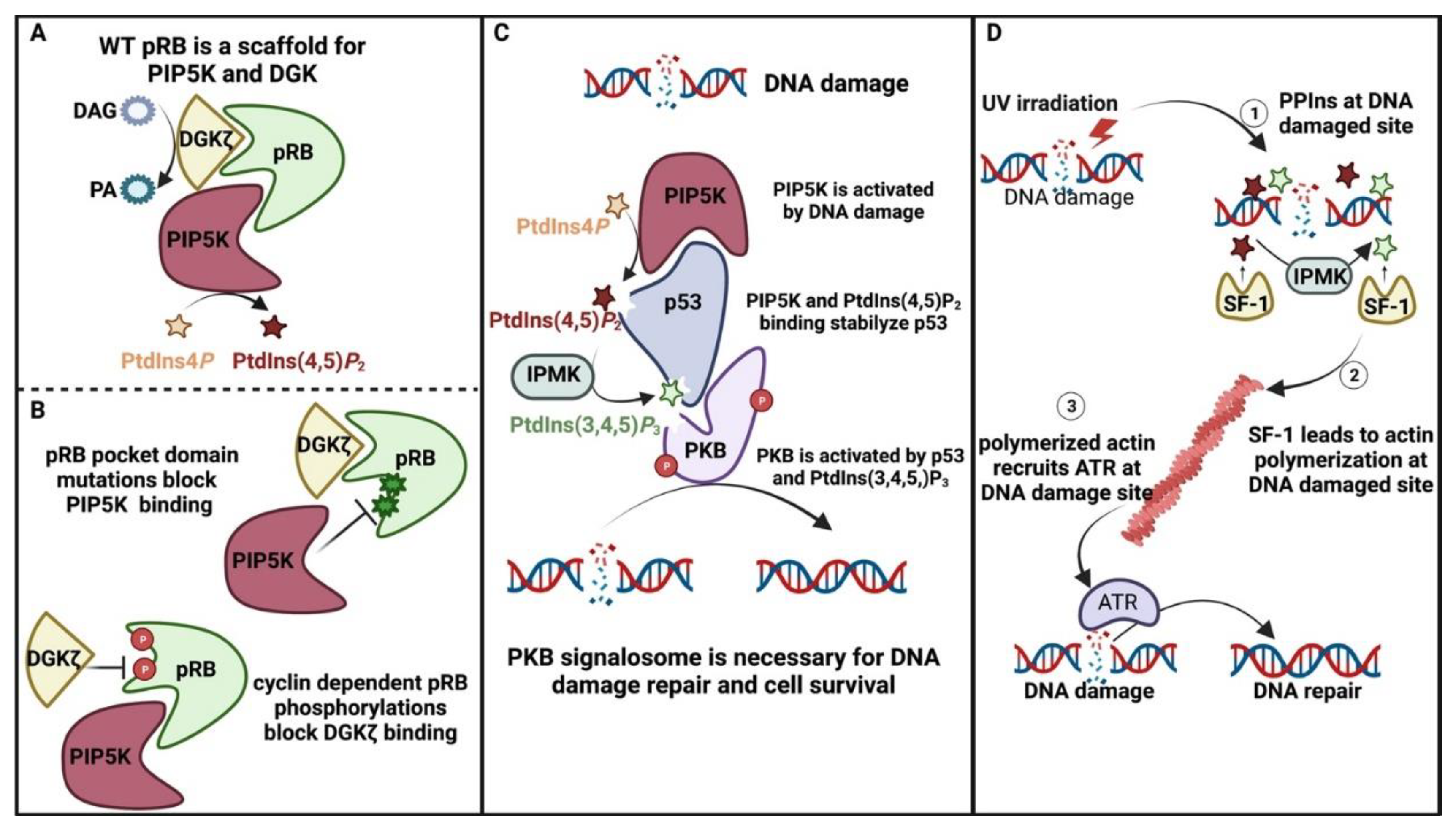
4.4. Nuclear PPIns and Their Role in Protecting the Genome
4.5. Nuclear Speckle and mRNA Machinery
5. Concluding Notes
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Balla, T.; Szentpetery, Z.; Kim, Y.J.; Dingjan, I.; Linders, P.T.A.; Verboogen, D.R.J.; Revelo, N.H.; ter Beest, M.; Bogaart, G.v.D.; Sbrissa, D.; et al. Phosphoinositide Signaling: New Tools and Insights. Physiology 2009, 24, 231–244. [Google Scholar] [CrossRef]
- Balla, T. Phosphoinositides: Tiny Lipids With Giant Impact on Cell Regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef]
- Morelec-Coulon, M.J.; Faure, M. Glycerol-inositol-phosphatidic acid. IV. Molecular structure. Bull. Soc. Chim. Biol. 1958, 40, 1307–1313. [Google Scholar]
- Hawthorne, J.N. The inositol phospholipids. J. Lipid Res. 1960, 1, 255–280. [Google Scholar] [CrossRef] [PubMed]
- Hawthorne, J.N.; Kemp, P.; Ellis, R.B. Phosphoinositides. 2. The inositol 1-phosphate structure in liver phosphatidylinositol. Biochem. J. 1960, 75, 501. [Google Scholar] [CrossRef] [PubMed]
- Irvine, R.F. Nuclear lipid signalling. Nat. Rev. Mol. Cell Biol. 2003, 4, 349–361. [Google Scholar] [CrossRef]
- Lemmon, M.A. Phosphoinositide Recognition Domains. Traffic 2003, 4, 201–213. [Google Scholar] [CrossRef]
- Lemmon, M. Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 2007, 74, 81–93. [Google Scholar] [CrossRef]
- Hammond, G.R.; Balla, T. Polyphosphoinositide binding domains: Key to inositol lipid biology. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2015, 1851, 746–758. [Google Scholar] [CrossRef]
- Ferguson, K.M.; Lemmon, M.A.; Schlessinger, J.; Sigler, P.B. Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 1995, 83, 1037–1046. [Google Scholar] [CrossRef]
- Harlan, J.E.; Hajduk, P.J.; Yoon, H.S.; Fesik, S.W. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature 1994, 371, 168–170. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Reyes-Ordoñez, A.; Compagnone, M.A.; Moreno, J.F.; Leslie, B.J.; Ha, T.; Chen, J. Redefining the specificity of phosphoinositide-binding by human PH domain-containing proteins. Nat. Commun. 2021, 12, 4339. [Google Scholar] [CrossRef] [PubMed]
- Stauffer, T.P.; Ahn, S.; Meyer, T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 1998, 8, 343–346. [Google Scholar] [CrossRef]
- Guillou, H.; Lécureuil, C.; Anderson, K.A.; Suire, S.; Ferguson, G.J.; Ellson, C.D.; Gray, A.; Divecha, N.; Hawkins, P.T.; Stephens, L.R. Use of the GRP1 PH domain as a tool to measure the relative levels of PtdIns(3,4,5)P3 through a protein-lipid overlay approach. J. Lipid Res. 2007, 48, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.; Anderson, K.E.; Juvin, V.; Smith, T.S.; Karpe, F.; Wakelam, M.J.O.; Stephens, L.R.; Hawkins, P.T. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods 2011, 8, 267–272. [Google Scholar] [CrossRef]
- Milne, S.B.; Ivanova, P.T.; DeCamp, D.; Hsueh, R.C.; Brown, H.A. A targeted mass spectrometric analysis of phosphatidylinositol phosphate species. J. Lipid Res. 2005, 46, 1796–1802. [Google Scholar] [CrossRef]
- Vadnal, R.E.; Parthasarathy, R. The identification of a novel inositol lipid, phosphatidylinostiol trisphosphate (PIP3), in rat cerebrum using in vivo techniques. Biochem. Biophys. Res. Commun. 1989, 163, 995–1001. [Google Scholar] [CrossRef]
- Lands, W.E. Metabolism of glycerolipids. 2. The enzymatic acylation of lysolecithin. J. Biol. Chem. 1960, 235, 2233–2237. [Google Scholar] [CrossRef]
- Murphy, R.C.; Folco, G. Lysophospholipid acyltransferases and leukotriene biosynthesis: Intersection of the Lands cycle and the arachidonate PI cycle. J. Lipid Res. 2019, 60, 219–226. [Google Scholar] [CrossRef]
- Anderson, K.E.; Kielkowska, A.; Durrant, T.N.; Juvin, V.; Clark, J.; Stephens, L.R.; Hawkins, P.T. Lysophosphatidylinositol-Acyltransferase-1 (LPIAT1) Is Required to Maintain Physiological Levels of PtdIns and PtdInsP2 in the Mouse. PLoS ONE 2013, 8, e58425. [Google Scholar] [CrossRef]
- Barneda, D.; Cosulich, S.; Stephens, L.; Hawkins, P. How is the acyl chain composition of phosphoinositides created and does it matter? Biochem. Soc. Trans. 2019, 47, 1291–1305. [Google Scholar] [CrossRef] [PubMed]
- Patki, V.; Virbasius, J.; Lane, W.S.; Toh, B.-H.; Shpetner, H.S.; Corvera, S. Identification of an early endosomal protein regulated by phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA 1997, 94, 7326–7330. [Google Scholar] [CrossRef] [PubMed]
- Fiume, R.; Keune, W.J.; Faenza, I.; Bultsma, Y.; Ramazzotti, G.; Jones, D.R.; Martelli, A.M.; Somner, L.; Follo, M.Y.; Divecha, N. Nuclear phosphoinositides: Location, regulation and function. Phosphoinosit. II Divers. Biol. Funct. 2012, 59, 335–361. [Google Scholar]
- Di Paolo, P.G.; De, C.P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Giuriato, S.; Blero, D.; Robaye, B.; Bruyns, C.; Payrastre, B.; Erneux, C. SHIP2 overexpression strongly reduces the proliferation rate of K562 erythroleukemia cell line. Biochem. Biophys. Res. Commun. 2002, 296, 106–110. [Google Scholar] [CrossRef]
- Clément, S.; Krause, U.; Desmedt, F.; Tanti, J.-F.; Behrends, J.; Pesesse, X.; Sasaki, T.; Penninger, J.; Doherty, M.; Malaisse, W.; et al. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature 2001, 409, 92–97. [Google Scholar] [CrossRef]
- Luo, J.-M.; Yoshida, H.; Komura, S.; Ohishi, N.; Pan, L.; Shigeno, K.; Hanamura, I.; Miura, K.; Iida, S.; Ueda, R.; et al. Possible dominant-negative mutation of the SHIP gene in acute myeloid leukemia. Leukemia 2003, 17, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Marion, E.; Kaisaki, P.J.; Pouillon, V.; Gueydan, C.; Levy, J.C.; Bodson, A.; Krzentowski, G.; Daubresse, J.-C.; Mockel, J.; Behrends, J.; et al. The Gene INPPL1, Encoding the Lipid Phosphatase SHIP2, Is a Candidate for Type 2 Diabetes In Rat and Man. Diabetes 2002, 51, 2012–2017. [Google Scholar] [CrossRef]
- Halstead, J.R.; Jalink, K.; Divecha, N. An emerging role for PtdIns(4,5)P2-mediated signalling in human disease. Trends Pharmacol. Sci. 2005, 26, 654–660. [Google Scholar] [CrossRef]
- McCrea, H.J.; De Camilli, P.; Inoue, K.; Ishibe, S.; Silswal, N.; Parelkar, N.K.; Wacker, M.J.; Brotto, M.; Andresen, J. Mutations in Phosphoinositide Metabolizing Enzymes and Human Disease. Physiology 2009, 24, 8–16. [Google Scholar] [CrossRef]
- Carrat, G.R.; Haythorne, E.; Tomas, A.; Haataja, L.; Müller, A.; Arvan, P.; Piunti, A.; Cheng, K.; Huang, M.; Pullen, T.J.; et al. The type 2 diabetes gene product STARD10 is a phosphoinositide-binding protein that controls insulin secretory granule biogenesis. Mol. Metab. 2020, 40, 101015. [Google Scholar] [CrossRef] [PubMed]
- Bridges, D.; Saltiel, A.R. Phosphoinositides in Insulin Action and Diabetes. In Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2012; p. 362. [Google Scholar]
- Kavran, J.M.; Klein, D.E.; Lee, A.; Falasca, M.; Isakoff, S.J.; Skolnik, E.Y.; Lemmon, M.A. Specificity and Promiscuity in Phosphoinositide Binding by Pleckstrin Homology Domains. J. Biol. Chem. 1998, 273, 30497–30508. [Google Scholar] [CrossRef] [PubMed]
- Paterson, H.F.; Savopoulos, J.W.; Perisic, O.; Cheung, R.; Ellis, M.V.; Williams, R.L.; Katan, M. Phospholipase C δ1 requires a pleckstrin homology domain for interaction with the plasma membrane. Biochem. J. 1995, 312, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Yagisawa, H.; Sakuma, K.; Paterson, H.F.; Cheung, R.; Allen, V.; Hirata, H.; Watanabe, Y.; Hirata, M.; Williams, R.L.; Katan, M. Replacements of single basic amino acids in the pleckstrin homology domain of phospholipase C-delta1 alter the ligand binding, phospholipase activity, and interaction with the plasma membrane. J. Biol. Chem. 1998, 273, 417–424. [Google Scholar] [CrossRef]
- Kwon, I.-S.; Lee, K.-H.; Choi, J.W.; Ahn, J.-Y. PI(3,4,5)P3 regulates the interaction between Akt and B23 in the nucleus. BMB Rep. 2010, 43, 127–132. [Google Scholar] [CrossRef]
- Kanai, F.; Liu, H.; Field, S.; Akbary, H.; Matsuo, T.; Brown, G.E.; Cantley, L.; Yaffe, M.B. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nature 2001, 3, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Ellson, C.D.; Andrews, S.; Stephens, L.R.; Hawkins, P.T. The PX domain: A new phosphoinositide-binding module. J. Cell Sci. 2002, 115, 1099–1105. [Google Scholar] [CrossRef]
- Chandra, M.; Chin, Y.K.-Y.; Mas, C.; Feathers, J.R.; Paul, B.; Datta, S.; Chen, K.-E.; Jia, X.; Yang, Z.; Norwood, S.J.; et al. Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities. Nat. Commun. 2019, 10, 1528. [Google Scholar] [CrossRef]
- Gozani, O.; Karuman, P.; Jones, D.R.; Ivanov, D.; Cha, J.; Lugovskoy, A.A.; Baird, C.L.; Zhu, H.; Field, S.J.; Lessnick, S.L.; et al. The PHD Finger of the Chromatin-Associated Protein ING2 Functions as a Nuclear Phosphoinositide Receptor. Cell 2003, 114, 99–111. [Google Scholar] [CrossRef]
- Kaadige, M.R.; Ayer, D.E. The Polybasic Region That Follows the Plant Homeodomain Zinc Finger 1 of Pf1 Is Necessary and Sufficient for Specific Phosphoinositide Binding. J. Biol. Chem. 2006, 281, 28831–28836. [Google Scholar] [CrossRef]
- Stenmark, H.; Aasland, R.; Driscoll, P. The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett. 2002, 513, 77–84. [Google Scholar] [CrossRef]
- Itoh, T.; Koshiba, S.; Kigawa, T.; Kikuchi, A.; Yokoyama, S.; Takenawa, T. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science 2001, 291, 1047–1051. [Google Scholar] [CrossRef] [PubMed]
- Ford, M.G.; Pearse, B.M.; Higgins, M.K.; Vallis, Y.; Owen, D.J.; Gibson, A.; Hopkins, C.R.; Evans, P.R.; McMahon, H.T. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 2001, 291, 1051–1055. [Google Scholar] [CrossRef]
- Lewis, A.E.; Sommer, L.; Arntzen, M.; Strahm, Y.; Morrice, N.A.; Divecha, N.; D’Santos, C.S. Identification of Nuclear Phosphatidylinositol 4,5-Bisphosphate-Interacting Proteins by Neomycin Extraction. Mol. Cell. Proteom. 2011, 10, S1–S15. [Google Scholar] [CrossRef]
- Brown, D.A. PIP2Clustering: From model membranes to cells. Chem. Phys. Lipids 2015, 192, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Sztacho, M.; Šalovská, B.; Červenka, J.; Balaban, C.; Hoboth, P.; Hozák, P. Limited Proteolysis-Coupled Mass Spectrometry Identifies Phosphatidylinositol 4,5-Bisphosphate Effectors in Human Nuclear Proteome. Cells 2021, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Santagata, S.; Boggon, T.J.; Baird, C.L.; Gomez, C.A.; Zhao, J.; Shan, W.S.; Myszka, D.G.; Shapiro, L. G-Protein Signaling Through Tubby Proteins. Science 2001, 292, 2041–2050. [Google Scholar] [CrossRef]
- Quinn, K.V.; Behe, P.; Tinker, A. Monitoring changes in membrane phosphatidylinositol 4,5-bisphosphate in living cells using a domain from the transcription factor tubby. J. Physiol. 2008, 586, 2855–2871. [Google Scholar] [CrossRef]
- Doucet, C.M.; Hetzer, M.W. Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma 2010, 119, 469–477. [Google Scholar] [CrossRef]
- la Cour, T.; Kiemer, L.; Mølgaard, A.; Gupta, R.; Skriver, K.; Brunak, S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004, 17, 527–536. [Google Scholar] [CrossRef]
- Cokol, M.; Nair, R.; Rost, B. Finding nuclear localization signals. EMBO Rep. 2000, 1, 411–415. [Google Scholar] [CrossRef]
- Holmer, L.; Worman, H. Inner nuclear membrane proteins: Functions and targeting. Cell. Mol. Life Sci. 2001, 58, 1741–1747. [Google Scholar] [CrossRef] [PubMed]
- Misteli, T. Beyond the Sequence: Cellular Organization of Genome Function. Cell 2007, 128, 787–800. [Google Scholar] [CrossRef]
- Carrero, G.; Hendzel, M.; de Vries, G. Modelling the compartmentalization of splicing factors. J. Theor. Biol. 2006, 239, 298–312. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.; Crabtree, G.R. Chromatin remodelling during development. Nature 2010, 463, 474–484. [Google Scholar] [CrossRef]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Spector, D.; Lamond, A. Nuclear Speckles. Cold Spring Harb. Perspect. Biol. 2011, 3, a000646. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Kreth, G.; Koester, H.; Fink, R.H.A.; Heintzmann, R.; Cremer, M.; Solovei, I.; Zink, D.; Cremer, C. Chromosome Territories, Interchromatin Domain Compartment, and Nuclear Matrix: An Integrated View of the Functional Nuclear Architecture. Crit. Rev. Eukaryot. Gene Expr. 2000, 10, 38. [Google Scholar] [CrossRef]
- Osborne, S.; Thomas, C.; Gschmeissner, S.; Schiavo, G. Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J. Cell Sci. 2001, 114, 2501–2511. [Google Scholar] [CrossRef]
- Kalasova, I.; Fáberová, V.; Kalendová, A.; Yildirim, S.; Uličná, L.; Venit, T.; Hozák, P. Tools for visualization of phosphoinositides in the cell nucleus. Histochem. Cell Biol. 2016, 145, 485–496. [Google Scholar] [CrossRef]
- Divecha, N.; Banfić, H.; Irvine, R.F. Inositides and the nucleus and inositides in the nucleus. Cell 1993, 74, 405–407. [Google Scholar] [CrossRef]
- Divecha, N.; Banfic, H.; Irvine, R.F. The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor (for IGF-1) in the plasma membrane, and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kinase. EMBO J. 1991, 10, 3207–3214. [Google Scholar] [CrossRef] [PubMed]
- Banfić, H.; Žižak, M.; Divecha, N.; Irvine, R.F. Nuclear diacylglycerol is increased during cell proliferation in vivo. Biochem. J. 1993, 290, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Vann, L.R.; Wooding, P.F.; Irvine, R.F.; Divecha, N. Metabolism and possible compartmentalization of inositol lipids in isolated rat-liver nuclei. Biochem. J. 1997, 327, 569–576. [Google Scholar] [CrossRef]
- Cocco, L.; Gilmour, R.S.; Ognibene, A.; Letcher, A.J.; Manzoli, F.A.; Irvine, R.F. Synthesis of polyphosphoinositides in nuclei of Friend cells. Evidence for polyphosphoinositide metabolism inside the nucleus which changes with cell differentiation. Biochem. J. 1987, 248, 765–770. [Google Scholar] [CrossRef]
- Boronenkov, I.V.; Loijens, J.C.; Umeda, M.; Anderson, R.A. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol. Biol. Cell 1998, 9, 3547–3560. [Google Scholar] [CrossRef] [PubMed]
- De Vries, K.J.; Heinrichs, A.A.J.; Cunningham, E.; Brunink, F.; Westerman, J.; Somerharju, P.J.; Cockcroft, S.; Wirtz, K.W.A.; Snoek, G.T. An isoform of the phosphatidylinositol-transfer protein transfers sphingomyelin and is associated with the Golgi system. Biochem. J. 1995, 310, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Rubbini, S.; Cocco, L.; Manzoli, L.; Lutterman, J.; Billi, A.; Matteucci, A.; Wirtz, K. Phosphoinositide Signalling in Nuclei of Friend Cells: DMSO-Induced Differentiation Reduces the Association of Phosphatidylinositol-Transfer Protein with the Nucleus. Biochem. Biophys. Res. Commun. 1997, 230, 302–305. [Google Scholar] [CrossRef]
- de Vries, K.; Westerman, J.; Bastiaens, P.; Jovin, T.; Wirtz, K.; Snoek, G. Fluorescently Labeled Phosphatidylinositol Transfer Protein Isoforms (α and β), Microinjected into Fetal Bovine Heart Endothelial Cells, Are Targeted to Distinct Intracellular Sites. Exp. Cell Res. 1996, 227, 33–39. [Google Scholar] [CrossRef]
- Tribble, E.K.; Ivanova, P.T.; Grabon, A.; Alb, J.G.; Faenza, I.; Cocco, L.; Brown, H.A.; Bankaitis, V.A. Quantitative profiling of the endonuclear glycerophospholipidome of murine embryonic fibroblasts. J. Lipid Res. 2016, 57, 1492–1506. [Google Scholar] [CrossRef]
- Carrillo, N.D.; Chen, M.; Cryns, V.L.; Anderson, R.A. Lipid transfer proteins initiate nuclear phosphoinositide signaling. bioRxiv 2023. [Google Scholar] [CrossRef]
- Prasanth, K.V.; Sacco-Bubulya, P.A.; Prasanth, S.G.; Spector, D.L. Sequential Entry of Components of Gene Expression Machinery into Daughter Nuclei. Mol. Biol. Cell 2003, 14, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Sobol, M.; Yildirim, S.; Philimonenko, V.V.; Marášek, P.; Castaño, E.; Hozák, P. UBF complexes with phosphatidylinositol 4,5-bisphosphate in nucleolar organizer regions regardless of ongoing RNA polymerase I activity. Nucleus 2013, 4, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.D.; Wells, W.W. Phosphorylation of rat liver nuclear envelopes. I. Characterization of in vitro protein phosphorylation. J. Biol. Chem. 1983, 258, 9360–9367. [Google Scholar] [CrossRef]
- Shah, Z.H.; Jones, D.R.; Sommer, L.; Foulger, R.; Bultsma, Y.; D’Santos, C.; Divecha, N. Nuclear phosphoinositides and their impact on nuclear functions. FEBS J. 2013, 280, 6295–6310. [Google Scholar] [CrossRef]
- Gaullier, J.-M.; Simonsen, A.; D’Arrigo, A.; Bremnes, B.; Stenmark, H.; Aasland, R. FYVE fingers bind PtdIns(3)P. Nature 1998, 394, 432–433. [Google Scholar] [CrossRef]
- Stenmark, H.; Aasland, R. FYVE-finger proteins--effectors of an inositol lipid. J. Cell Sci. 1999, 112, 4175–4183. [Google Scholar] [CrossRef]
- Gillooly, D.J.; Morrow, I.C.; Lindsay, M.; Gould, R.; Bryant, N.J.; Gaullier, J.; Parton, R.G.; Stenmark, H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000, 19, 4577–4588. [Google Scholar] [CrossRef]
- Norris, F.A.; Majerus, P.W. Hydrolysis of phosphatidylinositol 3,4-bisphosphate by inositol polyphosphate 4-phosphatase isolated by affinity elution chromatography. J. Biol. Chem. 1994, 269, 8716–8720. [Google Scholar] [CrossRef]
- Norris, F.A.; Auethavekiat, V.; Majerus, P.W. The Isolation and Characterization of cDNA Encoding Human and Rat Brain Inositol Polyphosphate 4-Phosphatase. J. Biol. Chem. 1995, 270, 16128–16133. [Google Scholar] [CrossRef] [PubMed]
- Devereaux, K.; Dall’armi, C.; Alcazar-Roman, A.; Ogasawara, Y.; Zhou, X.; Wang, F.; Yamamoto, A.; De Camilli, P.; Di Paolo, G. Regulation of Mammalian Autophagy by Class II and III PI 3-Kinases through PI3P Synthesis. PLoS ONE 2013, 8, e76405. [Google Scholar] [CrossRef] [PubMed]
- Franco, I.; Gulluni, F.; Campa, C.C.; Costa, C.; Margaria, J.P.; Ciraolo, E.; Martini, M.; Monteyne, D.; De Luca, E.; Germena, G.; et al. PI3K Class II α Controls Spatially Restricted Endosomal PtdIns3P and Rab11 Activation to Promote Primary Cilium Function. Dev. Cell 2014, 28, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Gulluni, F.; De Santis, M.C.; Margaria, J.P.; Martini, M.; Hirsch, E. Class II PI3K Functions in Cell Biology and Disease. Trends Cell Biol. 2019, 29, 339–359. [Google Scholar] [CrossRef]
- Didichenko, S.A.; Thelen, M. Phosphatidylinositol 3-Kinase C2α Contains a Nuclear Localization Sequence and Associates with Nuclear Speckles. J. Biol. Chem. 2001, 276, 48135–48142. [Google Scholar] [CrossRef] [PubMed]
- Višnjić, D.; Crljen, V.; Ćurić, J.; Batinić, D.; Volinia, S.; Banfić, H. The activation of nuclear phosphoinositide 3-kinase C2β in all-trans-retinoic acid-differentiated HL-60 cells. FEBS Lett. 2002, 529, 268–274. [Google Scholar] [CrossRef]
- Banfic, H.; Visnjic, D.; Mise, N.; Balakrishnan, S.; Deplano, S.; Korchev, Y.E.; Domin, J. Epidermal growth factor stimulates translocation of the class II phosphoinositide 3-kinase PI3K-C2β to the nucleus. Biochem. J. 2009, 422, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.T.; Einav, S.; Asquith, C.R.M. PIKfyve: A lipid kinase target for COVID-19, cancer and neurodegenerative disorders. Nat. Rev. Drug Discov. 2021, 20, 730. [Google Scholar] [CrossRef]
- Clague, M.J.; Lorenzo, Ó. The Myotubularin Family of Lipid Phosphatases. Traffic 2005, 6, 1063–1069. [Google Scholar] [CrossRef]
- Robinson, F.L.; Dixon, J.E. Myotubularin phosphatases: Policing 3-phosphoinositides. Trends Cell Biol. 2006, 16, 403–412. [Google Scholar] [CrossRef]
- Divecha, N.; Letcher, A.J.; Banfic, H.H.; Rhee, S.G.; Irvine, R.F. Changes in the components of a nuclear inositide cycle during differentiation in murine erythroleukaemia cells. Biochem. J. 1995, 312, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Fáberová, V.; Kalasová, I.; Krausová, A.; Hozák, P. Super-Resolution Localisation of Nuclear PI(4)P and Identification of Its Interacting Proteome. Cells 2020, 9, 1191. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Manford, A.; Xia, T.; Saxena, A.; Stefan, C.; Hu, F.; Emr, S. Crystal structure of the Yeast Sac1: Implications for its phosphoinositide phosphatase function. EMBO J. 2010, 29, 1489–1498. [Google Scholar] [CrossRef]
- Zewe, J.P.; Wills, R.C.; Sangappa, S.; Goulden, B.D.; Hammond, G.R. SAC1 degrades its lipid substrate PtdIns4P in the endoplasmic reticulum to maintain a steep chemical gradient with donor membranes. eLife 2018, 7, e35588. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.H.; Letcher, A.J.; D’Santos, C.S.; Halstead, J.R.; Irvine, R.F.; Divecha, N. Inositol lipids are regulated during cell cycle progression in the nuclei of murine erythroleukaemia cells. Biochem. J. 2001, 357, 905–910. [Google Scholar] [CrossRef]
- Clarke, J.H.; Irvine, R.F. Evolutionarily conserved structural changes in phosphatidylinositol 5-phosphate 4-kinase (PI5P4K) isoforms are responsible for differences in enzyme activity and localization. Biochem. J. 2013, 454, 49–57. [Google Scholar] [CrossRef]
- Divecha, N.; Brooksbank, C.E.L.; Irvine, R.F. Purification and characterization of phosphatidylinositol 4-phosphate 5-kinases. Biochem. J. 1992, 288, 637–642. [Google Scholar] [CrossRef]
- Brooksbank, C.E.L.; Hutchings, A.; Butcher, G.W.; Irvine, R.F.; Divecha, N. Monoclonal antibodies to phosphatidylinositol 4-phosphate 5-kinase: Distribution and intracellular localization of the C isoform. Biochem. J. 1993, 291, 77–82. [Google Scholar] [CrossRef]
- Bazenet, C.E.; Ruano, A.R.; Brockman, J.L.; Anderson, R.A. The human erythrocyte contains two forms of phosphatidylinositol-4-phosphate 5-kinase which are differentially active toward membranes. J. Biol. Chem. 1990, 265, 18012–18022. [Google Scholar] [CrossRef]
- Rameh, L.E.; Tolias, K.F.; Duckworth, B.C.; Cantley, L.C. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature 1997, 390, 192–196. [Google Scholar] [CrossRef]
- Divecha, N.; Truong, O.; Hsuan, J.J.; Hinchliffe, K.A.; Irvine, R.F. The cloning and sequence of the C isoform of PtdIns4P 5-kinase. Biochem. J. 1995, 309, 715–719. [Google Scholar] [CrossRef]
- Castellino, A.M.; Parker, G.J.; Boronenkov, I.V.; Anderson, R.A.; Chao, M.V. A Novel Interaction between the Juxtamembrane Region of the p55 Tumor Necrosis Factor Receptor and Phosphatidylinositol-4-phosphate 5-Kinase. J. Biol. Chem. 1997, 272, 5861–5870. [Google Scholar] [CrossRef] [PubMed]
- Boronenkov, I.V.; Anderson, R.A. The Sequence of Phosphatidylinositol-4-phosphate 5-Kinase Defines a Novel Family of Lipid Kinases. J. Biol. Chem. 1995, 270, 2881–2884. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.R.; Divecha, N. Linking lipids to chromatin. Curr. Opin. Genet. Dev. 2004, 14, 196–202. [Google Scholar] [CrossRef]
- Ciruela, A.; Hinchliffe, K.A.; Divecha, N.; Irvine, R.F. Nuclear targeting of the beta isoform of type II phosphatidylinositol phosphate kinase (phosphatidylinositol 5-phosphate 4-kinase) by its alpha-helix 7. Biochem. J. 2000, 346, 587–591. [Google Scholar] [CrossRef]
- Hasegawa, J.; Strunk, B.S.; Weisman, L.S. PI5P and PI(3,5)P2: Minor, but Essential Phosphoinositides. Cell Struct. Funct. 2017, 42, 49–60. [Google Scholar] [CrossRef]
- Poli, A.; Zaurito, A.E.; Abdul-Hamid, S.; Fiume, R.; Faenza, I.; Divecha, N. Phosphatidylinositol 5 Phosphate (PI5P): From Behind the Scenes to the Front (Nuclear) Stage. Int. J. Mol. Sci. 2019, 20, 2080. [Google Scholar] [CrossRef] [PubMed]
- Bulley, S.J.; Droubi, A.; Clarke, J.H.; Anderson, K.E.; Stephens, L.R.; Hawkins, P.T.; Irvine, R.F. In B cells, phosphatidylinositol 5-phosphate 4-kinase–α synthesizes PI(4,5)P2 to impact mTORC2 and Akt signaling. Proc. Natl. Acad. Sci. USA 2016, 113, 10571–10576. [Google Scholar] [CrossRef]
- Lundquist, M.R.; Goncalves, M.D.; Loughran, R.M.; Possik, E.; Vijayaraghavan, T.; Yang, A.; Pauli, C.; Ravi, A.; Verma, A.; Yang, Z.; et al. Phosphatidylinositol-5-Phosphate 4-Kinases Regulate Cellular Lipid Metabolism By Facilitating Autophagy. Mol. Cell 2018, 70, 531–544.e9. [Google Scholar] [CrossRef]
- Vicinanza, M.; Korolchuk, V.; Ashkenazi, A.; Puri, C.; Menzies, F.M.; Clarke, J.; Rubinsztein, D.C. PI(5)P Regulates Autophagosome Biogenesis. Mol. Cell 2015, 57, 219–234. [Google Scholar] [CrossRef]
- Poli, A.; Pennacchio, F.A.; Ghisleni, A.; di Gennaro, M.; Lecacheur, M.; Nastały, P.; Crestani, M.; Pramotton, F.M.; Iannelli, F.; Beznusenko, G.; et al. PIP4K2B is mechanoresponsive and controls heterochromatin-driven nuclear softening through UHRF1. Nat. Commun. 2023, 14, 1432. [Google Scholar] [CrossRef] [PubMed]
- Poli, A.; Abdul-Hamid, S.; Zaurito, A.E.; Campagnoli, F.; Bevilacqua, V.; Sheth, B.; Fiume, R.; Pagani, M.; Abrignani, S.; Divecha, N. PIP4Ks impact on PI3K, FOXP3, and UHRF1 signaling and modulate human regulatory T cell proliferation and immunosuppressive activity. Proc. Natl. Acad. Sci. USA 2021, 118, e2010053118. [Google Scholar] [CrossRef] [PubMed]
- Blind, R.D.; Sablin, E.P.; Kuchenbecker, K.M.; Chiu, H.-J.; Deacon, A.M.; Das, D.; Fletterick, R.J.; Ingraham, H.A. The signaling phospholipid PIP 3 creates a new interaction surface on the nuclear receptor SF-1. Proc. Natl. Acad. Sci. USA 2014, 111, 15054–15059. [Google Scholar] [CrossRef]
- Ndamukong, I.; Jones, D.R.; Lapko, H.; Divecha, N.; Avramova, Z. Phosphatidylinositol 5-Phosphate Links Dehydration Stress to the Activity of ARABIDOPSIS TRITHORAX-LIKE Factor ATX1. PLoS ONE 2010, 5, e13396. [Google Scholar] [CrossRef]
- Sbrissa, D.; Ikonomov, O.C.; Shisheva, A. PIKfyve, a Mammalian Ortholog of Yeast Fab1p Lipid Kinase, Synthesizes 5-Phosphoinositides. J. Biol. Chem. 1999, 274, 21589–21597. [Google Scholar] [CrossRef]
- Schaletzky, J.; Dove, S.K.; Short, B.; Lorenzo, O.; Clague, M.J.; Barr, F.A. Phosphatidylinositol-5-Phosphate Activation and Conserved Substrate Specificity of the Myotubularin Phosphatidylinositol 3-Phosphatases. Curr. Biol. 2003, 13, 504–509. [Google Scholar] [CrossRef]
- Zolov, S.N.; Bridges, D.; Zhang, Y.; Lee, W.W.; Riehle, E.; Verma, R.; Lenk, G.M.; Converso-Baran, K.; Weide, T.; Albin, R.L.; et al. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. Natl. Acad. Sci. USA 2012, 109, 17472–17477. [Google Scholar] [CrossRef]
- Ungewickell, A.; Hugge, C.; Kisseleva, M.; Chang, S.C.; Zou, J.; Feng, Y.; Galyov, E.E.; Wilson, M.; Majerus, P.W. The identification and characterization of two phosphatidylinositol-4,5-bisphosphate 4-phosphatases. Proc. Natl. Acad. Sci. USA 2005, 102, 18854–18859. [Google Scholar] [CrossRef]
- Zou, J.; Marjanovic, J.; Kisseleva, M.V.; Wilson, M.; Majerus, P.W. Type I phosphatidylinositol-4,5-bisphosphate 4-phosphatase regulates stress-induced apoptosis. Proc. Natl. Acad. Sci. USA 2007, 104, 16834–16839. [Google Scholar] [CrossRef] [PubMed]
- Posor, Y.; Eichhorn-Gruenig, M.; Puchkov, D.; Schöneberg, J.; Ullrich, A.; Lampe, A.; Müller, R.; Zarbakhsh, S.; Gulluni, F.; Hirsch, H.; et al. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature 2013, 499, 233–237. [Google Scholar] [CrossRef]
- Alliouachene, S.; Bilanges, B.; Chicanne, G.; Anderson, K.E.; Pearce, W.; Ali, K.; Valet, C.; Posor, Y.; Low, P.C.; Chaussade, C.; et al. Inactivation of the Class II PI3K-C2β Potentiates Insulin Signaling and Sensitivity. Cell Rep. 2015, 13, 1881–1894. [Google Scholar] [CrossRef] [PubMed]
- Yokogawa, T.; Nagata, S.; Nishio, Y.; Tsutsumi, T.; Ihara, S.; Shirai, R.; Morita, K.; Umeda, M.; Shirai, Y.; Saitoh, N.; et al. Evidence that 3′-phosphorylated polyphosphoinositides are generated at the nuclear surface: Use of immunostaining technique with monoclonal antibodies specific for PI 3,4-P2. FEBS Lett. 2000, 473, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Marat, A.L.; Wallroth, A.; Lo, W.T.; Müller, R.; Norata, G.D.; Falasca, M.; Schultz, C.; Haucke, V. mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science 2017, 356, 968–972. [Google Scholar] [CrossRef] [PubMed]
- Gozzelino, L.; De Santis, M.C.; Gulluni, F.; Hirsch, E.; Martini, M. PI(3,4)P2 Signaling in Cancer and Metabolism. Front. Oncol. 2020, 10, 360. [Google Scholar] [CrossRef]
- Kerr, W.G. Inhibitor and activator: Dual functions for SHIP in immunity and cancer. Ann. N. Y. Acad. Sci. 2010, 1217, 1–17. [Google Scholar] [CrossRef]
- Krystal, G.; Damen, J.E.; Helgason, C.D.; Huber, M.; Hughes, M.R.; Kalesnikoff, J.; Lam, V.; Rosten, P.; Ware, M.D.; Yew, S.; et al. SHIPs ahoy. Int. J. Biochem. Cell Biol. 1999, 31, 1007–1010. [Google Scholar] [CrossRef]
- Ooms, L.M.; Binge, L.C.; Davies, E.M.; Rahman, P.; Conway, J.R.; Gurung, R.; Ferguson, D.T.; Papa, A.; Fedele, C.G.; Vieusseux, J.L.; et al. The Inositol Polyphosphate 5-Phosphatase PIPP Regulates AKT1-Dependent Breast Cancer Growth and Metastasis. Cancer Cell 2015, 28, 155–169. [Google Scholar] [CrossRef]
- Norris, F.A.; Atkins, R.C.; Majerus, P.W. The cDNA Cloning and Characterization of Inositol Polyphosphate 4-Phosphatase Type II. J. Biol. Chem. 1997, 272, 23859–23864. [Google Scholar] [CrossRef]
- Watt, S.A.; Kular, G.; Fleming, I.N.; Downes, C.P.; Lucoqc, J.M. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C δ1. Biochem. J. 2002, 363, 657–666. [Google Scholar] [CrossRef]
- Miyazawa, A.; Umeda, M.; Horikoshi, T.; Yanagisawa, K.; Yoshioka, T.; Inoue, K. Production and characterization of monoclonal antibodies that bind to phosphatidylinositol 4,5-bisphosphate. Mol. Immunol. 1988, 25, 1025–1031. [Google Scholar]
- Fukami, K.; Matsuoka, K.; Nakanishi, O.; Yamakawa, A.; Kawai, S.; Takenawa, T. Antibody to phosphatidylinositol 4,5-bisphosphate inhibits oncogene-induced mitogenesis. Proc. Natl. Acad. Sci. USA 1988, 85, 9057–9061. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.L.; Steel, J.; Prestwich, G.D.; Schiavo, G. Generation of phosphatidylinositol-specific antibodies and their characterization. Biochem. Soc. Trans. 1999, 27, 648–652. [Google Scholar] [CrossRef]
- Yildirim, S.; Castano, E.; Sobol, M.; Philimonenko, V.V.; Dzijak, R.; Venit, T.; Hozák, P. Involvement of PIP2 in RNA Polymerase I transcription. J. Cell Sci. 2013, 126, 2730–2739. [Google Scholar] [CrossRef]
- Anderson, R.A.; Boronenkov, I.V.; Doughman, S.D.; Kunz, J.; Loijens, J.C. Phosphatidylinositol Phosphate Kinases, a Multifaceted Family of Signaling Enzymes. J. Biol. Chem. 1999, 274, 9907–9910. [Google Scholar] [CrossRef] [PubMed]
- Mellman, D.L.; Gonzales, M.L.; Song, C.; Barlow, C.A.; Wang, P.; Kendziorski, C.; Anderson, R.A. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 2008, 451, 1013–1017. [Google Scholar] [CrossRef]
- Ishihara, H.; Shibasaki, Y.; Kizuki, N.; Wada, T.; Yazaki, Y.; Asano, T.; Oka, Y. Type I phosphatidylinositol-4-phosphate 5-kinases. Cloning of the third isoform and deletion/substitution analysis of members of this novel lipid kinase family. J. Biol. Chem. 1998, 273, 8741–8748. [Google Scholar] [CrossRef] [PubMed]
- Loijens, J.C.; Boronenkov, I.V.; Parker, G.J.; Anderson, R.A. The phosphatidylinositol 4-phosphate 5-kinase family. Adv. Enzym. Regul. 1996, 36, 115–140. [Google Scholar] [CrossRef]
- van den Bout, I.; Divecha, N. PIP5K-driven PtdIns(4,5)P2 synthesis: Regulation and cellular functions. J. Cell Sci. 2009, 122, 3837–3850. [Google Scholar] [CrossRef]
- Tolias, K.F.; Rameh, L.E.; Ishihara, H.; Shibasaki, Y.; Chen, J.; Prestwich, G.D.; Cantley, L.C.; Carpenter, C.L. Type I Phosphatidylinositol-4-phosphate 5-Kinases Synthesize the Novel Lipids Phosphatidylinositol 3,5-Bisphosphate and Phosphatidylinositol 5-Phosphate. J. Biol. Chem. 1998, 273, 18040–18046. [Google Scholar] [CrossRef]
- Liu, Y.; Bankaitis, V.A. Phosphoinositide phosphatases in cell biology and disease. Prog. Lipid Res. 2010, 49, 201–217. [Google Scholar] [CrossRef]
- Ooms, L.M.; Horan, K.A.; Rahman, P.; Seaton, G.; Gurung, R.; Kethesparan, D.S.; Mitchell, C.A. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem. J. 2009, 419, 29–49. [Google Scholar] [CrossRef]
- Dyson, J.M.; Fedele, C.G.; Davies, E.M.; Becanovic, J.; Mitchell, C.A. Phosphoinositide Phosphatases: Just as Important as the Kinases. Phosphoinosit. I Enzym. Synth. Degrad. 2012, 58, 215–279. [Google Scholar] [CrossRef]
- Martelli, A.M.; Gilmour, R.S.; Bertagnolo, V.; Neri, L.M. Nuclear localization and signaling activity of phosphoinositidase C b in Swiss 3T3 cells. Nature 1992, 358, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Song, M.S.; Salmena, L.; Pandolfi, P.P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012, 13, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Hollander, M.C.; Blumenthal, G.M.; Dennis, P.A. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer 2011, 11, 289–301. [Google Scholar] [CrossRef]
- Lindsay, Y.; McCoull, D.; Davidson, L.; Leslie, N.R.; Fairservice, A.; Gray, A.; Lucocq, J.; Downes, C.P. Localization of agonist-sensitive PtdIns(3,4,5)P3 reveals a nuclear pool that is insensitive to PTEN expression. J. Cell Sci. 2006, 119, 5160–5168. [Google Scholar] [CrossRef]
- Edimo, W.E.; Vanderwinden, J.M.; Erneux, C. SHIP2 signalling at the plasma membrane, in the nucleus and at focal contacts. Adv. Biol. Regul. 2013, 53, 28–37. [Google Scholar] [CrossRef]
- Foukas, L.C.; Berenjeno, I.M.; Gray, A.; Khwaja, A.; Vanhaesebroeck, B. Activity of any class IA PI3K isoform can sustain cell proliferation and survival. Proc. Natl. Acad. Sci. USA 2010, 107, 11381–11386. [Google Scholar] [CrossRef]
- Davis, W.J.; Lehmann, P.Z.; Li, W. Nuclear PI3K signaling in cell growth and tumorigenesis. Front. Cell Dev. Biol. 2015, 3, 24. [Google Scholar] [CrossRef]
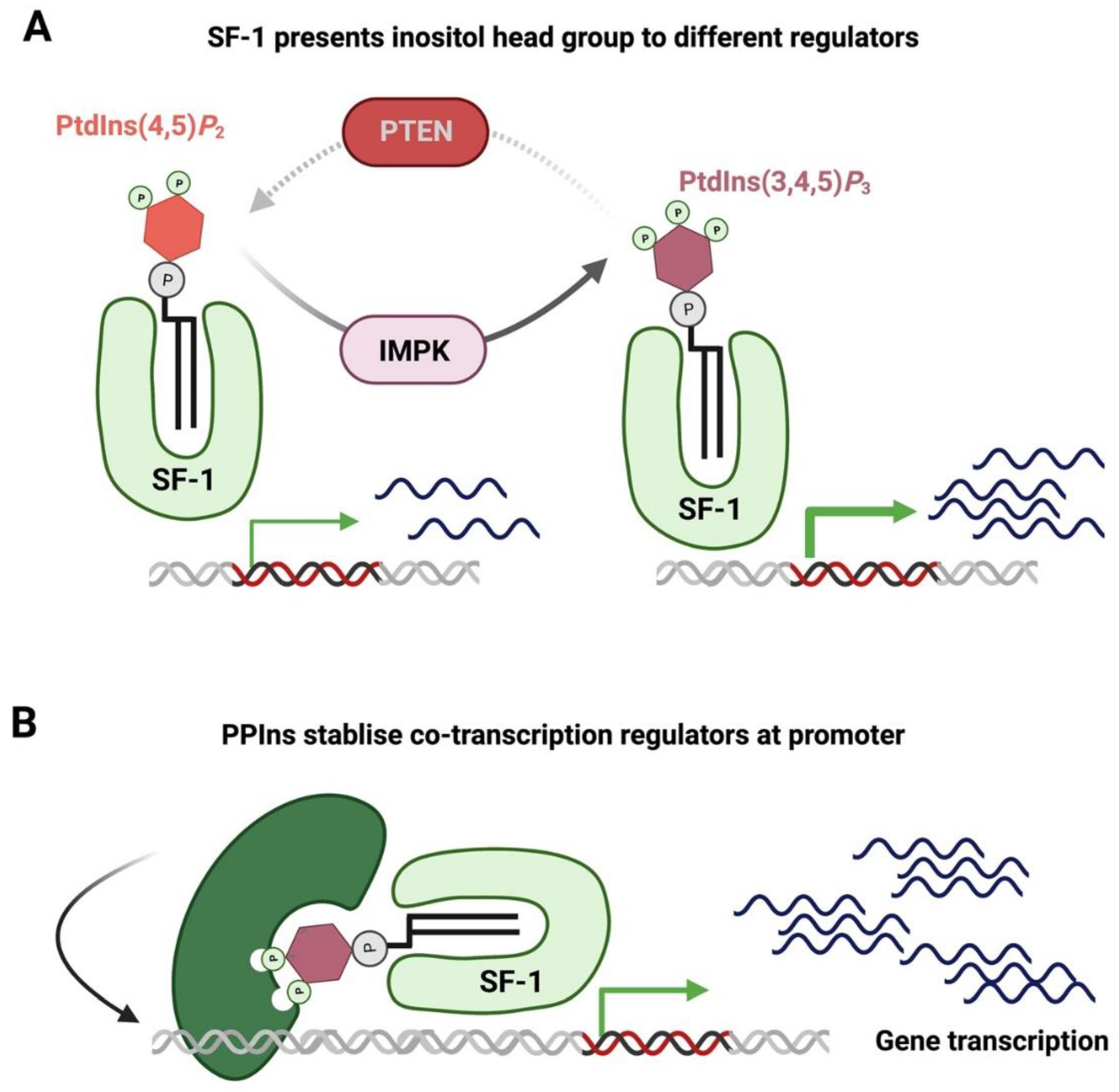
- Resnick, A.C.; Snowman, A.M.; Kang, B.N.; Hurt, K.J.; Snyder, S.H.; Saiardi, A. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc. Natl. Acad. Sci. USA 2005, 102, 12783–12788. [Google Scholar] [CrossRef]
- Blind, R.D.; Suzawa, M.; Ingraham, H.A. Direct Modification and Activation of a Nuclear Receptor–PIP2 Complex by the Inositol Lipid Kinase IPMK. Sci. Signal. 2012, 5, ra44. [Google Scholar] [CrossRef] [PubMed]
- Edimo, W.E.; Derua, R.; Janssens, V.; Nakamura, T.; Vanderwinden, J.M.; Waelkens, E.; Erneux, C. Evidence of SHIP2 Ser132 phosphorylation, its nuclear localization and stability. Biochem. J. 2011, 439, 391–404. [Google Scholar] [CrossRef]
- Chung, J.-H.; Eng, C. Nuclear-Cytoplasmic Partitioning of Phosphatase and Tensin Homologue Deleted on Chromosome 10 (PTEN) Differentially Regulates the Cell Cycle and Apoptosis. Cancer Res. 2005, 65, 8096–8100. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-L.; Sheng, X.; Hortobagyi, Z.K.; Mao, Z.; Gallick, G.E.; Yung, W.K.A. Nuclear PTEN-Mediated Growth Suppression Is Independent of Akt Down-Regulation. Mol. Cell. Biol. 2005, 25, 6211–6224. [Google Scholar] [CrossRef]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Hayes, J.J.; Pruss, D.; Wolffe, A.P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 1993, 72, 73–84. [Google Scholar] [CrossRef]
- Biel, M.; Wascholowski, V.; Giannis, A. Epigenetics—An Epicenter of Gene Regulation: Histones and Histone-Modifying Enzymes. Angew. Chem. Int. Ed. 2005, 44, 3186–3216. [Google Scholar] [CrossRef]
- McKay, L.M.; Carpenter, B.; Roberts, S.G. Regulation of the Wilms’ tumour suppressor protein transcriptional activation domain. Oncogene 1999, 18, 6546–6554. [Google Scholar] [CrossRef]
- Roberts, S.G. Transcriptional regulation by WT1 in development. Curr. Opin. Genet. Dev. 2005, 15, 542–547. [Google Scholar] [CrossRef]
- Carpenter, B.; Hill, K.J.; Charalambous, M.; Wagner, K.J.; Lahiri, D.; James, D.I.; Andersen, J.S.; Schumacher, V.; Royer-Pokora, B.; Mann, M.; et al. BASP1 Is a Transcriptional Cosuppressor for the Wilms’ Tumor Suppressor Protein WT1. Mol. Cell. Biol. 2004, 24, 537–549. [Google Scholar] [CrossRef]
- Toska, E.; Campbell, H.A.; Shandilya, J.; Goodfellow, S.J.; Shore, P.; Medler, K.F.; Roberts, S.G. Repression of Transcription by WT1-BASP1 Requires the Myristoylation of BASP1 and the PIP2-Dependent Recruitment of Histone Deacetylase. Cell Rep. 2012, 2, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.K.; Shearn, A. The Direct Interaction Between ASH2, a Drosophila Trithorax Group Protein, and SKTL, a Nuclear Phosphatidylinositol 4-Phosphate 5-Kinase, Implies a Role for Phosphatidylinositol 4,5-Bisphosphate in Maintaining Transcriptionally Active Chromatin. Genetics 2004, 167, 1213–1223. [Google Scholar] [CrossRef]
- Visa, N.; Percipalle, P. Nuclear Functions of Actin. Cold Spring Harb. Perspect. Biol. 2010, 2, a000620. [Google Scholar] [CrossRef]
- Rando, O.J.; Zhao, K.; Janmey, P.; Crabtree, G.R. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc. Natl. Acad. Sci. USA 2002, 99, 2824–2829. [Google Scholar] [CrossRef]
- Batista, N.J.; Desai, S.G.; Perez, A.M.; Finkelstein, A.; Radigan, R.; Singh, M.; Landman, A.; Drittel, B.; Abramov, D.; Ahsan, M.; et al. The Molecular and Cellular Basis of Hutchinson–Gilford Progeria Syndrome and Potential Treatments. Genes 2023, 14, 602. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Fukami, K.; Watanabe, Y.; Ozaki, C.; Takenawa, T. Phosphatidylinositol 4,5-bisphosphate reverses the inhibition of RNA transcription caused by histone H1. JBIC J. Biol. Inorg. Chem. 1998, 251, 281–287. [Google Scholar] [CrossRef]
- Gelato, K.A.; Tauber, M.; Ong, M.S.; Winter, S.; Hiragami-Hamada, K.; Sindlinger, J.; Lemak, A.; Bultsma, Y.; Houliston, S.; Schwarzer, D.; et al. Accessibility of Different Histone H3-Binding Domains of UHRF1 Is Allosterically Regulated by Phosphatidylinositol 5-Phosphate. Mol. Cell 2014, 54, 905–919. [Google Scholar] [CrossRef]
- Stijf-Bultsma, Y.; Sommer, L.; Tauber, M.; Baalbaki, M.; Giardoglou, P.; Jones, D.R.; Gelato, K.A.; van Pelt, J.; Shah, Z.; Rahnamoun, H.; et al. The Basal Transcription Complex Component TAF3 Transduces Changes in Nuclear Phosphoinositides into Transcriptional Output. Mol. Cell 2015, 58, 453–467. [Google Scholar] [CrossRef] [PubMed]
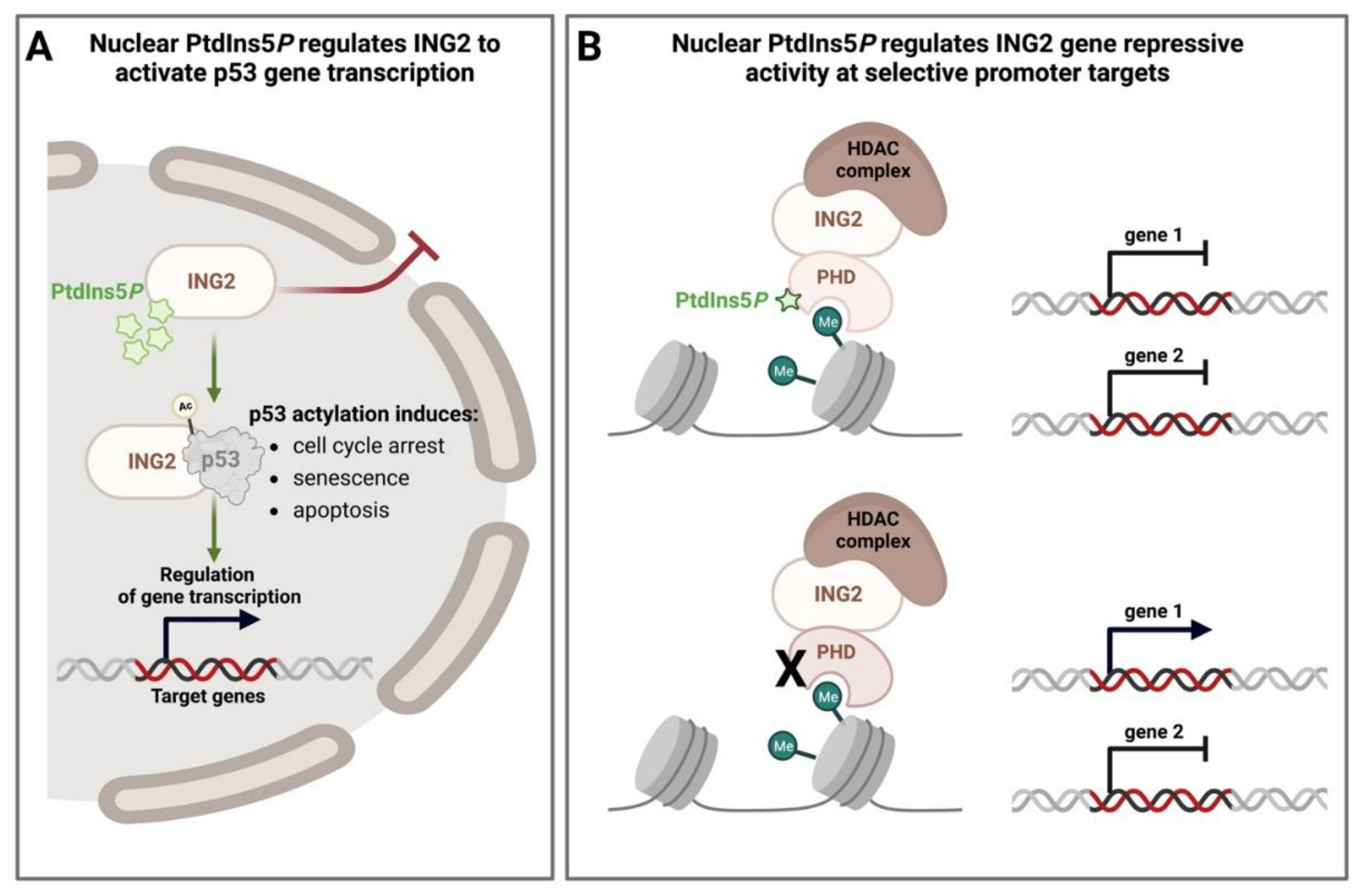
- Peña, P.V.; Davrazou, F.; Shi, X.; Walter, K.L.; Verkhusha, V.V.; Gozani, O.; Zhao, R.; Kutateladze, T.G. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 2006, 442, 100–103. [Google Scholar] [CrossRef]
- Li, H.; Ilin, S.; Wang, W.; Duncan, E.M.; Wysocka, J.; Allis, C.D.; Patel, D.J. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 2006, 442, 91–95. [Google Scholar] [CrossRef]
- Feng, X.; Hara, Y.; Riabowol, K. Different HATS of the ING1 gene family. Trends Cell Biol. 2002, 12, 532–538. [Google Scholar] [CrossRef]
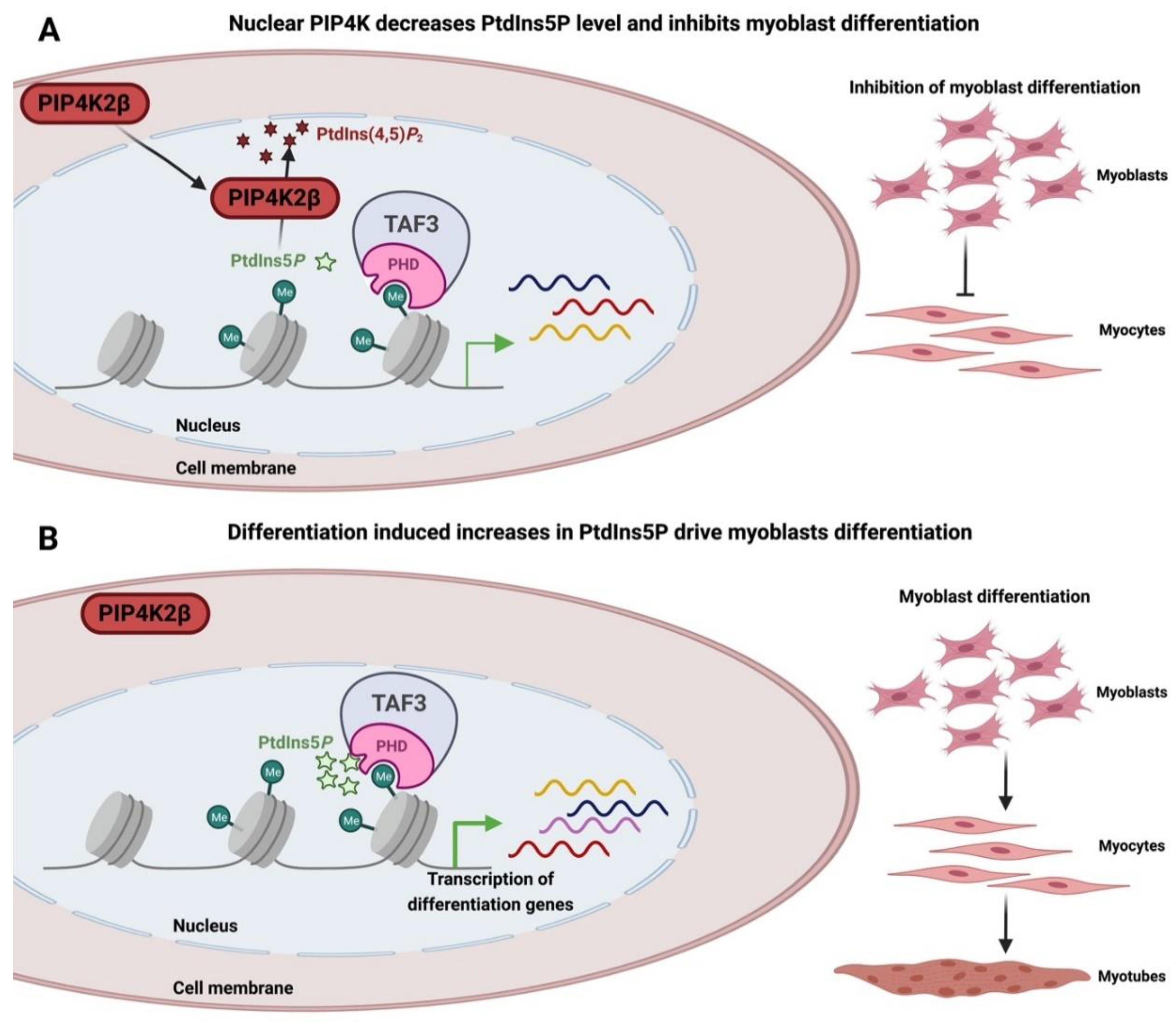
- Jones, D.R.; Bultsma, Y.; Keune, W.-J.; Halstead, J.R.; Elouarrat, D.; Mohammed, S.; Heck, A.J.; D’Santos, C.S.; Divecha, N. Nuclear PtdIns5P as a Transducer of Stress Signaling: An In Vivo Role for PIP4Kbeta. Mol. Cell 2006, 23, 685–695. [Google Scholar] [CrossRef]
- Bua, D.J.; Martin, G.M.; Binda, O.; Gozani, O. Nuclear phosphatidylinositol-5-phosphate regulates ING2 stability at discrete chromatin targets in response to DNA damage. Sci. Rep. 2013, 3, 2137. [Google Scholar] [CrossRef]
- Reed, S.M.; Quelle, D.E. p53 Acetylation: Regulation and Consequences. Cancers 2014, 7, 30–69. [Google Scholar] [CrossRef]
- Nagashima, M.; Shiseki, M.; Miura, K.; Hagiwara, K.; Linke, S.P.; Pedeux, R.; Wang, X.W.; Yokota, J.; Riabowol, K.; Harris, C.C. DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proc. Natl. Acad. Sci. USA 2001, 98, 9671–9676. [Google Scholar] [CrossRef]
- Liu, Z.; Scannell, D.R.; Eisen, M.B.; Tjian, R. Control of Embryonic Stem Cell Lineage Commitment by Core Promoter Factor, TAF3. Cell 2011, 146, 720–731. [Google Scholar] [CrossRef]
- Deato, M.D.E.; Marr, M.T.; Sottero, T.; Inouye, C.; Hu, P.; Tjian, R. MyoD Targets TAF3/TRF3 to Activate Myogenin Transcription. Mol. Cell 2008, 32, 96–105. [Google Scholar] [CrossRef]
- Yao, J.; Fetter, R.D.; Hu, P.; Betzig, E.; Tjian, R. Subnuclear segregation of genes and core promoter factors in myogenesis. Genes Dev. 2011, 25, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.; Eswara, K.; Yerkesh, Z.; Kharchenko, V.; Zandarashvili, L.; Szczepski, K.; Bensaddek, D.; Jaremko, Ł.; Black, B.E.; Fischle, W. Molecular basis of hUHRF1 allosteric activation for synergistic histone modification binding by PI5P. Sci. Adv. 2022, 8, eabl9461. [Google Scholar] [CrossRef] [PubMed]
- Relav, L.; Doghman-Bouguerra, M.; Ruggiero, C.; Muzzi, J.C.D.; Figueiredo, B.C.; Lalli, E. Steroidogenic Factor 1, a Goldilocks Transcription Factor from Adrenocortical Organogenesis to Malignancy. Int. J. Mol. Sci. 2023, 24, 3585. [Google Scholar] [CrossRef] [PubMed]
- Krylova, I.N.; Sablin, E.P.; Moore, J.; Xu, R.X.; Waitt, G.M.; MacKay, J.A.; Juzumiene, D.; Bynum, J.M.; Madauss, K.; Montana, V.; et al. Structural Analyses Reveal Phosphatidyl Inositols as Ligands for the NR5 Orphan Receptors SF-1 and LRH-1. Cell 2005, 120, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Choi, M.; Cavey, G.; Daugherty, J.; Suino, K.; Kovach, A.; Bingham, N.C.; Kliewer, S.A.; Xu, H. Crystallographic Identification and Functional Characterization of Phospholipids as Ligands for the Orphan Nuclear Receptor Steroidogenic Factor-1. Mol. Cell 2005, 17, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-H.; Hariharan, A.; Bastianello, G.; Toyama, Y.; Shivashankar, G.V.; Foiani, M.; Sheetz, M.P. DNA damage causes rapid accumulation of phosphoinositides for ATR signaling. Nat. Commun. 2017, 8, 2118. [Google Scholar] [CrossRef]
- Sobol, M.; Krausová, A.; Yildirim, S.; Kalasová, I.; Fáberová, V.; Vrkoslav, V.; Philimonenko, V.; Marášek, P.; Pastorek, L.; Čapek, M.; et al. Nuclear phosphatidylinositol 4,5-bisphosphate islets contribute to efficient RNA polymerase II-dependent transcription. J. Cell Sci. 2018, 131, jcs211094. [Google Scholar] [CrossRef]
- Gavgani, F.M.; Karlsson, T.; Tangen, I.L.; Morovicz, A.P.; Arnesen, V.S.; Turcu, D.C.; Ninzima, S.; Spang, K.; Krakstad, C.; Guillermet-Guibert, J.; et al. Nuclear upregulation of class I phosphoinositide 3-kinase p110β correlates with high 47S rRNA levels in cancer cells. J. Cell Sci. 2021, 134, jcs246090. [Google Scholar] [CrossRef] [PubMed]
- Dyson, N.J. RB1: A prototype tumor suppressor and an enigma. Genes Dev. 2016, 30, 1492–1502. [Google Scholar] [CrossRef]
- Kruiswijk, F.; Labuschagne, C.F.; Vousden, K.H. p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 2015, 16, 393–405. [Google Scholar] [CrossRef]
- Divecha, N.; Roefs, M.; Los, A.; Halstead, J.; Bannister, A.; D’Santos, C. Type I PIPkinases Interact with and Are Regulated by the Retinoblastoma Susceptibility Gene Product—pRB. Curr. Biol. 2002, 12, 582–587. [Google Scholar] [CrossRef]
- Los, A.P.; Vinke, F.P.; de Widt, J.; Topham, M.K.; van Blitterswijk, W.J.; Divecha, N. The Retinoblastoma Family Proteins Bind to and Activate Diacylglycerol Kinaseζ. J. Biol. Chem. 2006, 281, 858–866. [Google Scholar] [CrossRef]
- Los, A.P.; de Widt, J.; van Blitterswijk, W.J.; Divecha, N. Is there a role for diacylglycerol kinase-ζ in cell cycle regulation? Adv. Enzym. Regul. 2008, 48, 31–39. [Google Scholar] [CrossRef]
- Choi, S.; Chen, M.; Cryns, V.L.; Anderson, R.A. A nuclear phosphoinositide kinase complex regulates p53. Nature 2019, 21, 462–475. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Choi, S.; Wen, T.; Chen, C.; Thapa, N.; Lee, J.H.; Cryns, V.L.; Anderson, R.A. A p53–phosphoinositide signalosome regulates nuclear AKT activation. Nature 2022, 24, 1099–1113. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.N.; Franke, T.F.; Bellacosa, A.; Datta, K.; Gonzalez-Portal, M.E.; Taguchi, T.; Testa, J.R.; Tsichlis, P.N. The proteins encoded by c-akt and v-akt differ in post-translational modification, subcellular localization and oncogenic potential. Oncogene 1993, 8, 1957–1963. [Google Scholar] [PubMed]
- Neri, L.M.; Milani, D.; Bertolaso, L.; Stroscio, M.; Bertagnolo, V.; Capitani, S. Nuclear translocation of phosphatidylinositol 3-kinase in rat pheochromocytoma PC 12 cells after treatment with nerve growth factor. Cell. Mol. Biol. 1994, 40, 619–626. [Google Scholar]
- Nguyen, T.L.X.; Choi, J.W.; Lee, S.B.; Ye, K.; Woo, S.-D.; Lee, K.-H.; Ahn, J.-Y. Akt phosphorylation is essential for nuclear translocation and retention in NGF-stimulated PC12 cells. Biochem. Biophys. Res. Commun. 2006, 349, 789–798. [Google Scholar] [CrossRef]
- Stracker, T.H.; Petrini, J.H.J. The MRE11 complex: Starting from the ends. Nat. Rev. Mol. Cell Biol. 2011, 12, 90–103. [Google Scholar] [CrossRef]
- Branzei, D.; Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008, 9, 297–308. [Google Scholar] [CrossRef]
- Jones, D.R.; Foulger, R.; Keune, W.; Bultsma, Y.; Divecha, N. PtdIns5 P is an oxidative stress-induced second messenger that regulates PKB activation. FASEB J. 2012, 27, 1644–1656. [Google Scholar] [CrossRef]
- Keune, W.-J.; Jones, D.R.; Bultsma, Y.; Sommer, L.; Zhou, X.Z.; Lu, K.P.; Divecha, N. Regulation of Phosphatidylinositol-5-Phosphate Signaling by Pin1 Determines Sensitivity to Oxidative Stress. Sci. Signal. 2012, 5, ra86. [Google Scholar] [CrossRef]
- Bunce, M.W.; Boronenkov, I.V.; Anderson, R.A. Coordinated Activation of the Nuclear Ubiquitin Ligase Cul3-SPOP by the Generation of Phosphatidylinositol 5-Phosphate. J. Biol. Chem. 2008, 283, 8678–8686. [Google Scholar] [CrossRef]
- Xu, L.; Wei, Y.; Reboul, J.; Vaglio, P.; Shin, T.-H.; Vidal, M.; Elledge, S.J.; Harper, J.W. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 2003, 425, 316–321. [Google Scholar] [CrossRef]
- Geyer, R.; Wee, S.; Anderson, S.; Yates, J.; Wolf, D.A. BTB/POZ Domain Proteins Are Putative Substrate Adaptors for Cullin 3 Ubiquitin Ligases. Mol. Cell 2003, 12, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-M.; Wu, H.-L.; Xia, Q.-D.; Zhou, P.; Wang, S.-G.; Yu, X.; Hu, J. Novel insights into the SPOP E3 ubiquitin ligase: From the regulation of molecular mechanisms to tumorigenesis. Biomed. Pharmacother. 2022, 149, 112882. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Jang, S.-W.; Ye, K. Akt phosphorylation and nuclear phosphoinositide association mediate mRNA export and cell proliferation activities by ALY. Proc. Natl. Acad. Sci. USA 2008, 105, 8649–8654. [Google Scholar] [CrossRef]
- Mohan, N.; AP, S.; Francis, N.; Anderson, R.; Laishram, R.S. Phosphorylation regulates the Star-PAP-PIPKIα interaction and directs specificity toward mRNA targets. Nucleic Acids Res. 2015, 43, 7005–7020. [Google Scholar] [CrossRef]
- Gonzales, M.L.; Mellman, D.L.; Anderson, R.A. CKIα Is Associated with and Phosphorylates Star-PAP and Is Also Required for Expression of Select Star-PAP Target Messenger RNAs. J. Biol. Chem. 2008, 283, 12665–12673. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Chen, Y.; Gholamalamdari, O.; Wang, Y.; Ma, J.; Belmont, A.S. TSA-seq reveals a largely conserved genome organization relative to nuclear speckles with small position changes tightly correlated with gene expression changes. Genome Res. 2020, 31, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Xu, A.; Suh, P.-G.; Marmy-Conus, N.; Pearson, R.B.; Seok, O.Y.; Cocco, L.; Gilmour, R.S. Phosphorylation of Nuclear Phospholipase C β1 by Extracellular Signal-Regulated Kinase Mediates the Mitogenic Action of Insulin-Like Growth Factor I. Mol. Cell. Biol. 2001, 21, 2981–2990. [Google Scholar] [CrossRef]
- Xu, A.; Wang, Y.; Xu, L.Y.; Gilmour, R.S. Protein Kinase C α-mediated Negative Feedback Regulation Is Responsible for the Termination of Insulin-like Growth Factor I-induced Activation of Nuclear Phospholipase C β1 in Swiss 3T3 Cells. J. Biol. Chem. 2001, 276, 14980–14986. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vidalle, M.C.; Sheth, B.; Fazio, A.; Marvi, M.V.; Leto, S.; Koufi, F.-D.; Neri, I.; Casalin, I.; Ramazzotti, G.; Follo, M.Y.; et al. Nuclear Phosphoinositides as Key Determinants of Nuclear Functions. Biomolecules 2023, 13, 1049. https://doi.org/10.3390/biom13071049
Vidalle MC, Sheth B, Fazio A, Marvi MV, Leto S, Koufi F-D, Neri I, Casalin I, Ramazzotti G, Follo MY, et al. Nuclear Phosphoinositides as Key Determinants of Nuclear Functions. Biomolecules. 2023; 13(7):1049. https://doi.org/10.3390/biom13071049
Chicago/Turabian StyleVidalle, Magdalena C., Bhavwanti Sheth, Antonietta Fazio, Maria Vittoria Marvi, Stefano Leto, Foteini-Dionysia Koufi, Irene Neri, Irene Casalin, Giulia Ramazzotti, Matilde Y. Follo, and et al. 2023. "Nuclear Phosphoinositides as Key Determinants of Nuclear Functions" Biomolecules 13, no. 7: 1049. https://doi.org/10.3390/biom13071049
APA StyleVidalle, M. C., Sheth, B., Fazio, A., Marvi, M. V., Leto, S., Koufi, F.-D., Neri, I., Casalin, I., Ramazzotti, G., Follo, M. Y., Ratti, S., Manzoli, L., Gehlot, S., Divecha, N., & Fiume, R. (2023). Nuclear Phosphoinositides as Key Determinants of Nuclear Functions. Biomolecules, 13(7), 1049. https://doi.org/10.3390/biom13071049







