Evaluating Current Molecular Techniques and Evidence in Assessing Microbiome in Placenta-Related Health and Disorders in Pregnancy
Abstract
1. Introduction
2. Pregnancy
2.1. Physiology of Pregnancy and Microbiome
2.2. Microbiome and Maternal Immune System
2.3. Human Placenta
2.4. Healthy Placental Microbiome
3. Placenta-Related Syndromes
4. Methodology of Placental Microbiome Assessment
5. The Influence of the Method of Delivery on the Results
6. Placenta-Related Diseases and Microbiome
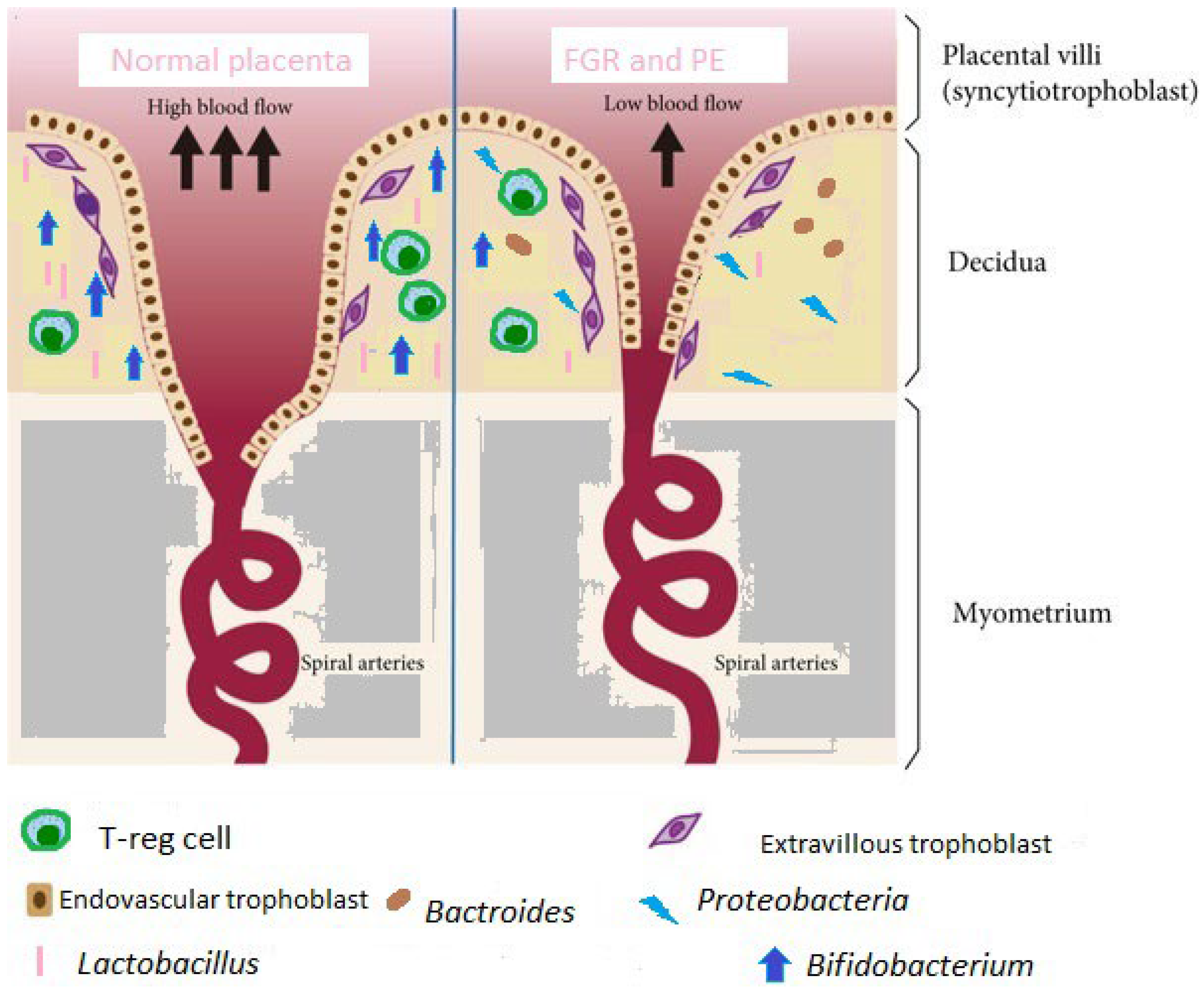
6.1. FGR
6.2. Preeclampsia
7. Potential Modifying Factors of Gene Transcription
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| BW | birth weight |
| CSTs | community state types |
| CS el | elective cesarean section |
| CS sp | spontaneous cesarean section |
| FGR | fetal growth restriction |
| HMP | Human Microbiome Project |
| IL | interleukin |
| LPS | endotoxin |
| OTUs | operational taxonomic units |
| PlGF | placental growth factor |
| PRRs | pathogen recognition receptors |
| sFLT | soluble fms-like tyrosine kinase-1 |
| SMURF | Short Multiple Regions Framework |
| TLRs | Toll-like receptors |
| VD | vaginal delivery |
| WGS | whole genome sequencing |
References
- Fleischmann, R.D.; Adams, M.D.; White, O.; Clayton, R.A.; Kirkness, E.F.; Kerlavage, A.R.; Bult, C.J.; Tomb, J.F.; Dougherty, B.A.; Merrick, J.M.; et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 1995, 269, 496–512. [Google Scholar] [CrossRef]
- Fraser, C.M.; Gocayne, J.D.; White, O.; Adams, M.D.; Clayton, R.A.; Fleischmann, R.D.; Bult, C.J.; Kerlavage, A.R.; Sutton, G.; Kelley, J.M.; et al. The Minimal Gene Complement of Mycoplasma genitalium. Science 1995, 270, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Sun, C. A marine bacterial community capable of degrading poly(ethylene terephthalate) and polyethylene. J. Hazard. Mater. 2021, 416, 125928. [Google Scholar] [CrossRef] [PubMed]
- McGuire, A.L.; Gabriel, S.; Tishkoff, S.A.; Wonkam, A.; Chakravarti, A.; Furlong, E.E.M.; Treutlein, B.; Meissner, A.; Chang, H.Y.; López-Bigas, N.; et al. The road ahead in genetics and genomics. Nat. Rev. Genet. 2020, 21, 581–596. [Google Scholar] [CrossRef] [PubMed]
- Amon, P.; Sanderson, I. What is the microbiome? Arch. Dis. Child. -Educ. Pr. Ed. 2017, 102, 257–260. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Macpherson, A.J.; Harris, N.L. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004, 4, 478–485. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 626. [Google Scholar] [CrossRef]
- Jost, M.; Wehkamp, U. The Skin Microbiome and Influencing Elements in Cutaneous T-Cell Lymphomas. Cancers 2022, 14, 1324. [Google Scholar] [CrossRef]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef] [PubMed]
- The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology: This report is based on a colloquium, “The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology,” sponsored by the American Academy of Microbiology and held 11–13 October 2002, in Longboat Key, Florida. Washington (DC): American Society for Microbiology. 2004. Available online: https://www.ncbi.nlm.nih.gov/books/NBK563534/ (accessed on 20 March 2023).
- Gilbert, S.F. A holobiont birth narrative: The epigenetic transmission of the human microbiome. Front. Genet. 2014, 5, 282. [Google Scholar] [CrossRef] [PubMed]
- Koren, O.; Goodrich, J.K.; Cullender, T.C.; Spor, A.; Laitinen, K.; Bäckhed, H.K.; Gonzalez, A.; Werner, J.J.; Angenent, L.T.; Knight, R.; et al. Host Remodeling of the Gut Microbiome and Metabolic Changes during Pregnancy. Cell 2012, 150, 470–480. [Google Scholar] [CrossRef]
- Prince, A.L.; Chu, D.M.; Seferovic, M.D.; Antony, K.M.; Ma, J.; Aagaard, K.M. The Perinatal Microbiome and Pregnancy: Moving Beyond the Vaginal Microbiome. Cold Spring Harb. Perspect. Med. 2015, 5, a023051. [Google Scholar] [CrossRef]
- Nuriel-Ohayon, M.; Neuman, H.; Ziv, O.; Belogolovski, A.; Barsheshet, Y.; Bloch, N.; Uzan, A.; Lahav, R.; Peretz, A.; Frishman, S.; et al. Progesterone Increases Bifidobacterium Relative Abundance during Late Pregnancy. Cell Rep. 2019, 27, 730–736.e3. [Google Scholar] [CrossRef] [PubMed]
- Fuhler, G. The immune system and microbiome in pregnancy. Best Pr. Res. Clin. Gastroenterol. 2020, 44–45, 101671. [Google Scholar] [CrossRef]
- Zhou, Y.; Qi, H.; Yin, N. Adaptations and alterations of maternal microbiota: From physiology to pathology. Med. Microecol. 2021, 9, 100045. [Google Scholar] [CrossRef]
- Koren, O.; Knights, D.; Gonzalez, A.; Waldron, L.; Segata, N.; Knight, R.; Huttenhower, C.; Ley, R.E. A Guide to Enterotypes across the Human Body: Meta-Analysis of Microbial Community Structures in Human Microbiome Datasets. PLoS Comput. Biol. 2013, 9, e1002863. [Google Scholar] [CrossRef]
- Wang, J.; Shi, Z.-H.; Yang, J.; Wei, Y.; Wang, X.-Y.; Zhao, Y.-Y. Gut microbiota dysbiosis in preeclampsia patients in the second and third trimesters. Chin. Med. J. 2020, 133, 1057–1065. [Google Scholar] [CrossRef]
- DiGiulio, D.B.; Callahan, B.J.; McMurdie, P.J.; Costello, E.K.; Lyell, D.J.; Robaczewska, A.; Sun, C.L.; Goltsman, D.S.A.; Wong, R.J.; Shaw, G.; et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. USA 2015, 112, 11060–11065. [Google Scholar] [CrossRef]
- MacIntyre, D.A.; Chandiramani, M.; Lee, Y.S.; Kindinger, L.; Smith, A.; Angelopoulos, N.; Lehne, B.; Arulkumaran, S.; Brown, R.; Teoh, T.G.; et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zakis, D.R.; Paulissen, E.; Kornete, L.; Kaan, A.M.; Nicu, E.A.; Zaura, E. The evidence for placental microbiome and its composition in healthy pregnancies: A systematic review. J. Reprod. Immunol. 2021, 149, 103455. [Google Scholar] [CrossRef] [PubMed]
- Gomez de Agüero, M.; Ganal-Vonarburg, S.C.; Fuhrer, T.; Rupp, S.; Uchimura, Y.; Li, H.; Steinert, A.; Heikenwalder, M.; Hapfelmeier, S.; Sauer, U.; et al. The maternal microbiota drives early postnatal innate immune development. Science 2016, 351, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Toldi, G.; Treszl, A.; Vasarhelyi, B. T Lymphocyte Characteristics and Immune Tolerance During Human Pregnancy. In Autoimmune Disorders—Pathogenetic Aspects; InTechEBOOK; 2011; ISBN 978-953-51-6537-8. Available online: https://doi.org/10.5772/20087 (accessed on 1 May 2023).
- Nunez, N.; Réot, L.; Menu, E. Neonatal Immune System Ontogeny: The Role of Maternal Microbiota and Associated Factors. How Might the Non-Human Primate Model Enlighten the Path? Vaccines 2021, 9, 584. [Google Scholar] [CrossRef]
- Gonzalez-Perez, G.; Hicks, A.L.; Tekieli, T.M.; Radens, C.M.; Williams, B.L.; Lamousé-Smith, E.S.N. Maternal Antibiotic Treatment Impacts Development of the Neonatal Intestinal Microbiome and Antiviral Immunity. J. Immunol. 2016, 196, 3768–3779. [Google Scholar] [CrossRef] [PubMed]
- Pichlmair, A. RIG-I-Mediated antiviral responses to single-stranded RNA searing 5′-phosphates. Science 2006, 314, 997–1001. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009, 21, 317–337. [Google Scholar] [CrossRef]
- Cinel, I.; Opal, S.M. Molecular biology of inflammation and sepsis: A primer. Crit. Care Med. 2009, 37, 291–304. [Google Scholar] [CrossRef]
- Wanke, I.; Steffen, H.; Christ, C.; Krismer, B.; Götz, F.; Peschel, A.; Schaller, M.; Schittek, B. Skin Commensals Amplify the Innate Immune Response to Pathogens by Activation of Distinct Signaling Pathways. J. Investig. Dermatol. 2011, 131, 382–390. [Google Scholar] [CrossRef]
- Lathrop, S.K.; Bloom, S.M.; Rao, S.M.; Nutsch, K.; Lio, C.-W.; Santacruz, N.; Peterson, D.A.; Stappenbeck, T.S.; Hsieh, C.-S. Peripheral education of the immune system by colonic commensal microbiota. Nature 2011, 478, 250–254. [Google Scholar] [CrossRef]
- Bilbo, S.D.; Levkoff, L.H.; Mahoney, J.H.; Watkins, L.R.; Rudy, J.W.; Maier, S.F. Neonatal Infection Induces Memory Impairments Following an Immune Challenge in Adulthood. Behav. Neurosci. 2005, 119, 293–301. [Google Scholar] [CrossRef]
- Perez-Muñoz, M.E.; Arrieta, M.-C.; Ramer-Tait, A.E.; Walter, J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome 2017, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Parnell, L.A.; Willsey, G.G.; Joshi, C.S.; Yin, Y.; Wargo, M.J.; Mysorekar, I.U. Functional characterization of Ralstonia insidiosa, a bona fide resident at the maternal-fetal interface. bioRxiv 2019, 721977. [Google Scholar] [CrossRef]
- Lauder, A.P.; Roche, A.M.; Sherrill-Mix, S.; Bailey, A.; Laughlin, A.L.; Bittinger, K.; Leite, R.; Elovitz, M.A.; Parry, S.; Bushman, F.D. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 2016, 4, 29. [Google Scholar] [CrossRef]
- Sterpu, I.; Fransson, E.; Hugerth, L.W.; Du, J.; Pereira, M.; Cheng, L.; Radu, S.A.; Calderón-Pérez, L.; Zha, Y.; Angelidou, P.; et al. No evidence for a placental microbiome in human pregnancies at term. Am. J. Obstet. Gynecol. 2021, 224, 296.e1–296.e23. [Google Scholar] [CrossRef] [PubMed]
- Mendz, G.L.; Kaakoush, N.O.; Quinlivan, J.A. Bacterial aetiological agents of intra-amniotic infections and preterm birth in pregnant women. Front. Cell. Infect. Microbiol. 2013, 3, 58. [Google Scholar] [CrossRef] [PubMed]
- Miko, E.; Csaszar, A.; Bodis, J.; Kovacs, K. The Maternal–Fetal Gut Microbiota Axis: Physiological Changes, Dietary Influence, and Modulation Possibilities. Life 2022, 12, 424. [Google Scholar] [CrossRef]
- Pessa-Morikawa, T.; Husso, A.; Kärkkäinen, O.; Koistinen, V.; Hanhineva, K.; Iivanainen, A.; Niku, M. Maternal microbiota-derived metabolic profile in fetal murine intestine, brain and placenta. BMC Microbiol. 2022, 22, 46. [Google Scholar] [CrossRef]
- Ziętek, M.; Celewicz, Z.; Szczuko, M. Short-Chain Fatty Acids, Maternal Microbiota and Metabolism in Pregnancy. Nutrients 2021, 13, 1244. [Google Scholar] [CrossRef]
- Chénard, T.; Prévost, K.; Dubé, J.; Massé, E. Immune System Modulations by Products of the Gut Microbiota. Vaccines 2020, 8, 461. [Google Scholar] [CrossRef]
- Amarasekara, R.; Jayasekara, R.W.; Senanayake, H.; Dissanayake, V.H.W. Microbiome of the placenta in pre-eclampsia supports the role of bacteria in the multifactorial cause of pre-eclampsia. J. Obstet. Gynaecol. Res. 2015, 41, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Lager, S.; de Goffau, M.; Sovio, U.; Peacock, S.J.; Parkhill, J.; Charnock-Jones, D.S.; Smith, G.C.S. Detecting eukaryotic microbiota with single-cell sensitivity in human tissue. Microbiome 2018, 6, 151. [Google Scholar] [CrossRef] [PubMed]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The Placenta Harbors a Unique Microbiome. Sci. Transl. Med. 2014, 6, 237ra65. [Google Scholar] [CrossRef] [PubMed]
- Younes, J.A.; Lievens, E.; Hummelen, R.; van der Westen, R.; Reid, G.; Petrova, M.I. Women and Their Microbes: The Unexpected Friendship. Trends Microbiol. 2018, 26, 16–32. [Google Scholar] [CrossRef]
- Gomez-Arango, L.F.; Barrett, H.L.; McIntyre, H.D.; Callaway, L.K.; Morrison, M.; Nitert, M.D. Contributions of the maternal oral and gut microbiome to placental microbial colonization in overweight and obese pregnant women. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Montenegro, D.A.; Borda, L.F.; Neuta, Y.; Gómez, L.A.; Castillo, D.M.; Loyo, D.; Lafaurie, G.I. Oral and uro-vaginal intra-amniotic infection in women with preterm delivery: A case-control study. J. Investig. Clin. Dent. 2019, 10, e12396. [Google Scholar] [CrossRef]
- Parnell, L.A.; Briggs, C.M.; Cao, B.; Delannoy-Bruno, O.; Schrieffer, A.E.; Mysorekar, I.U. Microbial communities in placentas from term normal pregnancy exhibit spatially variable profiles. Sci. Rep. 2017, 7, 11200. [Google Scholar] [CrossRef]
- de Goffau, M.C.; Lager, S.; Sovio, U.; Gaccioli, F.; Cook, E.; Peacock, S.J.; Parkhill, J.; Charnock-Jones, D.S.; Smith, G.C.S. Human placenta has no microbiome but can contain potential pathogens. Nature 2019, 572, 329–334. [Google Scholar] [CrossRef]
- Ríos-Covian, D.; Langella, P.; Martín, R. From Short- to Long-Term Effects of C-Section Delivery on Microbiome Establishment and Host Health. Microorganisms 2021, 9, 2122. [Google Scholar] [CrossRef]
- Kosińska-Kaczyńska, K. Placental Syndromes—A New Paradigm in Perinatology. Int. J. Environ. Res. Public Health 2022, 19, 7392. [Google Scholar] [CrossRef]
- Redman, C.W.G. Immunological aspects of pre-eclampsia. Bailliere’s Clin. Obstet. Gynaecol. 1992, 6, 601–615. [Google Scholar] [CrossRef]
- Roberts, J.; Redman, C. Pre-eclampsia: More than pregnancy-induced hypertension. Lancet 1993, 341, 1447–1451. [Google Scholar] [CrossRef]
- Wei, X.; Zhang, Y.; Wu, F.; Tian, F.; Lin, Y. The role of extravillous trophoblasts and uterine NK cells in vascular remodeling during pregnancy. Front. Immunol. 2022, 13, 3984. [Google Scholar] [CrossRef]
- Rao, V.A.; Kurian, N.K.; Rao, K.A. Cytokines, NK cells and regulatory T cell functions in normal pregnancy and reproductive failures. Am. J. Reprod. Immunol. 2023, 89, e13667. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowski, S.; Bednarek-Jędrzejek, M.; Kwiatkowska, E.; Cymbaluk-Płoska, A.; Torbè, A. Diagnosis of placental insufficiency independently of clinical presentations using sFlt-1/PLGF ratio, including SGA patients. Pregnancy Hypertens. 2021, 25, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Maric-Bilkan, C.; Abrahams, V.M.; Arteaga, S.S.; Bourjeily, G.; Conrad, K.P.; Catov, J.M.; Costantine, M.M.; Cox, B.; Garovic, V.; George, E.; et al. Research Recommendations from the National Institutes of Health Workshop on Predicting, Preventing, and Treating Preeclampsia. Hypertension 2019, 73, 757–766. [Google Scholar] [CrossRef]
- Kamphof, H.D.; Posthuma, S.; Gordijn, S.J.; Ganzevoort, W. Fetal Growth Restriction: Mechanisms, Epidemiology, and Management. Matern. Med. 2022, 4, 186–196. [Google Scholar] [CrossRef]
- Zhu, Y.-C.; Lin, L.; Li, B.-Y.; Li, X.-T.; Chen, D.-J.; Zhao, X.-L.; Cui, S.-H.; Ding, H.-J.; Ding, G.-F.; Meng, H.-X.; et al. Incidence and Clinical Features of Fetal Growth Restriction in 4 451 Women with Hypertensive Disorders of Pregnancy. Matern. Med. 2020, 2, 207–210. [Google Scholar] [CrossRef]
- Isaac, N.I.; Philippe, D.; Nicholas, A.; Raoult, D.; Eric, C. Metaproteomics of the human gut microbiota: Challenges and contributions to other OMICS. Clin. Mass Spectrom. 2019, 14, 18–30. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Khelaifia, S.; Alou, M.T.; Ndongo, S.; Dione, N.; Hugon, P.; Caputo, A.; Cadoret, F.; Traore, S.I.; Seck, E.H.; et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016, 7, 16203. [Google Scholar] [CrossRef]
- Schneider, T.; Riedel, K. Environmental proteomics: Analysis of structure and function of microbial communities. Proteomics 2010, 10, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J.; Devkota, S.; McCoy, K.D.; Relman, D.A.; Yassour, M.; Young, V.B. Lessons learned from the prenatal microbiome controversy. Microbiome 2021, 9, 8. [Google Scholar] [CrossRef]
- Seferovic, M.D.; Pace, R.M.; Carroll, M.; Belfort, B.; Major, A.M.; Chu, D.M.; Racusin, D.A.; Castro, E.C.; Muldrew, K.L.; Versalovic, J.; et al. Visualization of microbes by 16S in situ hybridization in term and preterm placentas without intraamniotic infection. Am. J. Obstet. Gynecol. 2019, 221, 146.e1–146.e23. [Google Scholar] [CrossRef]
- Theis, K.R.; Romero, R.; Winters, A.D.; Greenberg, J.M.; Gomez-Lopez, N.; Alhousseini, A.; Bieda, J.; Maymon, E.; Pacora, P.; Fettweis, J.M.; et al. Does the human placenta delivered at term have a microbiota? Results of cultivation, quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomics. Am. J. Obstet. Gynecol. 2019, 220, 267.e1–267.e39. [Google Scholar] [CrossRef] [PubMed]
- Prince, A.L.; Ma, J.; Kannan, P.S.; Alvarez, M.; Gisslen, T.; Harris, R.A.; Sweeney, E.L.; Knox, C.L.; Lambers, D.S.; Jobe, A.H.; et al. The placental membrane microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. Am. J. Obstet. Gynecol. 2016, 214, 627.e1–627.e16. [Google Scholar] [CrossRef] [PubMed]
- Lannon, S.M.R.; Adams Waldorf, K.M.; Fiedler, T.; Kapur, R.P.; Agnew, K.; Rajagopal, L.; Gravett, M.G.; Fredricks, D.N. Parallel detection of lactobacillus and bacterial vaginosis-associated bacterial DNA in the chorioamnion and vagina of pregnant women at term. J. Matern. Fetal Neonatal. Med. 2019, 32, 2702–2710. [Google Scholar] [CrossRef] [PubMed]
- Barak, S.; Oettinger-Barak, O.; Machtei, E.E.; Sprecher, H.; Ohel, G. Evidence of Periopathogenic Microorganisms in Placentas of Women with Preeclampsia. J. Periodontol. 2007, 78, 670–676. [Google Scholar] [CrossRef]
- Bassols, J.; Serino, M.; Carreras-Badosa, G.; Burcelin, R.; Blasco-Baque, V.; Lopez-Bermejo, A.; Fernandez-Real, J.-M. Gestational diabetes is associated with changes in placental microbiota and microbiome. Pediatr. Res. 2016, 80, 777–784. [Google Scholar] [CrossRef]
- Cahill-Ocallaghan, R.; Tan, S.; Dougan, G.; Gaora, P.; Pickard, D.; Kennea, N.; Sullivan, M.; Feldman, R.; Edwards, A.D. Universal DNA primers amplify bacterial DNA from human fetal membranes and link Fusobacterium nucleatum with prolonged preterm membrane rupture. Mol. Hum. Reprod. 2005, 11, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef] [PubMed]
- Jones, H.E.; Harris, K.; Azizia, M.; Bank, L.; Carpenter, B.; Hartley, J.C.; Klein, N.; Peebles, D. Differing Prevalence and Diversity of Bacterial Species in Fetal Membranes from Very Preterm and Term Labor. PLoS ONE 2009, 4, e8205. [Google Scholar] [CrossRef]
- Leiby, J.S.; McCormick, K.; Sherrill-Mix, S.; Clarke, E.L.; Kessler, L.R.; Taylor, L.J.; Hofstaedter, C.E.; Roche, A.M.; Mattei, L.M.; Bittinger, K.; et al. Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome 2018, 6, 196. [Google Scholar] [CrossRef]
- Zheng, J.; Xiao, X.; Zhang, Q.; Mao, L.; Yu, M.; Xu, J. The Placental Microbiome Varies in Association with Low Birth Weight in Full-Term Neonates. Nutrients 2015, 7, 6924–6937. [Google Scholar] [CrossRef]
- Zheng, J.; Xiao, X.; Zhang, Q.; Mao, L.; Yu, M.; Xu, J.; Wang, T. The Placental Microbiota Is Altered among Subjects with Gestational Diabetes Mellitus: A Pilot Study. Front. Physiol. 2017, 8, 675. [Google Scholar] [CrossRef]
- Zheng, J.; Xiao, X.-H.; Zhang, Q.; Mao, L.-L.; Yu, M.; Xu, J.-P.; Wang, T. Correlation of placental microbiota with fetal macrosomia and clinical characteristics in mothers and newborns. Oncotarget 2017, 8, 82314–82325. [Google Scholar] [CrossRef] [PubMed]
- Steel, J.H.; Malatos, S.; Kennea, N.; Edwards, A.D.; Miles, L.; Duggan, P.; Reynolds, P.R.; Feldman, R.G.; Sullivan, M.H.F. Bacteria and Inflammatory Cells in Fetal Membranes Do Not Always Cause Preterm Labor. Pediatr. Res. 2005, 57, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Vanterpool, S.F.; Been, J.V.; Houben, M.L.; Nikkels, P.G.J.; De Krijger, R.R.; Zimmermann, L.J.I.; Kramer, B.W.; Progulske-Fox, A.; Reyes, L. Porphyromonas gingivalis within Placental Villous Mesenchyme and Umbilical Cord Stroma Is Associated with Adverse Pregnancy Outcome. PLoS ONE 2016, 11, e0146157. [Google Scholar] [CrossRef]
- Castelino, M.; Eyre, S.; Moat, J.; Fox, G.; Martin, P.; Ho, P.; Upton, M.; Barton, A. Optimisation of methods for bacterial skin microbiome investigation: Primer selection and comparison of the 454 versus MiSeq platform. BMC Microbiol. 2017, 17, 23. [Google Scholar] [CrossRef]
- Teng, F.; Nair, S.S.D.; Zhu, P.; Li, S.; Huang, S.; Li, X.; Xu, J.; Yang, F. Impact of DNA extraction method and targeted 16S-rRNA hypervariable region on oral microbiota profiling. Sci. Rep. 2018, 19, 16321. [Google Scholar] [CrossRef] [PubMed]
- Olomu, I.N.; Pena-Cortes, L.C.; Long, R.A.; Vyas, A.; Krichevskiy, O.; Luellwitz, R.; Singh, P.; Mulks, M.H. Elimination of “kitome” and “splashome” contamination results in lack of detection of a unique placental microbiome. BMC Microbiol. 2020, 20, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Benny, P.A.; Al-Akwaa, F.M.; Dirkx, C.; Schlueter, R.J.; Wolfgruber, T.K.; Chern, I.Y.; Hoops, S.; Knights, D.; Garmire, L.X. Placentas delivered by pre-pregnant obese women have reduced abundance and diversity in the microbiome. FASEB J. 2021, 35, e21524. [Google Scholar] [CrossRef] [PubMed]
- Fuks, G.; Elgart, M.; Amir, A.; Zeisel, A.; Turnbaugh, P.J.; Soen, Y.; Shental, N. Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome 2018, 6, 17. [Google Scholar] [CrossRef]
- Stupak, A.; Gęca, T.; Kwaśniewska, A.; Mlak, R.; Piwowarczyk, P.; Nawrot, R.; Goździcka-Józefiak, A.; Kwaśniewski, W. Comparative Analysis of the Placental Microbiome in Pregnancies with Late Fetal Growth Restriction versus Physiological Pregnancies. Int. J. Mol. Sci. 2023, 24, 6922. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, J.; Shi, W.; Du, N.; Xu, X.; Zhang, Y.; Ji, P.; Zhang, F.; Jia, Z.; Wang, Y.; et al. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut 2018, 67, 1614–1625. [Google Scholar] [CrossRef]
- Shiozaki, A.; Yoneda, S.; Yoneda, N.; Yonezawa, R.; Matsubayashi, T.; Seo, G.; Saito, S. Intestinal Microbiota is Different in Women with Preterm Birth: Results from Terminal Restriction Fragment Length Polymorphism Analysis. PLoS ONE 2014, 9, e111374. [Google Scholar] [CrossRef]
- Gorczyca, K.; Obuchowska, A.; Kimber-Trojnar, Z.; Wierzchowska-Opoka, M.; Leszczyńska-Gorzelak, B. Changes in the Gut Microbiome and Pathologies in Pregnancy. Int. J. Environ. Res. Public Health 2022, 19, 9961. [Google Scholar] [CrossRef]
- O’Callaghan, J.; Turner, R.; Nitert, M.D.; Barrett, H.L.; Clifton, V.; Pelzer, E.S. Re-assessing microbiomes in the low-biomass reproductive niche. BJOG: Int. J. Obstet. Gynaecol. 2019, 127, 147–158. [Google Scholar] [CrossRef]
- Hu, J.; Benny, P.; Wang, M.; Ma, Y.; Lambertini, L.; Peter, I.; Xu, Y.; Lee, M.-J. Intrauterine Growth Restriction Is Associated with Unique Features of the Reproductive Microbiome. Reprod. Sci. 2020, 28, 828–837. [Google Scholar] [CrossRef]
- He, X.; Li, Z.; Li, X.; Zhao, H.; Hu, Y.; Han, W.; Wang, C.; Yin, C.; Chen, Y. The fecal microbiota of gravidas with fetal growth restriction newborns characterized by metagenomic sequencing. Curr. Res. Transl. Med. 2023, 71, 103354. [Google Scholar] [CrossRef]
- Wang, J.; Gu, X.; Yang, J.; Wei, Y.; Zhao, Y. Gut Microbiota Dysbiosis and Increased Plasma LPS and TMAO Levels in Patients with Preeclampsia. Front. Cell. Infect. Microbiol. 2019, 9, 409. [Google Scholar] [CrossRef]
- Chang, Y.; Chen, Y.; Zhou, Q.; Wang, C.; Chen, L.; Di, W.; Zhang, Y. Short-chain fatty acids accompanying changes in the gut microbiome contribute to the development of hypertension in patients with preeclampsia. Clin. Sci. 2020, 134, 289–302. [Google Scholar] [CrossRef]
- Chen, X.; Li, P.; Liu, M.; Zheng, H.; He, Y.; Chen, M.-X.; Tang, W.; Yue, X.; Huang, Y.; Zhuang, L.; et al. Gut dysbiosis induces the development of pre-eclampsia through bacterial translocation. Gut 2020, 69, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.-J.; Li, S.-H.; Li, S.-C.; Zhong, Z.-C.; Duan, H.-L.; Tian, C.; Li, H.; He, W.; Chen, M.-C.; He, T.-W.; et al. Early-Onset Preeclampsia Is Associated With Gut Microbial Alterations in Antepartum and Postpartum Women. Front. Cell. Infect. Microbiol. 2019, 9, 224. [Google Scholar] [CrossRef]
- Kok, D.E.; Steegenga, W.T.; Smid, E.J.; Zoetendal, E.G.; Ulrich, C.M.; Kampman, E. Bacterial folate biosynthesis and colorectal cancer risk: More than just a gut feeling. Crit. Rev. Food Sci. Nutr. 2020, 60, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Crider, K.S.; Yang, T.P.; Berry, R.J.; Bailey, L.B. Folate and DNA Methylation: A Review of Molecular Mechanisms and the Evidence for Folate’s Role. Adv. Nutr. Int. Rev. J. 2012, 3, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Davie, J.R. Inhibition of Histone Deacetylase Activity by Butyrate. J. Nutr. 2003, 133, 2485S–2493S. [Google Scholar] [CrossRef]
- Cortese, R.; Lu, L.; Yu, Y.; Ruden, D.; Claud, E.C. Epigenome-Microbiome crosstalk: A potential new paradigm influencing neonatal susceptibility to disease. Epigenetics 2016, 11, 205–215. [Google Scholar] [CrossRef]
- Januar, V.; Desoye, G.; Novakovic, B.; Cvitic, S.; Saffery, R. Epigenetic regulation of human placental function and pregnancy outcome: Considerations for causal inference. Am. J. Obstet. Gynecol. 2015, 213, S182–S196. [Google Scholar] [CrossRef]

| First Trimester [14,15,19] | Second Trimester [7,20] | Third Trimester [7,20] | |
|---|---|---|---|
| Phyla | Bacteroidetes, Firmicutes | Proteobacteria, Bacteroidetes, Actinobacteria, Tenericutes, Firmicutes, Bifidobacteriaceae, Enterobacteriaceae | Faecalibacterium, Actinobacteria, Bifidobacterium, Enterobacteriaceae, Streptococcus, Proteobacteria |
| Method | Studies |
|---|---|
| WGS | Aagaard et al. [45] |
| 16S RNA gene sequencing | Amarasekara et al. [43], de Goffu et al. [50], Parnall et al. [49], Aagaard et al. [45], Seferovic et al. [64], Theis et al. [65], Bassols et al. [66], Cahill et al. [67], Collado et al. [68], Jones et al. [69], Leiby et al. [70], Zheng et al. [71,72,73]. |
| 16S RNA FISH | Seferovic et al. [64], Cahill et al. [67], Steel et al. [74]. |
| Immunofluorescence | Vanterpool et al. [75]. |
| Metagenomics | De Goffu et al. [50], Aagaard et al. [45], Prince et al. [76], Theis et al. [65], Leiby et al. [70]. |
| Nested PCR | De Goffu et al. [50], Theis et al. [65]. |
| Histology techniques | Lannon et al. [77], Prince et al. [76], Sefrovic et al. [64]. |
| SMURF | Fucks et al. [78]. |
| LC–ESI-MS/MS | Stupak et al. [79]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stupak, A.; Kwaśniewski, W. Evaluating Current Molecular Techniques and Evidence in Assessing Microbiome in Placenta-Related Health and Disorders in Pregnancy. Biomolecules 2023, 13, 911. https://doi.org/10.3390/biom13060911
Stupak A, Kwaśniewski W. Evaluating Current Molecular Techniques and Evidence in Assessing Microbiome in Placenta-Related Health and Disorders in Pregnancy. Biomolecules. 2023; 13(6):911. https://doi.org/10.3390/biom13060911
Chicago/Turabian StyleStupak, Aleksandra, and Wojciech Kwaśniewski. 2023. "Evaluating Current Molecular Techniques and Evidence in Assessing Microbiome in Placenta-Related Health and Disorders in Pregnancy" Biomolecules 13, no. 6: 911. https://doi.org/10.3390/biom13060911
APA StyleStupak, A., & Kwaśniewski, W. (2023). Evaluating Current Molecular Techniques and Evidence in Assessing Microbiome in Placenta-Related Health and Disorders in Pregnancy. Biomolecules, 13(6), 911. https://doi.org/10.3390/biom13060911






