Pharmaceuticals Promoting Premature Termination Codon Readthrough: Progress in Development
Abstract
1. Introduction
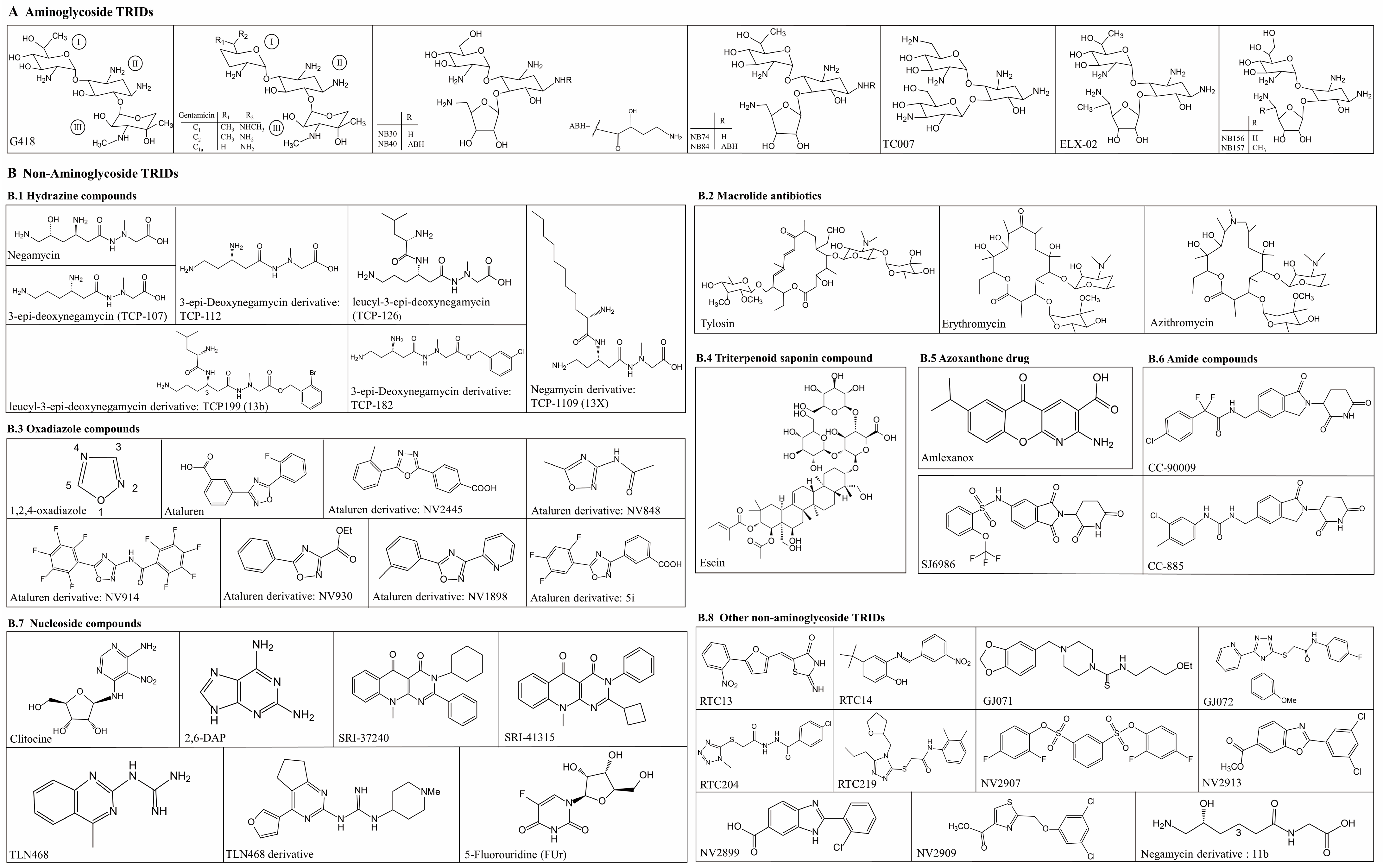
2. Aminoglycosides TRIDs
2.1. G418
2.2. Gentamicin
2.3. Aminoglycoside Derivatives
2.3.1. NB30 and NB54
2.3.2. NB74 and NB84
2.3.3. TC007
2.3.4. ELX-02
2.3.5. Other Aminoglycoside Derivatives
3. Non-Aminoglycoside TRIDs
3.1. Hydrazine Compounds—Negamycin and Its Derivatives
3.2. Macrolide Antibiotics—Tylosin, Erythromycin and Azithromycin
3.3. Oxadiazole Compounds—Ataluren and Its Derivatives
3.4. Triterpenoid Saponin Compound-Escin
3.5. Azoxanthone Drug-Amlexanox
3.6. Amide Compounds-CC-90009, CC-885 and SJ6986
3.7. Nucleoside Compounds
3.7.1. Clitocine
3.7.2. H7 and 2,6-DAP
3.7.3. SRI-37240 and SRI-41315
3.7.4. 2-Guanidino-Quinazoline
3.7.5. 5-Fluorouridine (FUr)
3.8. Other Non-Aminoglycoside TRIDs
4. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mort, M.; Ivanov, D.; Cooper, D.N.; Chuzhanova, N.A. A Meta-Analysis of Nonsense Mutations Causing Human Genetic Disease. Hum. Mutat. 2008, 29, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Singer, R.H. Cellular Variability of Nonsense-Mediated MRNA Decay. Nat. Commun. 2021, 12, 7203. [Google Scholar] [CrossRef] [PubMed]
- Palma, M.; Lejeune, F. Deciphering the Molecular Mechanism of Stop Codon Readthrough. Biol. Rev. Camb. Philos. Soc. 2021, 96, 310–329. [Google Scholar] [CrossRef]
- Martins-Dias, P.; Romão, L. Nonsense Suppression Therapies in Human Genetic Diseases. Cell. Mol. Life Sci. 2021, 78, 4677–4701. [Google Scholar] [CrossRef]
- Floquet, C.; Hatin, I.; Rousset, J.-P.; Bidou, L. Statistical Analysis of Readthrough Levels for Nonsense Mutations in Mammalian Cells Reveals a Major Determinant of Response to Gentamicin. PLoS Genet. 2012, 8, e1002608. [Google Scholar] [CrossRef] [PubMed]
- Dabrowski, M.; Bukowy-Bieryllo, Z.; Zietkiewicz, E. Advances in Therapeutic Use of a Drug-Stimulated Translational Readthrough of Premature Termination Codons. Mol. Med. 2018, 24, 25. [Google Scholar] [CrossRef]
- Lombardi, S.; Testa, M.F.; Pinotti, M.; Branchini, A. Molecular Insights into Determinants of Translational Readthrough and Implications for Nonsense Suppression Approaches. Int. J. Mol. Sci. 2020, 21, 9449. [Google Scholar] [CrossRef]
- Nguyen, L.S.; Wilkinson, M.F.; Gecz, J. Nonsense-Mediated MRNA Decay: Inter-Individual Variability and Human Disease. Neurosci. Biobehav. Rev. 2014, 46, 175–186. [Google Scholar] [CrossRef]
- Lee, H.-L.R.; Dougherty, J.P. Pharmaceutical Therapies to Recode Nonsense Mutations in Inherited Diseases. Pharmacol. Ther. 2012, 136, 227–266. [Google Scholar] [CrossRef]
- Nagel-Wolfrum, K.; Möller, F.; Penner, I.; Baasov, T.; Wolfrum, U. Targeting Nonsense Mutations in Diseases with Translational Read-Through-Inducing Drugs (TRIDs). BioDrugs 2016, 30, 49–74. [Google Scholar] [CrossRef]
- Goldmann, T.; Overlack, N.; Möller, F.; Belakhov, V.; van Wyk, M.; Baasov, T.; Wolfrum, U.; Nagel-Wolfrum, K. A Comparative Evaluation of NB30, NB54 and PTC124 in Translational Read-through Efficacy for Treatment of an USH1C Nonsense Mutation. EMBO Mol. Med. 2012, 4, 1186–1199. [Google Scholar] [CrossRef]
- Morais, P.; Adachi, H.; Yu, Y.-T. Suppression of Nonsense Mutations by New Emerging Technologies. Int. J. Mol. Sci. 2020, 21, 4394. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.F.; Mogg, A.E. Suppression of a Nonsense Mutation in Mammalian Cells in Vivo by the Aminoglycoside Antibiotics G-418 and Paromomycin. Nucleic. Acids Res. 1985, 13, 6265–6272. [Google Scholar] [CrossRef] [PubMed]
- Keeling, K.M.; Xue, X.; Gunn, G.; Bedwell, D.M. Therapeutics Based on Stop Codon Readthrough. Annu. Rev. Genom. Hum. Genet. 2014, 15, 371–394. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.R.; Puglisi, J.D. Structural Origins of Aminoglycoside Specificity for Prokaryotic Ribosomes. J. Mol. Biol. 2001, 306, 1037–1058. [Google Scholar] [CrossRef]
- Bidou, L.; Allamand, V.; Rousset, J.-P.; Namy, O. Sense from Nonsense: Therapies for Premature Stop Codon Diseases. Trends Mol. Med. 2012, 18, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Schilff, M.; Sargsyan, Y.; Hofhuis, J.; Thoms, S. Stop Codon Context-Specific Induction of Translational Readthrough. Biomolecules 2021, 11, 1006. [Google Scholar] [CrossRef]
- Baradaran-Heravi, A.; Bauer, C.C.; Pickles, I.B.; Hosseini-Farahabadi, S.; Balgi, A.D.; Choi, K.; Linley, D.M.; Beech, D.J.; Roberge, M.; Bon, R.S. Nonselective TRPC Channel Inhibition and Suppression of Aminoglycoside-Induced Premature Termination Codon Readthrough by the Small Molecule AC1903. J. Biol. Chem. 2022, 298, 101546. [Google Scholar] [CrossRef]
- Prokhorova, I.; Altman, R.B.; Djumagulov, M.; Shrestha, J.P.; Urzhumtsev, A.; Ferguson, A.; Chang, C.-W.T.; Yusupov, M.; Blanchard, S.C.; Yusupova, G. Aminoglycoside Interactions and Impacts on the Eukaryotic Ribosome. Proc. Natl. Acad. Sci. USA 2017, 114, E10899–E10908. [Google Scholar] [CrossRef]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef]
- Brasell, E.J.; Chu, L.; El Kares, R.; Seo, J.H.; Loesch, R.; Iglesias, D.M.; Goodyer, P. The Aminoglycoside Geneticin Permits Translational Readthrough of the CTNS W138X Nonsense Mutation in Fibroblasts from Patients with Nephropathic Cystinosis. Pediatr. Nephrol. 2019, 34, 873–881. [Google Scholar] [CrossRef]
- Wangen, J.R.; Green, R. Stop Codon Context Influences Genome-Wide Stimulation of Termination Codon Readthrough by Aminoglycosides. Elife 2020, 9, e52611. [Google Scholar] [CrossRef]
- Friesen, W.J.; Johnson, B.; Sierra, J.; Zhuo, J.; Vazirani, P.; Xue, X.; Tomizawa, Y.; Baiazitov, R.; Morrill, C.; Ren, H.; et al. The Minor Gentamicin Complex Component, X2, Is a Potent Premature Stop Codon Readthrough Molecule with Therapeutic Potential. PLoS ONE 2018, 13, e0206158. [Google Scholar] [CrossRef]
- Ng, M.Y.; Zhang, H.; Weil, A.; Singh, V.; Jamiolkowski, R.; Baradaran-Heravi, A.; Roberge, M.; Jacobson, A.; Friesen, W.; Welch, E.; et al. New in Vitro Assay Measuring Direct Interaction of Nonsense Suppressors with the Eukaryotic Protein Synthesis Machinery. ACS Med. Chem. Lett. 2018, 9, 1285–1291. [Google Scholar] [CrossRef]
- Mosallaei, D.; Hao, M.; Antaya, R.J.; Levian, B.; Kwong, A.; Cogan, J.; Hamilton, C.; Schwieger-Briel, A.; Tan, C.; Tang, X.; et al. Molecular and Clinical Outcomes After Intravenous Gentamicin Treatment for Patients With Junctional Epidermolysis Bullosa Caused by Nonsense Variants. JAMA Dermatol. 2022, 158, 366–374. [Google Scholar] [CrossRef]
- Martínez-Santamaría, L.; Maseda, R.; de Arriba, M.D.C.; Membrilla, J.A.; Sigüenza, A.I.; Mascías, J.; García, M.; Quintana, L.; Esteban-Rodríguez, I.; Hernández-Fernández, C.P.; et al. Evaluation of Systemic Gentamicin as Translational Readthrough Therapy for a Patient with Epidermolysis Bullosa Simplex with Muscular Dystrophy Owing to PLEC1 Pathogenic Nonsense Variants. JAMA Dermatol. 2022, 158, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Jones, J.R.; Lanier, J.; Keeling, K.M.; Lindsey, J.R.; Tousson, A.; Bebök, Z.; Whitsett, J.A.; Dey, C.R.; Colledge, W.H.; et al. Aminoglycoside Suppression of a Premature Stop Mutation in a Cftr−/− Mouse Carrying a Human CFTR-G542X Transgene. J. Mol. Med. 2002, 80, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Wilschanski, M.; Yahav, Y.; Yaacov, Y.; Blau, H.; Bentur, L.; Rivlin, J.; Aviram, M.; Bdolah-Abram, T.; Bebok, Z.; Shushi, L.; et al. Gentamicin-Induced Correction of CFTR Function in Patients with Cystic Fibrosis and CFTR Stop Mutations. N. Engl. J. Med. 2003, 349, 1433–1441. [Google Scholar] [CrossRef]
- Sermet-Gaudelus, I.; Renouil, M.; Fajac, A.; Bidou, L.; Parbaille, B.; Pierrot, S.; Davy, N.; Bismuth, E.; Reinert, P.; Lenoir, G.; et al. In Vitro Prediction of Stop-Codon Suppression by Intravenous Gentamicin in Patients with Cystic Fibrosis: A Pilot Study. BMC Med. 2007, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Barton-Davis, E.R.; Cordier, L.; Shoturma, D.I.; Leland, S.E.; Sweeney, H.L. Aminoglycoside Antibiotics Restore Dystrophin Function to Skeletal Muscles of Mdx Mice. J. Clin. Investig. 1999, 104, 375–381. [Google Scholar] [CrossRef]
- Wagner, K.R.; Hamed, S.; Hadley, D.W.; Gropman, A.L.; Burstein, A.H.; Escolar, D.M.; Hoffman, E.P.; Fischbeck, K.H. Gentamicin Treatment of Duchenne and Becker Muscular Dystrophy Due to Nonsense Mutations. Ann. Neurol. 2001, 49, 706–711. [Google Scholar] [CrossRef]
- Politano, L.; Nigro, G.; Nigro, V.; Piluso, G.; Papparella, S.; Paciello, O.; Comi, L.I. Gentamicin Administration in Duchenne Patients with Premature Stop Codon. Preliminary Results. Acta Myol. 2003, 22, 15–21. [Google Scholar]
- Malik, V.; Rodino-Klapac, L.R.; Viollet, L.; Wall, C.; King, W.; Al-Dahhak, R.; Lewis, S.; Shilling, C.J.; Kota, J.; Serrano-Munuera, C.; et al. Gentamicin-Induced Readthrough of Stop Codons in Duchenne Muscular Dystrophy. Ann. Neurol. 2010, 67, 771–780. [Google Scholar] [CrossRef]
- James, P.D.; Raut, S.; Rivard, G.E.; Poon, M.-C.; Warner, M.; McKenna, S.; Leggo, J.; Lillicrap, D. Aminoglycoside Suppression of Nonsense Mutations in Severe Hemophilia. Blood 2005, 106, 3043–3048. [Google Scholar] [CrossRef]
- Shiozuka, M.; Wagatsuma, A.; Kawamoto, T.; Sasaki, H.; Shimada, K.; Takahashi, Y.; Nonomura, Y.; Matsuda, R. Transdermal Delivery of a Readthrough-Inducing Drug: A New Approach of Gentamicin Administration for the Treatment of Nonsense Mutation-Mediated Disorders. J. Biochem. 2010, 147, 463–470. [Google Scholar] [CrossRef]
- Wang, S.; Yang, Z.; Liu, Y.; Zhao, M.-T.; Zhao, J.; Zhang, H.; Liu, Z.-Y.; Wang, X.-L.; Ma, L.; Yang, Y.-H. Application of Topical Gentamicin—A New Era in the Treatment of Genodermatosis. World J. Pediatr. 2021, 17, 568–575. [Google Scholar] [CrossRef]
- Ohguchi, Y.; Nomura, T.; Suzuki, S.; Takeda, M.; Miyauchi, T.; Mizuno, O.; Shinkuma, S.; Fujita, Y.; Nemoto, O.; Ono, K.; et al. Gentamicin-Induced Readthrough and Nonsense-Mediated MRNA Decay of SERPINB7 Nonsense Mutant Transcripts. J. Investig. Dermatol. 2018, 138, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yu, X.; Pan, C.; Wang, Y.; Han, J.; Yao, Z.; Li, M. Effect of Gentamicin Ointment in Patients with Nagashima-Type Palmoplantar Keratosis: A Double-Blind Vehicle-Controlled Study. Acta Derm. Venereol. 2021, 101, adv00392. [Google Scholar] [CrossRef] [PubMed]
- Woodley, D.T.; Cogan, J.; Hou, Y.; Lyu, C.; Marinkovich, M.P.; Keene, D.; Chen, M. Gentamicin Induces Functional Type VII Collagen in Recessive Dystrophic Epidermolysis Bullosa Patients. J. Clin. Investig. 2017, 127, 3028–3038. [Google Scholar] [CrossRef] [PubMed]
- Kwong, A.; Cogan, J.; Hou, Y.; Antaya, R.; Hao, M.; Kim, G.; Lincoln, V.; Chen, Q.; Woodley, D.T.; Chen, M. Gentamicin Induces Laminin 332 and Improves Wound Healing in Junctional Epidermolysis Bullosa Patients with Nonsense Mutations. Mol. Ther. 2020, 28, 1327–1338. [Google Scholar] [CrossRef]
- Li, Y.; Shen, J.; Liang, J.; Zheng, L.; Chen, F.; Yao, Z.; Li, M. Gentamicin Induces COL17A1 Nonsense Mutation Readthrough in Junctional Epidermolysis Bullosa. J. Dermatol. 2020, 47, e82–e83. [Google Scholar] [CrossRef]
- Hung, J.-H.; Hou, P.-C.; Huang, F.-C.; Hsu, C.-K. Topical Gentamicin Ointment Induces LAMB3 Nonsense Mutation Readthrough and Improves Corneal Erosions in a Patient with Junctional Epidermolysis Bullosa. Clin. Exp. Ophthalmol. 2021, 49, 309–312. [Google Scholar] [CrossRef]
- Kellermayer, R.; Szigeti, R.; Keeling, K.M.; Bedekovics, T.; Bedwell, D.M. Aminoglycosides as Potential Pharmacogenetic Agents in the Treatment of Hailey–Hailey Disease. J. Investig. Dermatol. 2006, 126, 229–231. [Google Scholar] [CrossRef]
- Peled, A.; Samuelov, L.; Sarig, O.; Bochner, R.; Malki, L.; Pavlovsky, M.; Pichinuk, E.; Weil, M.; Sprecher, E. Treatment of Hereditary Hypotrichosis Simplex of the Scalp with Topical Gentamicin. Br. J. Dermatol. 2020, 183, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Hammersen, J.; Neuner, A.; Wild, F.; Schneider, H. Attenuation of Severe Generalized Junctional Epidermolysis Bullosa by Systemic Treatment with Gentamicin. Dermatology 2019, 235, 315–322. [Google Scholar] [CrossRef]
- Baradaran-Heravi, A.; Balgi, A.D.; Zimmerman, C.; Choi, K.; Shidmoossavee, F.S.; Tan, J.S.; Bergeaud, C.; Krause, A.; Flibotte, S.; Shimizu, Y.; et al. Novel Small Molecules Potentiate Premature Termination Codon Readthrough by Aminoglycosides. Nucleic Acids Res. 2016, 44, 6583–6598. [Google Scholar] [CrossRef]
- Rabea, S.M.; Baradaran-Heravi, A.; Balgi, A.D.; Krause, A.; Farahabadi, S.H.; Roberge, M.; Grierson, D.S. 2-Aminothiazole-4-Carboxamides Enhance Readthrough of Premature Termination Codons by Aminoglycosides. ACS Med. Chem. Lett. 2019, 10, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, M.W.; Gerak, C.A.N.; Chow, C.C.T.; Rastelli, E.J.; Elmore, K.E.; Stahl, F.; Hosseini-Farahabadi, S.; Baradaran-Heravi, A.; Coltart, D.M.; Roberge, M. The Antimalarial Drug Mefloquine Enhances TP53 Premature Termination Codon Readthrough by Aminoglycoside G418. PLoS ONE 2019, 14, e0216423. [Google Scholar] [CrossRef]
- Hosseini-Farahabadi, S.; Baradaran-Heravi, A.; Zimmerman, C.; Choi, K.; Flibotte, S.; Roberge, M. Small Molecule Y-320 Stimulates Ribosome Biogenesis, Protein Synthesis, and Aminoglycoside-Induced Premature Termination Codon Readthrough. PLoS Biol. 2021, 19, e3001221. [Google Scholar] [CrossRef] [PubMed]
- Shulman, E.; Belakhov, V.; Wei, G.; Kendall, A.; Meyron-Holtz, E.G.; Ben-Shachar, D.; Schacht, J.; Baasov, T. Designer Aminoglycosides That Selectively Inhibit Cytoplasmic Rather than Mitochondrial Ribosomes Show Decreased Ototoxicity. J. Biol. Chem. 2014, 289, 2318–2330. [Google Scholar] [CrossRef]
- Nudelman, I.; Rebibo-Sabbah, A.; Cherniavsky, M.; Belakhov, V.; Hainrichson, M.; Chen, F.; Schacht, J.; Pilch, D.S.; Ben-Yosef, T.; Baasov, T. Development of Novel Aminoglycoside (NB54) with Reduced Toxicity and Enhanced Suppression of Disease-Causing Premature Stop Mutations. J. Med. Chem. 2009, 52, 2836–2845. [Google Scholar] [CrossRef]
- Rebibo-Sabbah, A.; Nudelman, I.; Ahmed, Z.M.; Baasov, T.; Ben-Yosef, T. In Vitro and Ex Vivo Suppression by Aminoglycosides of PCDH15 Nonsense Mutations Underlying Type 1 Usher Syndrome. Hum. Genet. 2007, 122, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Rowe, S.M.; Sloane, P.; Tang, L.P.; Backer, K.; Mazur, M.; Buckley-Lanier, J.; Nudelman, I.; Belakhov, V.; Bebok, Z.; Schwiebert, E.; et al. Suppression of CFTR Premature Termination Codons and Rescue of CFTR Protein and Function by the Synthetic Aminoglycoside NB54. J. Mol. Med. 2011, 89, 1149–1161. [Google Scholar] [CrossRef]
- Brendel, C.; Belakhov, V.; Werner, H.; Wegener, E.; Gärtner, J.; Nudelman, I.; Baasov, T.; Huppke, P. Readthrough of Nonsense Mutations in Rett Syndrome: Evaluation of Novel Aminoglycosides and Generation of a New Mouse Model. J. Mol. Med. 2011, 89, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Belakhov, V.; Kandasamy, J.; Baasov, T.; Li, S.-C.; Li, Y.-T.; Bedwell, D.M.; Keeling, K.M. The Designer Aminoglycoside NB84 Significantly Reduces Glycosaminoglycan Accumulation Associated with MPS I-H in the Idua-W392X Mouse. Mol. Genet. Metab. 2012, 105, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Nudelman, I.; Glikin, D.; Smolkin, B.; Hainrichson, M.; Belakhov, V.; Baasov, T. Repairing Faulty Genes by Aminoglycosides: Development of New Derivatives of Geneticin (G418) with Enhanced Suppression of Diseases-Causing Nonsense Mutations. Bioorg. Med. Chem. 2010, 18, 3735–3746. [Google Scholar] [CrossRef]
- Gunn, G.; Dai, Y.; Du, M.; Belakhov, V.; Kandasamy, J.; Schoeb, T.R.; Baasov, T.; Bedwell, D.M.; Keeling, K.M. Long-Term Nonsense Suppression Therapy Moderates MPS I-H Disease Progression. Mol. Genet. Metab. 2014, 111, 374–381. [Google Scholar] [CrossRef]
- Chang, C.-W.T.; Hui, Y.; Elchert, B.; Wang, J.; Li, J.; Rai, R. Pyranmycins, a Novel Class of Aminoglycosides with Improved Acid Stability: The SAR of D-Pyranoses on Ring III of Pyranmycin. Org. Lett. 2002, 4, 4603–4606. [Google Scholar] [CrossRef]
- Mattis, V.B.; Rai, R.; Wang, J.; Chang, C.-W.T.; Coady, T.; Lorson, C.L. Novel Aminoglycosides Increase SMN Levels in Spinal Muscular Atrophy Fibroblasts. Hum. Genet. 2006, 120, 589–601. [Google Scholar] [CrossRef]
- Mattis, V.B.; Ebert, A.D.; Fosso, M.Y.; Chang, C.-W.; Lorson, C.L. Delivery of a Read-through Inducing Compound, TC007, Lessens the Severity of a Spinal Muscular Atrophy Animal Model. Hum. Mol. Genet. 2009, 18, 3906–3913. [Google Scholar] [CrossRef]
- Mattis, V.B.; Fosso, M.Y.; Chang, C.-W.; Lorson, C.L. Subcutaneous Administration of TC007 Reduces Disease Severity in an Animal Model of SMA. BMC Neurosci. 2009, 10, 142. [Google Scholar] [CrossRef]
- Kerem, E. ELX-02: An Investigational Read-through Agent for the Treatment of Nonsense Mutation-Related Genetic Disease. Expert Opin. Investig. Drugs 2020, 29, 1347–1354. [Google Scholar] [CrossRef]
- Bidou, L.; Bugaud, O.; Belakhov, V.; Baasov, T.; Namy, O. Characterization of New-Generation Aminoglycoside Promoting Premature Termination Codon Readthrough in Cancer Cells. RNA Biol. 2017, 14, 378–388. [Google Scholar] [CrossRef]
- Crawford, D.K.; Mullenders, J.; Pott, J.; Boj, S.F.; Landskroner-Eiger, S.; Goddeeris, M.M. Targeting G542X CFTR Nonsense Alleles with ELX-02 Restores CFTR Function in Human-Derived Intestinal Organoids. J. Cyst. Fibros. 2021, 20, 436–442. [Google Scholar] [CrossRef] [PubMed]
- de Poel, E.; Spelier, S.; Suen, S.W.F.; Kruisselbrink, E.; Graeber, S.Y.; Mall, M.A.; Weersink, E.J.M.; van der Eerden, M.M.; Koppelman, G.H.; van der Ent, C.K.; et al. Functional Restoration of CFTR Nonsense Mutations in Intestinal Organoids. J. Cyst. Fibros. 2022, 21, 246–253. [Google Scholar] [CrossRef]
- Brasell, E.J.; Chu, L.L.; Akpa, M.M.; Eshkar-Oren, I.; Alroy, I.; Corsini, R.; Gilfix, B.M.; Yamanaka, Y.; Huertas, P.; Goodyer, P. The Novel Aminoglycoside, ELX-02, Permits CTNSW138X Translational Read-through and Restores Lysosomal Cystine Efflux in Cystinosis. PLoS ONE 2019, 14, e0223954. [Google Scholar] [CrossRef] [PubMed]
- Crawford, D.K.; Alroy, I.; Sharpe, N.; Goddeeris, M.M.; Williams, G. ELX-02 Generates Protein via Premature Stop Codon Read-Through without Inducing Native Stop Codon Read-Through Proteins. J. Pharmacol. Exp. Ther. 2020, 374, 264–272. [Google Scholar] [CrossRef]
- Leubitz, A.; Frydman-Marom, A.; Sharpe, N.; van Duzer, J.; Campbell, K.C.M.; Vanhoutte, F. Safety, Tolerability, and Pharmacokinetics of Single Ascending Doses of ELX-02, a Potential Treatment for Genetic Disorders Caused by Nonsense Mutations, in Healthy Volunteers. Clin. Pharmacol. Drug Dev. 2019, 8, 984–994. [Google Scholar] [CrossRef]
- Haverty, T.; Wyatt, D.J.; Porter, K.M.; Leubitz, A.; Banks, K.; Goodyer, P.; Hu, M. Phase 1 Renal Impairment Trial Results Supports Targeted Individualized Dosing of ELX-02 in Patients With Nephropathic Cystinosis. J. Clin. Pharmacol. 2021, 61, 923–931. [Google Scholar] [CrossRef]
- Leubitz, A.; Vanhoutte, F.; Hu, M.; Porter, K.; Gordon, E.; Tencer, K.; Campbell, K.; Banks, K.; Haverty, T. A Randomized, Double-Blind, Placebo-Controlled, Multiple Dose Escalation Study to Evaluate the Safety and Pharmacokinetics of ELX-02 in Healthy Subjects. Clin. Pharmacol. Drug Dev. 2021, 10, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Sabbavarapu, N.M.; Shavit, M.; Degani, Y.; Smolkin, B.; Belakhov, V.; Baasov, T. Design of Novel Aminoglycoside Derivatives with Enhanced Suppression of Diseases-Causing Nonsense Mutations. ACS Med. Chem. Lett. 2016, 7, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.J.; Kang, W.; Kwon, Y.; Oh, J.W.; Jung, H.; Seo, M.; Seol, Y.; Wi, J.B.; Ban, Y.H.; Yoon, Y.J.; et al. Chemo-Enzymatic Synthesis of Pseudo-Trisaccharide Aminoglycoside Antibiotics with Enhanced Nonsense Read-through Inducer Activity. Chem. Med. Chem. 2022, 18, e202200497. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, A.; Hamada, K.; Hayashi, Y. Chemotherapeutics Overcoming Nonsense Mutation-Associated Genetic Diseases: Medicinal Chemistry of Negamycin. J. Antibiot. 2018, 71, 205–214. [Google Scholar] [CrossRef]
- Arakawa, M.; Shiozuka, M.; Nakayama, Y.; Hara, T.; Hamada, M.; Kondo, S.; Ikeda, D.; Takahashi, Y.; Sawa, R.; Nonomura, Y.; et al. Negamycin Restores Dystrophin Expression in Skeletal and Cardiac Muscles of Mdx Mice. J. Biochem. 2003, 134, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, S.J.; Blaha, G.; Moore, P.B. Negamycin Binds to the Wall of the Nascent Chain Exit Tunnel of the 50S Ribosomal Subunit. Antimicrob. Agents Chemother. 2007, 51, 4462–4465. [Google Scholar] [CrossRef]
- Taguchi, A.; Hamada, K.; Kotake, M.; Shiozuka, M.; Nakaminami, H.; Pillaiyar, T.; Takayama, K.; Yakushiji, F.; Noguchi, N.; Usui, T.; et al. Discovery of Natural Products Possessing Selective Eukaryotic Readthrough Activity: 3-Epi-Deoxynegamycin and Its Leucine Adduct. Chem. Med. Chem. 2014, 9, 2233–2237. [Google Scholar] [CrossRef]
- Hamada, K.; Omura, N.; Taguchi, A.; Baradaran-Heravi, A.; Kotake, M.; Arai, M.; Takayama, K.; Taniguchi, A.; Roberge, M.; Hayashi, Y. New Negamycin-Based Potent Readthrough Derivative Effective against TGA-Type Nonsense Mutations. ACS Med. Chem. Lett. 2019, 10, 1450–1456. [Google Scholar] [CrossRef]
- Otani, Y.; Taguchi, A.; Hamada, K.; Hayashi, Y.; Yamaguchi, Y.; Baba, H. Influence of Novel Readthrough Agents on Myelin Protein Zero Translation in the Peripheral Nervous System. Neuropharmacology 2022, 211, 109059. [Google Scholar] [CrossRef]
- Thompson, J.; Pratt, C.A.; Dahlberg, A.E. Effects of a Number of Classes of 50S Inhibitors on Stop Codon Readthrough during Protein Synthesis. Antimicrob. Agents Chemother. 2004, 48, 4889–4891. [Google Scholar] [CrossRef]
- Zilberberg, A.; Lahav, L.; Rosin-Arbesfeld, R. Restoration of APC Gene Function in Colorectal Cancer Cells by Aminoglycoside- and Macrolide-Induced Read-through of Premature Termination Codons. Gut 2010, 59, 496–507. [Google Scholar] [CrossRef]
- Caspi, M.; Firsow, A.; Rajkumar, R.; Skalka, N.; Moshkovitz, I.; Munitz, A.; Pasmanik-Chor, M.; Greif, H.; Megido, D.; Kariv, R.; et al. A Flow Cytometry-Based Reporter Assay Identifies Macrolide Antibiotics as Nonsense Mutation Read-through Agents. J. Mol. Med. 2016, 94, 469–482. [Google Scholar] [CrossRef]
- Osman, E.Y.; Washington, C.W.; Simon, M.E.; Megiddo, D.; Greif, H.; Lorson, C.L. Analysis of Azithromycin Monohydrate as a Single or a Combinatorial Therapy in a Mouse Model of Severe Spinal Muscular Atrophy. J. Neuromuscul. Dis. 2017, 4, 237–249. [Google Scholar] [CrossRef]
- Dabrowski, M.; Bukowy-Bieryllo, Z.; Jackson, C.L.; Zietkiewicz, E. Properties of Non-Aminoglycoside Compounds Used to Stimulate Translational Readthrough of PTC Mutations in Primary Ciliary Dyskinesia. Int. J. Mol. Sci. 2021, 22, 4923. [Google Scholar] [CrossRef]
- Campofelice, A.; Lentini, L.; Di Leonardo, A.; Melfi, R.; Tutone, M.; Pace, A.; Pibiri, I. Strategies against Nonsense: Oxadiazoles as Translational Readthrough-Inducing Drugs (TRIDs). Int. J. Mol. Sci. 2019, 20, 3329. [Google Scholar] [CrossRef]
- Welch, E.M.; Barton, E.R.; Zhuo, J.; Tomizawa, Y.; Friesen, W.J.; Trifillis, P.; Paushkin, S.; Patel, M.; Trotta, C.R.; Hwang, S.; et al. PTC124 Targets Genetic Disorders Caused by Nonsense Mutations. Nature 2007, 447, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Reeves, E.P.; Dunlea, D.M.; McQuillan, K.; O’Dwyer, C.A.; Carroll, T.P.; Saldova, R.; Akepati, P.R.; Wormald, M.R.; McElvaney, O.J.; Shutchaidat, V.; et al. Circulating Truncated Alpha-1 Antitrypsin Glycoprotein in Patient Plasma Retains Anti-Inflammatory Capacity. J. Immunol. 2019, 202, 2240–2253. [Google Scholar] [CrossRef] [PubMed]
- Eintracht, J.; Forsythe, E.; May-Simera, H.; Moosajee, M. Translational Readthrough of Ciliopathy Genes BBS2 and ALMS1 Restores Protein, Ciliogenesis and Function in Patient Fibroblasts. E Bio. Med. 2021, 70, 103515. [Google Scholar] [CrossRef]
- Bezzerri, V.; Lentini, L.; Api, M.; Busilacchi, E.M.; Cavalieri, V.; Pomilio, A.; Diomede, F.; Pegoraro, A.; Cesaro, S.; Poloni, A.; et al. Novel Translational Read-through-Inducing Drugs as a Therapeutic Option for Shwachman-Diamond Syndrome. Biomedicines 2022, 10, 886. [Google Scholar] [CrossRef]
- Pibiri, I.; Lentini, L.; Melfi, R.; Tutone, M.; Baldassano, S.; Ricco Galluzzo, P.; Di Leonardo, A.; Pace, A. Rescuing the CFTR Protein Function: Introducing 1,3,4-Oxadiazoles as Translational Readthrough Inducing Drugs. Eur. J. Med. Chem. 2018, 159, 126–142. [Google Scholar] [CrossRef] [PubMed]
- Pibiri, I.; Lentini, L.; Tutone, M.; Melfi, R.; Pace, A.; Di Leonardo, A. Exploring the Readthrough of Nonsense Mutations by Non-Acidic Ataluren Analogues Selected by Ligand-Based Virtual Screening. Eur. J. Med. Chem. 2016, 122, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Moosajee, M.; Tracey-White, D.; Smart, M.; Weetall, M.; Torriano, S.; Kalatzis, V.; da Cruz, L.; Coffey, P.; Webster, A.R.; Welch, E. Functional Rescue of REP1 Following Treatment with PTC124 and Novel Derivative PTC-414 in Human Choroideremia Fibroblasts and the Nonsense-Mediated Zebrafish Model. Hum. Mol. Genet. 2016, 25, 3416–3431. [Google Scholar] [CrossRef]
- Pibiri, I.; Lentini, L.; Melfi, R.; Gallucci, G.; Pace, A.; Spinello, A.; Barone, G.; Di Leonardo, A. Enhancement of Premature Stop Codon Readthrough in the CFTR Gene by Ataluren (PTC124) Derivatives. Eur. J. Med. Chem. 2015, 101, 236–244. [Google Scholar] [CrossRef]
- Mutyam, V.; Du, M.; Xue, X.; Keeling, K.M.; White, E.L.; Bostwick, J.R.; Rasmussen, L.; Liu, B.; Mazur, M.; Hong, J.S.; et al. Discovery of Clinically Approved Agents That Promote Suppression of Cystic Fibrosis Transmembrane Conductance Regulator Nonsense Mutations. Am. J. Respir. Crit. Care Med. 2016, 194, 1092–1103. [Google Scholar] [CrossRef]
- Benhabiles, H.; Gonzalez-Hilarion, S.; Amand, S.; Bailly, C.; Prévotat, A.; Reix, P.; Hubert, D.; Adriaenssens, E.; Rebuffat, S.; Tulasne, D.; et al. Optimized Approach for the Identification of Highly Efficient Correctors of Nonsense Mutations in Human Diseases. PLoS ONE 2017, 12, e0187930. [Google Scholar] [CrossRef]
- Sharma, J.; Du, M.; Wong, E.; Mutyam, V.; Li, Y.; Chen, J.; Wangen, J.; Thrasher, K.; Fu, L.; Peng, N.; et al. A Small Molecule That Induces Translational Readthrough of CFTR Nonsense Mutations by ERF1 Depletion. Nat. Commun. 2021, 12, 4358. [Google Scholar] [CrossRef]
- Bidou, L.; Bugaud, O.; Merer, G.; Coupet, M.; Hatin, I.; Chirkin, E.; Karri, S.; Demais, S.; François, P.; Cintrat, J.-C.; et al. 2-Guanidino-Quinazoline Promotes the Readthrough of Nonsense Mutations Underlying Human Genetic Diseases. Proc. Natl. Acad. Sci. USA 2022, 119, e2122004119. [Google Scholar] [CrossRef]
- Du, L.; Damoiseaux, R.; Nahas, S.; Gao, K.; Hu, H.; Pollard, J.M.; Goldstine, J.; Jung, M.E.; Henning, S.M.; Bertoni, C.; et al. Nonaminoglycoside Compounds Induce Readthrough of Nonsense Mutations. J. Exp. Med. 2009, 206, 2285–2297. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Friesen, W.J.; Tomizawa, Y.; Leszyk, J.D.; Zhuo, J.; Johnson, B.; Dakka, J.; Trotta, C.R.; Xue, X.; Mutyam, V.; et al. Ataluren Stimulates Ribosomal Selection of Near-Cognate TRNAs to Promote Nonsense Suppression. Proc. Natl. Acad. Sci. USA 2016, 113, 12508–12513. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.Y.; Li, H.; Ghelfi, M.D.; Goldman, Y.E.; Cooperman, B.S. Ataluren and Aminoglycosides Stimulate Read-through of Nonsense Codons by Orthogonal Mechanisms. Proc. Natl. Acad. Sci. USA 2021, 118, e2020599118. [Google Scholar] [CrossRef]
- Huang, S.; Bhattacharya, A.; Ghelfi, M.D.; Li, H.; Fritsch, C.; Chenoweth, D.M.; Goldman, Y.E.; Cooperman, B.S. Ataluren Binds to Multiple Protein Synthesis Apparatus Sites and Competitively Inhibits Release Factor-Dependent Termination. Nat. Commun. 2022, 13, 2413. [Google Scholar] [CrossRef] [PubMed]
- Tutone, M.; Pibiri, I.; Lentini, L.; Pace, A.; Almerico, A.M. Deciphering the Nonsense Readthrough Mechanism of Action of Ataluren: An in Silico Compared Study. ACS Med. Chem. Lett. 2019, 10, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Michorowska, S. Ataluren—Promising Therapeutic Premature Termination Codon Readthrough Frontrunner. Pharmaceuticals 2021, 14, 785. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.; Sacristan-Reviriego, A.; Perdigão, P.R.L.; Sai, H.; Georgiou, M.; Kalitzeos, A.; Carr, A.-J.F.; Coffey, P.J.; Michaelides, M.; Bainbridge, J.; et al. Investigation of PTC124-Mediated Translational Readthrough in a Retinal Organoid Model of AIPL1-Associated Leber Congenital Amaurosis. Stem Cell Rep. 2022, 17, 2187–2202. [Google Scholar] [CrossRef]
- Beryozkin, A.; Samanta, A.; Gopalakrishnan, P.; Khateb, S.; Banin, E.; Sharon, D.; Nagel-Wolfrum, K. Translational Read-Through Drugs (TRIDs) Are Able to Restore Protein Expression and Ciliogenesis in Fibroblasts of Patients with Retinitis Pigmentosa Caused by a Premature Termination Codon in FAM161A. Int. J. Mol. Sci. 2022, 23, 3541. [Google Scholar] [CrossRef]
- Vössing, C.; Owczarek-Lipska, M.; Nagel-Wolfrum, K.; Reiff, C.; Jüschke, C.; Neidhardt, J. Translational Read-Through Therapy of RPGR Nonsense Mutations. Int. J. Mol. Sci. 2020, 21, 8418. [Google Scholar] [CrossRef] [PubMed]
- Samanta, A.; Stingl, K.; Kohl, S.; Ries, J.; Linnert, J.; Nagel-Wolfrum, K. Ataluren for the Treatment of Usher Syndrome 2A Caused by Nonsense Mutations. Int. J. Mol. Sci. 2019, 20, 6274. [Google Scholar] [CrossRef]
- Torriano, S.; Erkilic, N.; Baux, D.; Cereso, N.; De Luca, V.; Meunier, I.; Moosajee, M.; Roux, A.-F.; Hamel, C.P.; Kalatzis, V. The Effect of PTC124 on Choroideremia Fibroblasts and IPSC-Derived RPE Raises Considerations for Therapy. Sci. Rep. 2018, 8, 8234. [Google Scholar] [CrossRef]
- Lee, Y.-K.; Lau, Y.-M.; Cai, Z.-J.; Lai, W.-H.; Wong, L.-Y.; Tse, H.-F.; Ng, K.-M.; Siu, C.-W. Modeling Treatment Response for Lamin A/C Related Dilated Cardiomyopathy in Human Induced Pluripotent Stem Cells. J. Am. Heart Assoc. 2017, 6, e005677. [Google Scholar] [CrossRef]
- Ramsden, C.M.; Nommiste, B.; Lane, A.R.; Carr, A.-J.F.; Powner, M.B.; Smart, M.J.K.; Chen, L.L.; Muthiah, M.N.; Webster, A.R.; Moore, A.T.; et al. Rescue of the MERTK Phagocytic Defect in a Human IPSC Disease Model Using Translational Read-through Inducing Drugs. Sci. Rep. 2017, 7, 51. [Google Scholar] [CrossRef]
- Zomer-van Ommen, D.D.; Vijftigschild, L.a.W.; Kruisselbrink, E.; Vonk, A.M.; Dekkers, J.F.; Janssens, H.M.; de Winter-de Groot, K.M.; van der Ent, C.K.; Beekman, J.M. Limited Premature Termination Codon Suppression by Read-through Agents in Cystic Fibrosis Intestinal Organoids. J. Cyst. Fibros. 2016, 15, 158–162. [Google Scholar] [CrossRef]
- Kosmidis, G.; Veerman, C.C.; Casini, S.; Verkerk, A.O.; van de Pas, S.; Bellin, M.; Wilde, A.A.M.; Mummery, C.L.; Bezzina, C.R. Readthrough-Promoting Drugs Gentamicin and PTC124 Fail to Rescue Nav1.5 Function of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Carrying Nonsense Mutations in the Sodium Channel Gene SCN5A. Circ. Arrhythm. Electrophysiol. 2016, 9, e004227. [Google Scholar] [CrossRef]
- Schwarz, N.; Carr, A.-J.; Lane, A.; Moeller, F.; Chen, L.L.; Aguilà, M.; Nommiste, B.; Muthiah, M.N.; Kanuga, N.; Wolfrum, U.; et al. Translational Read-through of the RP2 Arg120stop Mutation in Patient IPSC-Derived Retinal Pigment Epithelium Cells. Hum. Mol. Genet. 2015, 24, 972–986. [Google Scholar] [CrossRef]
- Lojewski, X.; Staropoli, J.F.; Biswas-Legrand, S.; Simas, A.M.; Haliw, L.; Selig, M.K.; Coppel, S.H.; Goss, K.A.; Petcherski, A.; Chandrachud, U.; et al. Human IPSC Models of Neuronal Ceroid Lipofuscinosis Capture Distinct Effects of TPP1 and CLN3 Mutations on the Endocytic Pathway. Hum. Mol. Genet. 2014, 23, 2005–2022. [Google Scholar] [CrossRef]
- Konstan, M.W.; VanDevanter, D.R.; Rowe, S.M.; Wilschanski, M.; Kerem, E.; Sermet-Gaudelus, I.; DiMango, E.; Melotti, P.; McIntosh, J.; De Boeck, K. Efficacy and Safety of Ataluren in Patients with Nonsense-Mutation Cystic Fibrosis Not Receiving Chronic Inhaled Aminoglycosides: The International, Randomized, Double-Blind, Placebo-Controlled Ataluren Confirmatory Trial in Cystic Fibrosis (ACT CF). J. Cyst. Fibros. 2020, 19, 595–601. [Google Scholar] [CrossRef]
- McDonald, C.M.; Campbell, C.; Torricelli, R.E.; Finkel, R.S.; Flanigan, K.M.; Goemans, N.; Heydemann, P.; Kaminska, A.; Kirschner, J.; Muntoni, F.; et al. Ataluren in Patients with Nonsense Mutation Duchenne Muscular Dystrophy (ACT DMD): A Multicentre, Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet 2017, 390, 1489–1498. [Google Scholar] [CrossRef]
- Ryan, N.J. Ataluren: First Global Approval. Drugs 2014, 74, 1709–1714. [Google Scholar] [CrossRef]
- McDonald, C.M.; Muntoni, F.; Penematsa, V.; Jiang, J.; Kristensen, A.; Bibbiani, F.; Goodwin, E.; Gordish-Dressman, H.; Morgenroth, L.; Werner, C.; et al. Ataluren Delays Loss of Ambulation and Respiratory Decline in Nonsense Mutation Duchenne Muscular Dystrophy Patients. J. Comp. Eff. Res. 2022, 11, 139–155. [Google Scholar] [CrossRef] [PubMed]
- Muntoni, F.; Desguerre, I.; Guglieri, M.; Osorio, A.N.; Kirschner, J.; Tulinius, M.; Buccella, F.; Elfring, G.; Werner, C.; Schilling, T.; et al. Ataluren Use in Patients with Nonsense Mutation Duchenne Muscular Dystrophy: Patient Demographics and Characteristics from the STRIDE Registry. J. Comp. Eff. Res. 2019, 8, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Mercuri, E.; Muntoni, F.; Osorio, A.N.; Tulinius, M.; Buccella, F.; Morgenroth, L.P.; Gordish-Dressman, H.; Jiang, J.; Trifillis, P.; Zhu, J.; et al. Safety and Effectiveness of Ataluren: Comparison of Results from the STRIDE Registry and CINRG DMD Natural History Study. J. Comp. Eff. Res. 2020, 9, 341–360. [Google Scholar] [CrossRef] [PubMed]
- STRIDE Data Show Translarna™ Delays Loss of Ambulation by More Than Five Years in Boys with Nonsense Mutation Duchenne Muscular Dystrophy. PTC Therapeutics, Inc. Available online: https://ir.ptcbio.com/news-releases/news-release-details/stride-data-show-translarnatm-delays-loss-ambulation-more-five (accessed on 12 March 2023).
- PTC Therapeutics Announces CHMP Recommendation of Translarna™ (Ataluren) Label Update for Non-Ambulatory Patients with Duchenne Muscular Dystrophy. PTC Therapeutics, Inc. Available online: https://ir.ptcbio.com/news-releases/news-release-details/ptc-therapeutics-announces-chmp-recommendation-translarnatm (accessed on 12 March 2023).
- Bitetti, I.; Mautone, C.; Bertella, M.; Manna, M.R.; Varone, A. Early Treatment with Ataluren of a 2-Year-Old Boy with Nonsense Mutation Duchenne Dystrophy. Acta Myol. 2021, 40, 184–186. [Google Scholar] [CrossRef]
- Pasca, L.; Gardani, A.; Paoletti, M.; Velardo, D.; Berardinelli, A. Good Response to the Late Treatment with Ataluren in a Boy with Duchenne Muscular Dystrophy: Could the Previous Mild Course of the Disease Have Affected the Outcome? Acta Myol. 2022, 41, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Devinsky, O.; King, L.; Bluvstein, J.; Friedman, D. Ataluren for Drug-resistant Epilepsy in Nonsense Variant-mediated Dravet Syndrome and CDKL5 Deficiency Disorder. Ann. Clin. Transl. Neurol. 2021, 8, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Fazzari, M.; Frasca, A.; Bifari, F.; Landsberger, N. Aminoglycoside Drugs Induce Efficient Read-through of CDKL5 Nonsense Mutations, Slightly Restoring Its Kinase Activity. RNA Biol. 2019, 16, 1414–1423. [Google Scholar] [CrossRef]
- PTC Therapeutics Reports Fourth Quarter and Full Year 2019 Financial Results and Provides a Corporate Update. PTC Therapeutics, Inc. Available online: https://ir.ptcbio.com/news-releases/news-release-details/ptc-therapeutics-reports-fourth-quarter-and-full-year-2019 (accessed on 12 March 2023).
- Auld, D.S.; Thorne, N.; Maguire, W.F.; Inglese, J. Mechanism of PTC124 Activity in Cell-Based Luciferase Assays of Nonsense Codon Suppression. Proc. Natl. Acad. Sci. USA 2009, 106, 3585–3590. [Google Scholar] [CrossRef] [PubMed]
- Auld, D.S.; Lovell, S.; Thorne, N.; Lea, W.A.; Maloney, D.J.; Shen, M.; Rai, G.; Battaile, K.P.; Thomas, C.J.; Simeonov, A.; et al. Molecular Basis for the High-Affinity Binding and Stabilization of Firefly Luciferase by PTC124. Proc. Natl. Acad. Sci. USA 2010, 107, 4878–4883. [Google Scholar] [CrossRef]
- McElroy, S.P.; Nomura, T.; Torrie, L.S.; Warbrick, E.; Gartner, U.; Wood, G.; McLean, W.H.I. A Lack of Premature Termination Codon Read-through Efficacy of PTC124 (Ataluren) in a Diverse Array of Reporter Assays. PLoS Biol. 2013, 11, e1001593. [Google Scholar] [CrossRef] [PubMed]
- Kuang, L.; Hashimoto, K.; Huang, E.J.; Gentry, M.S.; Zhu, H. Frontotemporal Dementia Non-Sense Mutation of Progranulin Rescued by Aminoglycosides. Hum. Mol. Genet. 2020, 29, 624–634. [Google Scholar] [CrossRef]
- Harmer, S.C.; Mohal, J.S.; Kemp, D.; Tinker, A. Readthrough of Long-QT Syndrome Type 1 Nonsense Mutations Rescues Function but Alters the Biophysical Properties of the Channel. Biochem. J. 2012, 443, 635–642. [Google Scholar] [CrossRef]
- Bolze, F.; Mocek, S.; Zimmermann, A.; Klingenspor, M. Aminoglycosides, but Not PTC124 (Ataluren), Rescue Nonsense Mutations in the Leptin Receptor and in Luciferase Reporter Genes. Sci. Rep. 2017, 7, 1020. [Google Scholar] [CrossRef]
- Gallelli, L. Escin: A Review of Its Anti-Edematous, Anti-Inflammatory, and Venotonic Properties. Drug Des. Devel. Ther. 2019, 13, 3425–3437. [Google Scholar] [CrossRef]
- Dosanjh, A.; Won, C.Y. Amlexanox: A Novel Therapeutic for Atopic, Metabolic, and Inflammatory Disease. Yale J. Biol. Med. 2020, 93, 759–763. [Google Scholar]
- Bailly, C. The Potential Value of Amlexanox in the Treatment of Cancer: Molecular Targets and Therapeutic Perspectives. Biochem. Pharmacol. 2022, 197, 114895. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Hilarion, S.; Beghyn, T.; Jia, J.; Debreuck, N.; Berte, G.; Mamchaoui, K.; Mouly, V.; Gruenert, D.C.; Déprez, B.; Lejeune, F. Rescue of Nonsense Mutations by Amlexanox in Human Cells. Orphanet. J. Rare Dis. 2012, 7, 58. [Google Scholar] [CrossRef]
- Wang, X.; Shan, X.; Gregory-Evans, K.; Gregory-Evans, C.Y. RNA-Based Therapies in Animal Models of Leber Congenital Amaurosis Causing Blindness. Precis. Clin. Med. 2020, 3, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, V.S.; Jiang, Q.; Prisco, M.; Gruber, C.; Hofbauer, J.P.; Chen, M.; Has, C.; Bruckner-Tuderman, L.; McGrath, J.A.; Uitto, J.; et al. Amlexanox Enhances Premature Termination Codon Read-through in COL7A1 and Expression of Full Length Type VII Collagen: Potential Therapy for Recessive Dystrophic Epidermolysis Bullosa. J. Investig. Dermatol. 2017, 137, 1842–1849. [Google Scholar] [CrossRef]
- Banning, A.; Schiff, M.; Tikkanen, R. Amlexanox Provides a Potential Therapy for Nonsense Mutations in the Lysosomal Storage Disorder Aspartylglucosaminuria. Biochim. Et Biophys. Acta (BBA)-Mol. Basis Dis. 2018, 1864, 668–675. [Google Scholar] [CrossRef]
- Huang, L.; Aghajan, M.; Quesenberry, T.; Low, A.; Murray, S.F.; Monia, B.P.; Guo, S. Targeting Translation Termination Machinery with Antisense Oligonucleotides for Diseases Caused by Nonsense Mutations. Nucleic Acid Ther. 2019, 29, 175–186. [Google Scholar] [CrossRef]
- Lee, R.E.; Lewis, C.A.; He, L.; Bulik-Sullivan, E.C.; Gallant, S.C.; Mascenik, T.M.; Dang, H.; Cholon, D.M.; Gentzsch, M.; Morton, L.C.; et al. Small-Molecule ERF3a Degraders Rescue CFTR Nonsense Mutations by Promoting Premature Termination Codon Readthrough. J. Clin. Investig. 2022, 132, e154571. [Google Scholar] [CrossRef] [PubMed]
- Baradaran-Heravi, A.; Balgi, A.D.; Hosseini-Farahabadi, S.; Choi, K.; Has, C.; Roberge, M. Effect of Small Molecule ERF3 Degraders on Premature Termination Codon Readthrough. Nucleic Acids Res. 2021, 49, 3692–3708. [Google Scholar] [CrossRef]
- Fuchs, O. Targeting Cereblon in Hematologic Malignancies. Blood Rev. 2022, 57, 100994. [Google Scholar] [CrossRef]
- Hansen, J.D.; Correa, M.; Alexander, M.; Nagy, M.; Huang, D.; Sapienza, J.; Lu, G.; LeBrun, L.A.; Cathers, B.E.; Zhang, W.; et al. CC-90009: A Cereblon E3 Ligase Modulating Drug That Promotes Selective Degradation of GSPT1 for the Treatment of Acute Myeloid Leukemia. J. Med. Chem. 2021, 64, 1835–1843. [Google Scholar] [CrossRef]
- Surka, C.; Jin, L.; Mbong, N.; Lu, C.-C.; Jang, I.S.; Rychak, E.; Mendy, D.; Clayton, T.; Tindall, E.; Hsu, C.; et al. CC-90009, a Novel Cereblon E3 Ligase Modulator, Targets Acute Myeloid Leukemia Blasts and Leukemia Stem Cells. Blood 2021, 137, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Friesen, W.J.; Trotta, C.R.; Tomizawa, Y.; Zhuo, J.; Johnson, B.; Sierra, J.; Roy, B.; Weetall, M.; Hedrick, J.; Sheedy, J.; et al. The Nucleoside Analog Clitocine Is a Potent and Efficacious Readthrough Agent. RNA 2017, 23, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-G.; Xiang, J.; Zeng, X.-L.; Li, X.; Wu, P.; Fung, K.P.; Liu, F.-Y. Clitocine Induces Apoptosis and Enhances the Lethality of ABT-737 in Human Colon Cancer Cells by Disrupting the Interaction of Mcl-1 and Bak. Cancer Lett. 2014, 355, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-G.; Ruan, F.; Zeng, X.-L.; Xiang, J.; Li, X.; Wu, P.; Fung, K.P.; Liu, F.-Y. Clitocine Potentiates TRAIL-Mediated Apoptosis in Human Colon Cancer Cells by Promoting Mcl-1 Degradation. Apoptosis 2016, 21, 1144–1157. [Google Scholar] [CrossRef]
- Busch, A.; Hertel, K.J. HEXEvent: A Database of Human EXon Splicing Events. Nucleic Acids Res. 2013, 41, D118–D124. [Google Scholar] [CrossRef]
- Trzaska, C.; Amand, S.; Bailly, C.; Leroy, C.; Marchand, V.; Duvernois-Berthet, E.; Saliou, J.-M.; Benhabiles, H.; Werkmeister, E.; Chassat, T.; et al. 2,6-Diaminopurine as a Highly Potent Corrector of UGA Nonsense Mutations. Nat. Commun. 2020, 11, 1509. [Google Scholar] [CrossRef]
- Guy, M.P.; Shaw, M.; Weiner, C.L.; Hobson, L.; Stark, Z.; Rose, K.; Kalscheuer, V.M.; Gecz, J.; Phizicky, E.M. Defects in TRNA Anticodon Loop 2′-O-Methylation Are Implicated in Nonsyndromic X-Linked Intellectual Disability Due to Mutations in FTSJ1. Hum. Mutat. 2015, 36, 1176–1187. [Google Scholar] [CrossRef]
- Leroy, C.; Spelier, S.; Essonghe, N.C.; Poix, V.; Kong, R.; Gizzi, P.; Bourban, C.; Amand, S.; Bailly, C.; Guilbert, R.; et al. Use of 2,6-Diaminopurine as a Potent Suppressor of UGA Premature Stop Codons in Cystic Fibrosis. Mol. Ther. 2023, 31, 970–985. [Google Scholar] [CrossRef]
- Komarova (Andreyanova), E.S.; Osterman, I.A.; Pletnev, P.I.; Ivanenkov, Y.A.; Majouga, A.G.; Bogdanov, A.A.; Sergiev, P.V. 2-Guanidino-Quinazolines as a Novel Class of Translation Inhibitors. Biochimie 2017, 133, 45–55. [Google Scholar] [CrossRef]
- Morrill, C.; Friesen, W.J.; Babu, S.; Baiazitov, R.Y.; Du, W.; Karloff, D.B.; Lee, C.-S.; Moon, Y.-C.; Ren, H.; Sierra, J.; et al. Guanidino Quinazolines and Pyrimidines Promote Readthrough of Premature Termination Codons in Cells with Native Nonsense Mutations. Bioorganic Med. Chem. Lett. 2022, 76, 128989. [Google Scholar] [CrossRef] [PubMed]
- Palomar-Siles, M.; Heldin, A.; Zhang, M.; Strandgren, C.; Yurevych, V.; van Dinter, J.T.; Engels, S.A.G.; Hofman, D.A.; Öhlin, S.; Meineke, B.; et al. Translational Readthrough of Nonsense Mutant TP53 by MRNA Incorporation of 5-Fluorouridine. Cell Death Dis. 2022, 13, 997. [Google Scholar] [CrossRef] [PubMed]
- Grem, J.L. 5-Fluorouracil: Forty-plus and Still Ticking. A Review of Its Preclinical and Clinical Development. Investig. New Drugs 2000, 18, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Ju, J.; Schmitz, J.C.; Song, B.; Kudo, K.; Chu, E. Regulation of P53 Expression in Response to 5-Fluorouracil in Human Cancer RKO Cells. Clin. Cancer Res. 2007, 13, 4245–4251. [Google Scholar] [CrossRef]
- Kayali, R.; Ku, J.-M.; Khitrov, G.; Jung, M.E.; Prikhodko, O.; Bertoni, C. Read-through Compound 13 Restores Dystrophin Expression and Improves Muscle Function in the Mdx Mouse Model for Duchenne Muscular Dystrophy. Hum. Mol. Genet. 2012, 21, 4007–4020. [Google Scholar] [CrossRef]
- Gatti, R.A. SMRT Compounds Correct Nonsense Mutations in Primary Immunodeficiency and Other Genetic Models. Ann. N. Y. Acad. Sci. 2012, 1250, 33–40. [Google Scholar] [CrossRef]
- Gómez-Grau, M.; Garrido, E.; Cozar, M.; Rodriguez-Sureda, V.; Domínguez, C.; Arenas, C.; Gatti, R.A.; Cormand, B.; Grinberg, D.; Vilageliu, L. Evaluation of Aminoglycoside and Non-Aminoglycoside Compounds for Stop-Codon Readthrough Therapy in Four Lysosomal Storage Diseases. PLoS ONE 2015, 10, e0135873. [Google Scholar] [CrossRef]
- Martorell, L.; Cortina, V.; Parra, R.; Barquinero, J.; Vidal, F. Variable Readthrough Responsiveness of Nonsense Mutations in Hemophilia A. Haematologica 2020, 105, 508–518. [Google Scholar] [CrossRef]
- Tarrasó, G.; Real-Martinez, A.; Parés, M.; Romero-Cortadellas, L.; Puigros, L.; Moya, L.; de Luna, N.; Brull, A.; Martín, M.A.; Arenas, J.; et al. Absence of p.R50X Pygm Read-through in McArdle Disease Cellular Models. Dis. Model. Mech. 2020, 13, dmm043281. [Google Scholar] [CrossRef]
- Ortuño-Costela, M.d.C.; Cerrada, V.; Moreno-Izquierdo, A.; García-Consuegra, I.; Laberthonnière, C.; Delourme, M.; Garesse, R.; Arenas, J.; Fuster García, C.; García García, G.; et al. Generation of the First Human In Vitro Model for McArdle Disease Based on IPSC Technology. Int. J. Mol. Sci. 2022, 23, 13964. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.E.; Ku, J.-M.; Du, L.; Hu, H.; Gatti, R.A. Synthesis and Evaluation of Compounds That Induce Readthrough of Premature Termination Codons. Bioorganic Med. Chem. Lett. 2011, 21, 5842–5848. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Jung, M.E.; Damoiseaux, R.; Completo, G.; Fike, F.; Ku, J.-M.; Nahas, S.; Piao, C.; Hu, H.; Gatti, R.A. A New Series of Small Molecular Weight Compounds Induce Read through of All Three Types of Nonsense Mutations in the ATM Gene. Mol. Ther. 2013, 21, 1653–1660. [Google Scholar] [CrossRef]
- Lavin, M.F. Generating SM(a)RTer Compounds for Translation Termination Suppression in A-T and Other Genetic Disorders. Mol. Ther. 2013, 21, 1650–1652. [Google Scholar] [CrossRef] [PubMed]
- Tutone, M.; Pibiri, I.; Perriera, R.; Campofelice, A.; Culletta, G.; Melfi, R.; Pace, A.; Almerico, A.M.; Lentini, L. Pharmacophore-Based Design of New Chemical Scaffolds as Translational Readthrough-Inducing Drugs (TRIDs). ACS Med. Chem. Lett. 2020, 11, 747–753. [Google Scholar] [CrossRef]
- Roy, B.; Leszyk, J.D.; Mangus, D.A.; Jacobson, A. Nonsense Suppression by Near-Cognate TRNAs Employs Alternative Base Pairing at Codon Positions 1 and 3. Proc. Natl. Acad. Sci. USA 2015, 112, 3038–3043. [Google Scholar] [CrossRef] [PubMed]
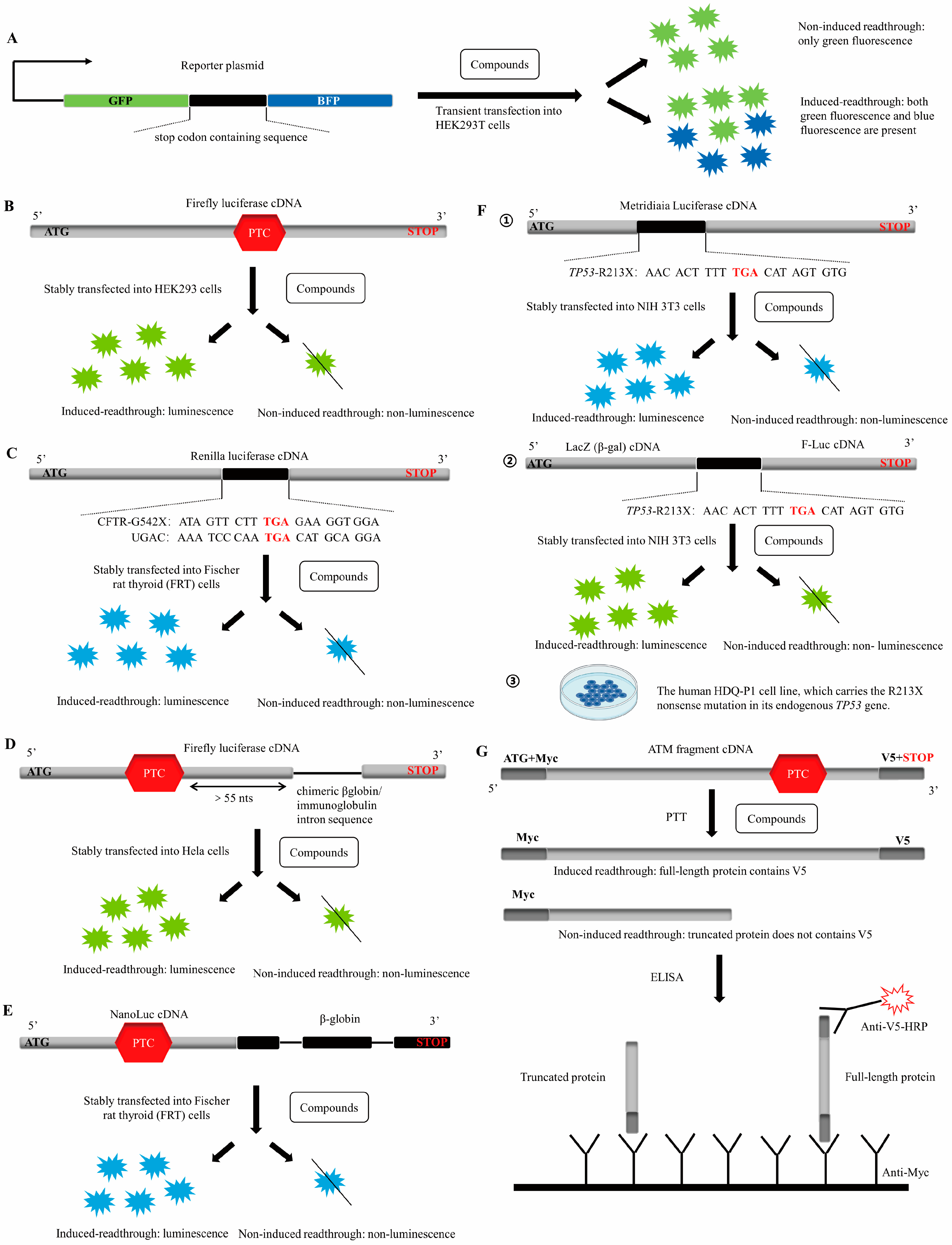
| Disease | Gene | Administration | Dose and Duration | Results |
|---|---|---|---|---|
| EBS-MD [26] | PLEC1 | Intravenous | n = 1; 7.5 mg/kg/d for 14 consecutive days; 2 treatment courses | Increased expression of plectin in the skin for at least 5 months, myalgia disappeared, and quality of life improved |
| JEB [25] | LAMA3/ LAMB3 | Intravenous | 1. n = 3 (7.5 mg/kg daily for 14 days); 2. n = 2 (10 mg/kg daily for 24 days) | All 5 patients exhibited increased laminin 332 in the dermal-epidermal junction, and improved wound closure |
| JEB [45] | LAMB3 | Intravenous | n = 5; 7.5 mg/kg/d for three weeks | a positive impact on skin fragility and daily life in four patients |
| JEB [42] | LAMB3 | Topical | n = 1; 0.3% gentamicin ointment once at bedtime | Conjunctival cells showed positive staining for laminin-332, and the amelioration of the corneal erosions, |
| NPPK [38] | SERPINB7 | Topical | n = 20; 0.1% or 0.3% gentamicin ointment once daily for 30 days | Significantly improve hyperkeratosis and foul smell |
| GS-JEB [40] | LAMA3/ LAMB3 | Topical | n = 3; 0.5% gentamicin ointment twice a day for 2 weeks | Increased expression of laminin 332 at the dermal-epidermal junction for at least 3 months, and improved wound closure. |
| JEB-gen intermed [41] | COL17A1 | Topical | n = 1; 0.3% gentamicin ointment once daily for 90 days | Improved wound closure and reduced blister formation. After drug withdrawal, there was no relapse for at least 2 months. |
| HSS [44] | CDSN | Topical | n = 4; 0.1% gentamicin ointment twice a day for 6 months | Significant hair growth and SALT score reduction |
| NPPK [37] | SERPINB7 | Topical | n = 5; 0.1% gentamicin ointment twice a day for 4 weeks | Suppressed hyperkeratosis did not improve the degree of erythema |
| RDEB [39] | COL17A1 | Topical; intradermal injection | n = 5; 0.1% gentamicin ointment 3 times daily for 2 weeks; 8 mg for 2 days | Both induce type VII collagen and anchoring fibrils; Topical: corrected dermal-epidermal separation, improved wound closure, and reduced blister formation. |
| HHD [43] | ATP2C1 | Topical | n = 1; 0.1% gentamicin ointment twice a day for 18 days | Far more effective in inducing remission in an HHD patient than an accepted topical disinfectant |
| Disease | Gene | Model | Conclusion |
|---|---|---|---|
| Leber congenital amaurosis type 4 [103] | AIPL1 | Retinal organoids | The increased level of full-length AIPL1 protein mediated by ataluren readthrough was not sufficient to restore rod PDE6 to the levels required to reduce cGMP |
| Retinitis pigmentosa [104] | FAM161A | Fibroblasts from six patients | Ataluren was able to restore FAM161A expression in FAM161A-mutated cells as well as its co-localization with α-tubulin along the microtubules. |
| Bardet-Biedl syndrome; Alström syndrome [87] | BBS2; ALMS1 | Patient-derived fibroblasts | Ataluren treatment recovered full-length BBS2 or ALMS1 protein expression and ciliary function. |
| Retinitis pigmentosa [105] | RPGR | Patient-derived fibroblasts | Applying ataluren restored RPGR at the cilium in approximately 8% of patient-derived cells |
| Usher syndrome [106] | USH2A | Patient-derived fibroblasts | Ataluren increased USH2A protein expression and the number of ciliated cells |
| Inherited retinal dystrophies [107] | CHM | Patient-derived fibroblasts; Patient iPSCs -derived RPE | Ataluren treatment induced a non-significant trend for functional rescue, which could not be improved by nonsense-mediated decay inhibition. |
| Lamin A/C (LMNA)-related cardiomyopathy [108] | LMNA | Patient iPSCs-derived cardiomyocytes | Ataluren treatment increased the production of full-length LMNA proteins in only the R225X mutant, not in other mutations. |
| Retinitis pigmentosa [109] | MERTK | Patient iPSCs -derived RPE | Following treatment with ataluren was able to restore the expression of MERTK. Furthermore, the ataluren treatment restored 12% of the phagocytic function. |
| Cystic fibrosis [110] | CFTR | Intestinal organoids | Functional restoration of CFTR by PTC124 could not be confirmed |
| Heart disease [111] | SCN5A | Patient iPSCs-derived cardiomyocytes | The authors did not observe the rescue of the electrophysiological phenotype in hiPSC-derived cardiomyocytes from the patients |
| Choroideremia [91] | CHM | Patient-derived fibroblasts | After treatment with ataluren, prenylation activity recovered, but no increase of REP1 protein was detected. |
| X-linked retinitis Pigmentosa [112] | RP2 | Patient iPSCs -derived RPE | After treatment with ataluren, up to 20% of endogenous, full-length RP2 protein can be restored. |
| Neuronal ceroid lipofuscinosis [113] | TPP1 | Patient iPSCs-derived neural progenitor cells | Nonsense suppression by PTC124 resulted in both an increase in TPP1 activity and an attenuation of neuropathology |
| Phase and Status | Duration | Age | Group and Dose | Results |
|---|---|---|---|---|
| IIa (NCT00264888) Completed | 2005.12–2007.05 | 5 years and older | 1. n = 6 (4, 4, 8 mg/kg/day) 2. n = 20 (10, 10, 20 mg/kg/day) 3. n = 12 (20, 20, 40 mg/kg/day) | 61% of subjects demonstrated positive changes in dystrophin expression after 28 days of treatment; Changes in Clinical Measures were not statistically significant; Ataluren was generally well tolerated. |
| IIb (NCT00592553) Completed | 2008.02–2009.12 | 5 years and older | 1. n = 57 (10, 10, 20 mg/kg/day) 2. n = 60 (20, 20, 40 mg/kg/day) 3. n = 57 (placebo) | 40 mg/kg/day slowed the rate of decline of walking ability (6MWD) after 48 weeks of treatment (ataluren performed better in a post-hoc analysis); 80 mg/kg/day had no activity; well tolerated. |
| III (NCT01826487) Completed | 2013.03–2014.08 | 7–16 years | 1. n = 115 (10, 10, 20 mg/kg/day) 2. n = 115 (placebo) | After 48 weeks of treatment, the change in 6MWD between ataluren-treated and placebo-treated patients in the intention-to-treat population was insignificant; but the benefit observed in patients with a baseline 6MWD of 300 m or more to less than 400 m supports the clinical benefit of ataluren. |
| III (NCT01557400) Completed | 2012.05–2018.01 | 12.8 ± 2.4 years | Single Group Assignment n = 94 (10, 10, 20 mg/kg/day) | After 240 weeks of treatment, ataluren plus standard of care delays disease progression and benefits ambulatory and non-ambulatory patients with nmDMD |
| II (NCT02819557) Completed | 2016.09–2018.09 | 2–5 years | Single Group Assignment n = 14 (10, 10, 20 mg/kg/day) | The safety and pharmacokinetic profile of ataluren in children from 2–5 years with nmDMD was consistent with that for older children; Clinical benefits were also observed at 52 weeks with ataluren. |
| III (NCT03179631) Ongoing | 2017.07–2023.07 | 5 years and older | Randomized, double-blind, placebo-controlled | The primary outcome to be assessed is the change slope in 6MWD over 72 weeks. Its estimated completion date is July 2023. |
| NCT02369731 Ongoing | 2015.04–2025.05 | 2 years and older | A long-term, multicenter, observational study | It evaluates the safety and effectiveness of ataluren in usual care. Its estimated completion date is May 2025. |
| II (NCT04336826) Ongoing | 2021.12–2022.12 | 6 months to 2 years | An open-label study | It evaluates the safety and pharmacokinetics of ataluren over 24 weeks. Its estimated completion date is December 2022. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Li, J.; Shi, W.; Nie, Z.; Zhang, S.; Ma, F.; Hu, J.; Chen, J.; Li, P.; Xie, X. Pharmaceuticals Promoting Premature Termination Codon Readthrough: Progress in Development. Biomolecules 2023, 13, 988. https://doi.org/10.3390/biom13060988
Li S, Li J, Shi W, Nie Z, Zhang S, Ma F, Hu J, Chen J, Li P, Xie X. Pharmaceuticals Promoting Premature Termination Codon Readthrough: Progress in Development. Biomolecules. 2023; 13(6):988. https://doi.org/10.3390/biom13060988
Chicago/Turabian StyleLi, Shan, Juan Li, Wenjing Shi, Ziyan Nie, Shasha Zhang, Fengdie Ma, Jun Hu, Jianjun Chen, Peiqiang Li, and Xiaodong Xie. 2023. "Pharmaceuticals Promoting Premature Termination Codon Readthrough: Progress in Development" Biomolecules 13, no. 6: 988. https://doi.org/10.3390/biom13060988
APA StyleLi, S., Li, J., Shi, W., Nie, Z., Zhang, S., Ma, F., Hu, J., Chen, J., Li, P., & Xie, X. (2023). Pharmaceuticals Promoting Premature Termination Codon Readthrough: Progress in Development. Biomolecules, 13(6), 988. https://doi.org/10.3390/biom13060988





