Abstract
Serotonin N-acetyltransferase (SNAT) functions as the penultimate or final enzyme in melatonin biosynthesis, depending on the substrate. The Escherichia coli orthologue of archaeal SNAT from Thermoplasma volcanium was identified as RimI (EcRimI), with 42% amino acid similarity to archaeal SNAT. EcRimI has been reported to be an N-acetyltransferase enzyme. Here, we investigated whether EcRimI also exhibits SNAT enzyme activity. To achieve this goal, we purified recombinant EcRimI and examined its SNAT enzyme kinetics. As expected, EcRimI showed SNAT activity toward various amine substrates including serotonin and 5-methoxytryptamine, with Km and Vmax values of 531 μM and 528 pmol/min/mg protein toward serotonin and 201 μM and 587 pmol/min/mg protein toward 5-methoxytryptamine, respectively. In contrast to the rimI mutant E. coli strain that showed no growth defect, the EcRimI overexpression strain exhibited a 2-fold higher growth rate than the control strain after 24 h incubation in nutrient-rich medium. The EcRimI overexpression strain produced more melatonin than the control strain in the presence of 5-methoxytryptamine. The enhanced growth effect of EcRimI overexpression was also observed under cadmium stress. The higher growth rate associated with EcRimI expression was attributed to increased protein N-acetyltransferase activity, increased synthesis of melatonin, or the combined effects of both.
1. Introduction
Melatonin is a universal molecule present in almost all living organisms, including bacteria, archaea, plants, and animals [1,2,3]. Its primary identified function is associated with its potent antioxidant activity, as one molecule of melatonin can scavenge free radicals in a reaction cascade consuming up to 10 free radicals [4,5]. In addition to its intrinsic antioxidant activity, melatonin plays an important biological role in a number of species-specific functions. For example, it is a well-known pineal hormone that regulates the circadian rhythm and seasonal reproduction in animals [6,7]. By contrast, in plants, it acts as a master regulator of growth and development, affecting seed germination [8,9], photomorphogenesis [10], flowering [11,12], senescence [13], and grain yield [14,15] in concert with numerous plant hormones [16,17]. It also plays a pivotal role in alleviating plant damage resulting from various environment stresses, including abiotic and biotic stresses, either through enhancement of antioxidant activities and various defense genes [18,19] or improvement of protein quality control [12,20]. Melatonin is synthesized by bacteria, and appears to be involved in detoxifying reactive oxygen species to prevent free radical attack [21,22,23], although it has also been reported to inhibit the growth of plant-pathogenic bacteria [24].
In melatonin biosynthesis, the aromatic amino acid tryptophan serves as the initial substrate and melatonin is synthesized via four enzymatic reactions with tryptophan 5-hydroxylase (TPH), aromatic amino acid decarboxylase (tryptophan decarboxylase), serotonin N-acetyltransferase (SNAT; also named arylakylamine N-acetyltransferase), and N-acetylserotonin O-methyltransferase (ASMT) in animals. Analogously, plants also employ four enzymes, but TPH is replaced with tryptamine 5-hydroxylase, which catalyzes tryptamine into serotonin [25]. Among these four enzymes, SNAT plays a key role in melatonin biosynthesis, functioning as either the penultimate or final enzyme of melatonin biosynthesis, depending on substrate, in both animals and plants [2,25]. SNAT acetylates serotonin and 5-methoxytryptamine into N-acetylserotonin and melatonin, respectively [26]. In 1995, the first SNAT gene was cloned from sheep using a cDNA expression library [27], and its orthologues have been cloned and characterized from a wide range of species including Gram-positive bacteria and fungi but not from higher plants, nematodes, or arthropods [28]. Later, the first plant SNAT gene was cloned from the rice GCN5-related N-acetyltransferase (GNAT) family [29]. As expected, the SNAT protein from rice did not exhibit sequence homology with that from animals. The possible ancestor of the plant SNAT gene has been cloned and characterized in cyanobacteria [30].
Despite reports of SNAT genes from a diverse array of species, the presence of SNAT in archaea has long remained a mystery. Surprisingly, the archaeal SNAT gene from Thermoplasma volcanium was recently cloned [3] and a human orthologue was discovered [31]. This archaeal SNAT also belongs to the GNAT family, but it is classified as a new clade of SNAT, distinct from those of animals and plants. In particular, the Gram-negative bacterium Escherichia coli had long been reported to produce melatonin [32], but no SNAT homolog genes have yet been described. However, based on the discovery of archaeal SNAT, we identified an orthologous gene in E. coli.
In this study, we selected E. coli RimI (EcRimI), an archaeal SNAT orthologue expressed in E. coli, and purified recombinant EcRimI. We found that EcRimI possessed SNAT enzyme activity. Furthermore, its overexpression of EcRimI in E. coli was functionally linked to enhanced melatonin biosynthesis in the presence of 5-methoxytryptamine. The EcRimI overexpression strain of E. coli exhibited enhanced growth at 28 and 37 °C compared to the control strain. We concluded that enhanced synthesis of melatonin in the EcRimI overexpression strain was in part responsible for the enhanced growth rate due to ameliorating of starvation and stationary-phase stress, although we cannot rule out a possible role of protein acetylation.
2. Materials and Methods
2.1. Synthesis of Escherichia coli RimI Gene
Based on the nucleotide sequence information of E. coli RimI (GenBank accession number WP_137442509), the full-length EcRimI gene with the length of 447 bp was custom-synthesized at Bioneer (Daejeon, Republic of Korea).
2.2. Escherichia coli Expression and Purification of Recombinant EcRimI Protein
The full-length synthetic EcRimI gene was amplified by PCR using primer set (RimI forward primer, 5′-GCC ATG GGA AAC ACG ATT TCT TCC CTC GAA-3′; RimI reverse primer, 5′-CTC GAG CAT ACT GAT TGG CAA CGC-3′) with a template plasmid containing EcRimI DNA such as pBHA-RimI, which was provided by Bioneer. The PCR product was ligated into the TA vector (RBC Bioscience, New Taipei City, Taiwan) followed by NcoI and XhoI digestion. The NcoI and XhoI insert of EcRimI was then ligated into the same restriction sites of the E. coli expression vector pET28b (Invitrogen, Carlsbad, CA, USA), leading to the generation of pET28b-RimI. The pET28b-RimI plasmid was transformed into E. coli strain BL21(DE3) (Invitrogen). An overnight culture (10 mL) grown in Lennox broth (LB) medium (10 g/L pancreatic digest of casein, 5 g/L yeast extract, and 5 g/L NaCl) containing antibiotic kanamycin (50 mg/L) was inoculated into 100 mL of Terrific broth (TB) medium [20 g/L Bacto-tryptone, 24 g/L Bacto-yeast extract, glycerol 4 mL/L, and phosphate buffer (0.017 M KH2PO4 and 0.072 M K2HPO4)] containing 50 mg/L kanamycin and incubated at 37 °C for 4 h, followed by the addition of 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG; Sigma, St. Louis, MO, USA). The culture was further grown at 24 °C and shaken at 180 rpm for 12 h. The purification procedure using affinity (Ni2+) chromatography was performed according to the manufacturer’s recommendations (Qiagen, Tokyo, Japan).
2.3. Homology Analysis
The analysis of amino acid sequence homology was carried out using the BLASTp tool in the non-redundant protein sequences databases at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/, accessed on 3 September 2019). Phylogenetic tree analysis was performed using the BLAST-Explorer program [33].
2.4. Measurement of Serotonin N-Acetyltransferase Enzyme Kinetics
The purified recombinant EcRimI protein (3 μg or varying concentrations) was incubated in a total volume of 100 µL containing 0.5 mM serotonin (or other substrates) and 0.5 mM acetyl-CoA in 100 mM potassium phosphate (pH 8.8 or varying pH) at 55 °C (or other temperatures) for 30 min. Enzymatic reaction products such as N-acetylserotonin, N-acetyltryptamine, N-acetyltyramine, and melatonin were subjected to high-performance liquid chromatography (HPLC) analysis as described previously [11]. Lineweaver–Burk plots were employed to calculate substrate affinity (Km) and the maximum reaction rate (Vmax) using two substrates such as serotonin and 5-methoxytryptamine. Protein concentrations were determined using the Bradford method and a protein assay dye (Bio-Rad, Hercules, CA, USA). The analysis was performed in triplicate.
2.5. Growth Measurement of Escherichia coli
A 1 mL seed culture that had been incubated overnight at 37 °C in Lennox broth (LB) medium (10 g/L pancreatic digest of casein, 5 g/L yeast extract, 5 g/L NaCl) containing 50 mg/L kanamycin was inoculated into 10 mL Terrific broth (TB) medium containing 50 mg/L kanamycin in 40 mL polypropylene conical tubes (SPL Life Science, Pocheon-si, Republic of Korea) and continuously cultured at various temperatures. The absorbance at 600 nm of the culture was measured using a spectrophotometer (MicroDigital Nabi, GyungGi, Republic of Korea) periodically until 24 h.
2.6. Melatonin Measurement in Escherichia coli
One hundred microliters of seed culture incubated overnight at 37 °C in LB medium containing 50 mg/L kanamycin was inoculated into 1 mL LB medium containing 50 mg/L kanamycin and incubated at 37 °C until the optical density of the E. coli culture at 600 nm (OD600) reached 1.0. After the addition of 1 mM isopropyl-β-D-thiogalactopyranoside and 1 mM 5-methoxytryptamine, the culture was grown and shaken at 250 rpm at varying temperatures such as 28, 37, and 42 °C for the indicated time periods. The resulting cultures were centrifuged at 11,500× g for 5 min and separated into cell pellet and medium (supernatant) fractions. The medium fractions (0.2 mL) were mixed with 0.2 mL of 100% methanol. The resulting 10 µL aliquots were subjected to high-performance liquid chromatography (HPLC) using a fluorescence detector system (Waters, Milford, MA, USA) as described previously [34].
2.7. Cadmium Treatment of Escherichia coli
One hundred microliters of seed culture incubated overnight at 37 °C in LB medium containing 50 mg/L kanamycin was inoculated into 1 mL LB medium containing 50 mg/L kanamycin and incubated at 37 °C until the optical density of the E. coli culture at 600 nm (OD600) reached either 0.5 or 1.0. After adding 1 or 5 mM cadmium chloride (CdCl2), the culture was grown at 37 °C for 20 h with shaking at 250 rpm. The absorbance of the resulting cultures was measured at 600 nm using a spectrophotometer (MicroDigital Nabi).
2.8. Statistical Analysis
The data were analyzed by analysis of variance using IBM SPSS Statistics 23 software (IBM Corp., Armonk, NY, USA). Means with different letters indicate significantly different values at p < 0.05 according to Tukey’s post hoc honestly significant difference (HSD) test. Data are presented as means ± standard deviations.
3. Results
3.1. Gene Selection and Synthesis of the Escherichia coli RimI Gene
The BLASTp program (http://www.ncbi.nih.gov/, accessed on 3 November 2019) revealed that the archaeal SNAT (TvSNAT) protein [3] had ~23% identity and ~42% similarity to the RimI protein of E. coli (EcRimI), which encodes the protein N-acetyltransferase enzyme (Figure 1A). EcRimI acetylates a number of proteins, including ribosomal protein S18 [35], translation elongation factor Tu [36], and other proteins [37]. The human protein N-acetyltransferase Naa50, an orthologue of archaeal SNAT, shows SNAT enzyme activity [31], indicating that E. coli RimI was likely to also exhibit SNAT activity. Phylogenetic analysis indicated that EcRimI belonged to the SNAT family and was positioned between the plant and archaeal SNAT subfamilies, but much closer to the archaeal subfamily than the plant SNAT subfamily (Figure 1B). The full-length EcRimI nucleotide sequence was synthesized in accordance with sequence information reported in the GenBank database (WP_137442509).

Figure 1.
(A) Amino acid sequence alignment of TvSNAT and E. coli RimI (SNAT). The conserved acetyl coenzyme A binding sites are underlined. Plus signs (+) denote similar amino acids. Dashes denote gaps. (B) Phylogenetic tree showing E. coli SNAT, archaeal orthologues and rice SNAT genes. The scale bar represents 0.3 substitutions per site. GenBank accession numbers are: TvSNAT (NC_002689), E. coli RimI (WP_137442509), rice SNAT1 (AK059369), rice SNAT2 (AK068156), human Naa50 (BAB14397), and Arabidopsis Naa50 (NM_121172).
3.2. Enzyme Kinetic Analysis of Recombinant EcRimI
The synthetic full-length EcRimI gene was cloned for expression as a fusion protein with a C-terminal hexa-histidine tag, followed by Ni2+ affinity purification, as illustrated in Figure 2A. The purified recombinant EcRimI protein was first investigated for the capacity to catalyze serotonin into N-acetylserotonin (NAS). As shown in Figure 2B, NAS was produced in vitro by recombinant EcRimI protein in a concentration-dependent manner. The optimal SNAT activity was observed at a temperature of 55 °C and half-peak activity occurred at 45 °C. SNAT activity at 37 °C was a quarter of that at 55 °C. No NAS was produced at 70 °C, which was consistent with archaeal SNAT [3]. EcRimI exhibited peak activity at pH 8.8, followed by pH 7.8 and pH 6.5, and very low SNAT activity was observed at pH 5.5. This pH preference of EcRimI SNAT activity was identical to that of archaeal SNAT, but differed from human Naa50, which has peak SNAT activity at pH 7.8 [3,31]. In addition to serotonin, several other amines were accepted as substrates. The maximum SNAT activity occurred with 5-methoxytryptamine, which was catalyzed into melatonin by the EcRimI enzyme, followed in rank order by tryptamine, serotonin, and tyramine. The substrate preference order of EcRimI differed from that of archaeal SNAT, in which tyramine is the optimal substrate and 5-methoxytryptamine is the least favorable substrate [3]. Tyramine has been reported as the optimal substrate for sheep SNAT and rice SNAT1 [38]. In terms of the possible acceptance of polyamines such as spermidine and octopamine as RimI substrates, we conducted an SNAT inhibition assay (0.5 mM serotonin) in the presence of each substrate at various concentrations to determine whether SNAT activity was affected by the presence of these polyamines. These inhibition analyses were performed because no standard compounds of N-acetylspermidine and N-acetyloctopamine are commercially available. As shown in Figure 3, spermidine did not inhibit SNAT activity at concentrations up to 1 mM under the 45 °C temperature condition. Similarly, octopamine slightly inhibited SNAT activity at 0.2 mM, but enhanced SNAT activity at 0.5 mM, shifting to 90% inhibition at 1 mM (Figure 3A). Under the 55 °C assay condition, spermidine enhanced SNAT activity, whereas octopamine did not alter SNAT activity except at 0.5 mM, where it showed a 50% inhibition rate (Figure 3B). Collectively, E. coli SNAT (EcSNAT or EcRimI) likely did not accept polyamines as substrates, in sharp contrast to archaeal SNAT, which accepts polyamines as substrates [3].
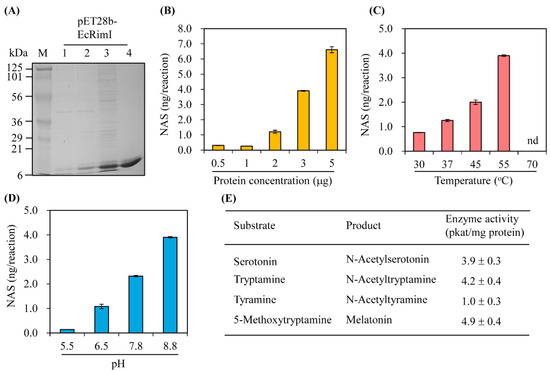
Figure 2.
Affinity purification and enzymatic characteristics of E. coli RimI (EcRimI) protein. (A) Purification of C-terminal 6× His-tagged EcRimI protein. E. coli BL21 (DE3) cells harboring the pET28b-EcRimI plasmid were induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) for 24 h at 24 °C. M, molecular mass standards; lane 1, total protein in 30 µL bacterial culture without IPTG; lane 2, total proteins in 30 µL bacterial culture with IPTG; lane 3, 10 µg soluble protein; lane 4, 5 µg protein purified through affinity chromatography. Protein samples were separated using 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue. Serotonin N-acetyltransferase enzyme activity as a function of (B) protein concentration, (C) temperature, (D) pH, and (E) substrate. Recombinant purified EcRimI (3 µg) was assayed for 0.5 h at 55 °C (varying temperature or pH) in the presence of 0.5 mM serotonin (or other substrate) and 0.5 mM acetyl coenzyme A, followed by high-performance liquid chromatography (HPLC) detection. NAS represents N-acetylserotonin. Values are means ± standard deviation (SD; n = 3).
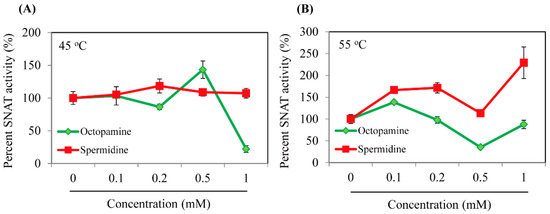
Figure 3.
Dose-dependent inhibition of SNAT enzyme activity of recombinant EcRimI by polyamines. (A) SNAT enzyme inhibition in the presence of either spermidine or octopamine at 45 °C. (B) SNAT enzyme inhibition in the presence of either spermidine or octopamine at 55 °C. SNAT enzyme activity was measured in the presence of various levels of polyamines and 0.5 mM serotonin. SNAT activity is expressed as a percentage relative to that in the absence of polyamines. Values are means ± SD (n = 3).
The Km and Vmax values of EcRimI toward serotonin as a substrate were 531 μM and 528 pmol/min/mg protein, respectively (Figure 4A). For the substrate 5-methoxytryptamine, EcRimI exhibited Km and Vmax values of 201 μM and 587 pmol/min/mg protein, respectively (Figure 4B). The catalytic efficiency (Vmax/Km) was 3-fold higher toward 5-methoxytryptamine than toward serotonin, suggesting that EcRimI preferred 5-methoxytryptamine to serotonin as a substrate for melatonin biosynthesis. The enzyme kinetics values of EcRimI were very similar to those of archaeal SNAT, supporting EcRimI as an archaeal SNAT orthologue. By contrast, human Naa50, a known archaeal SNAT orthologue, exhibited much higher Km and Vmax values for serotonin of 986 μM and 1800 pmol/min/mg protein, respectively. Based on the data presented above regarding the enzymatic properties of SNAT, EcRimI possessed SNAT activity, in accordance with archaeal SNAT and human Naa50 [3,31]. However, the exact mechanism through which EcRimI synthesizes melatonin in E. coli awaits further in-depth analysis, particularly in view of the dual roles of EcRimI in melatonin synthesis and protein N-acetylation.
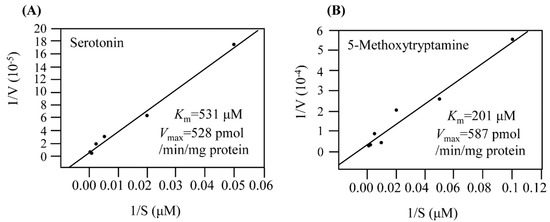
Figure 4.
Determination of Km and Vmax values for recombinant EcRimI toward the substrates (A) serotonin and (B) 5-methoxytryptamine. Km and Vmax values were determined using Lineweaver–Burk plots at 55 °C.
3.3. Escherichia coli Growth Curves for the EcRimI Overexpression Strain
The E. coli rimI mutant strain exhibits moderate growth retardation in minimal medium, but this growth difference is nonsignificant in nutrient-rich medium such as LB medium [36]. Importantly, as temperature strongly affects the solubility of the recombinant protein expressed in E. coli [39], we tested a wide range of temperatures for comparison of E. coli growth between the control (pET28b) and overexpression (pET28b-RimI) strains. In contrast to the previous study with the rimI mutant strain, we used nutrient-rich media such as Terrific broth (TB) medium to examine the growth effects of EcRimI overexpression. A significant growth disadvantage of the EcRimI overexpression strain relative to the control strain was apparent during the first 8 h of culture, but this growth retardation of the EcRimI overexpression strain was overcome at 12 h, and 2-fold higher growth was achieved at 24 h under the 28 °C culture temperature condition (Figure 5A). The growth recovery effect in the EcRimI overexpression strain was faster and more pronounced at the 37 °C culture temperature than at 28 °C. The EcRimI overexpression strain grew more slowly than the control during the first 4 h incubation and reached an equal growth rate comparable to the control strain at 6 h, then grew faster than the control strain from 8 h, and finally showed more than 2-fold higher growth than the control strain at 24 h (Figure 5B). At the 42 °C culture temperature, a similar growth disadvantage of the EcRimI overexpression strain was observed until 8 h, and OD600 values equal to those of the control strain were achieved at 12 h. At the end of 24 h of culturing at 42 °C, the growth advantage of EcRimI overexpression was marginal compared to that at 28 and 37 °C (Figure 5C). These data clearly indicated that EcRimI overexpression conferred a strong ability to enhance E. coli growth at the later stage of bacterial cultivation compared to the control strain, although whether these positive effects resulted from either protein N-acetylation or melatonin synthesis remained unclear. This mechanism should be investigated in the near future.

Figure 5.
Growth curves of control (pET28b) and EcRimI overexpression (pET28b-RimI) E. coli strains at (A) 28 °C, (B) 37 °C, and (C) 42 °C. Each point represents three independent replicates. E. coli was grown in nutrient-rich TB medium.
3.4. Melatonin Production in Escherichia coli
E. coli produces small amounts of melatonin (>1 ng/mL) [34]. To identify melatonin in E. coli, 5-methoxytryptamine, a direct substrate of melatonin production by SNAT, was added to the culture medium. After 24 h incubation in the presence of 1 mM 5-methoxytryptamine and 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at various temperatures, melatonin levels were quantified in the medium fraction of the E. coli culture, as the majority of melatonin was found in the medium fraction rather than in E. coli cells [34]. As shown in Figure 6A, the control E. coli strain (pET28b) also produced melatonin at concentrations of 383, 322, and 301 ng/mL at the incubation temperatures of 28, 37, and 42 °C, respectively. By contrast, the EcRimI overexpression E. coli strain (pET28b-RimI) produced melatonin at 617, 707, and 301 ng/mL at 28, 37, and 42 °C, respectively. Thus, the EcRimI overexpression strain produced melatonin at rates 1.6- and 2.2-fold higher than the control strain when E. coli was incubated at 28 and 37 °C, respectively. However, no differences in melatonin production between the EcRimI overexpression and control strains were observed at 42 °C culture temperature. These results were consistent with the growth curves at 42 °C. Thus, the optimal temperature for melatonin production in the EcRimI overexpression E. coli strain was 37 °C, followed by 28 °C. Time-course analysis of melatonin production was performed at the optimal temperature of 37 °C (Figure 6B). Melatonin production increased as the incubation time was prolonged. However, in the absence of 5-methoxytryptamine, the melatonin level was below our HPLC detection limit of 1 ng/mL. Our data suggested that E. coli could synthesize melatonin in the presence of 5-methoxytryptamine, which is present in the diet [40,41]. A biosynthetic pathway from serotonin to N-acetylserotonin and then to melatonin is also possible, but is unlikely due to the requirement of an additional enzyme, such as ASMT. These possibilities remain to be investigated in future research.
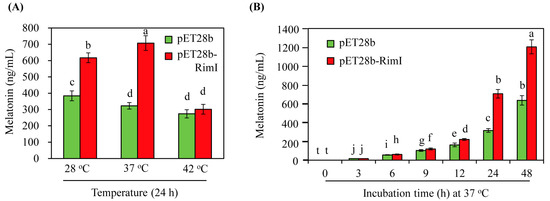
Figure 6.
(A) Melatonin production of the control (pET28b) and EcRimI overexpression (pET28b-RimI) strains at 24 h under various culture temperature conditions. (B) Time course of melatonin production by the control (pET28b) and EcRimI overexpression (pET28b-RimI) strains at 37 °C. Medium fractions were subjected to HPLC analysis for melatonin quantification. Different letters indicate significant differences from the wild type (Tukey’s honest significant difference (HSD) test; p < 0.05). Values are presented as means ± SD (n = 3).
3.5. Cadmium Response of the EcRimI Overexpression Strain
Cadmium treatment of E. coli inhibits antioxidant enzymes and lowers the levels of antioxidants such as glutathione, which decreases the scavenging of reactive oxygen species and leads to mechanical damage [42]. Correspondingly, adding glutathione alleviates damage from oxidative stress and increases the growth rate of E. coli [43]. In contrast to glutathione, melatonin treatment hampers the growth of Xanthomonas oryzae, a plant-pathogenic bacterium [24]. To assess the impacts of cadmium stress, the EcRimI overexpression strain was challenged with cadmium treatment. As shown in Figure 7, the EcRimI overexpression strain did not exhibit cadmium stress tolerance compared to the control. However, even in the presence of cadmium, the EcRimI overexpression strain of E. coli showed significantly enhanced growth relative to the control strain, regardless of the cell density. These data suggested that the growth enhancement associated with EcRimI overexpression was barely affected by heavy metal stress, indicating that EcRimI played a general role in improving E. coli growth through modulation of protein N-acetylation, melatonin biosynthesis, or both.
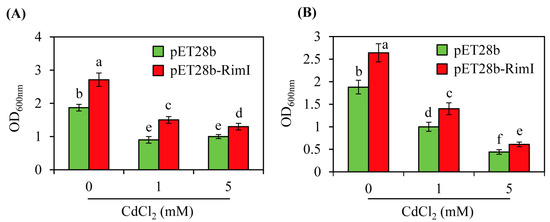
Figure 7.
Growth curves of the control (pET28b) and EcRimI overexpression (pET28b-RimI) E. coli strains in the presence of cadmium. (A) Cadmium was applied to a culture at OD600 = 1.0 and incubated for 20 h at 37 °C. (B) Cadmium was applied to a culture at OD600 = 0.5 and incubated for 20 h at 37 °C. The medium used in this experiment was LB (Lennox broth). Overnight seed cultures of 100 µL were inoculated into 1 mL fresh LB medium containing the antibiotic kanamycin (50 µg/mL) and grown at 37 °C until reaching OD600 = 1.0 or 0.5, followed by cadmium treatment for 20 h. Each point represents three independent replicates. Different letters indicate significant differences from the wild type (Tukey’s HSD test; p < 0.05).
4. Discussion
The biosynthetic pathways and biological roles of melatonin have been well documented since its discovery in the pineal gland of cows in 1958 [44] and in several plant species in 1995 [45,46]. Accordingly, increasing numbers of melatonin biosynthesis-related genes have been cloned and characterized from prokaryotes and eukaryotes through sequence homology-based cloning strategies using sheep SNAT [27,47]. Among such genes, SNAT has been most widely studied, as it plays a key role in melatonin biosynthesis in diverse organisms [48]. Consequently, many sheep SNAT orthologues have been cloned from human [49], yeast [50], Drosophila melanogaster [51], Chlamydomonas reinhardtii [52], and Xanthomonas oryzae [53] (Table 1). Similarly, plant SNAT genes have been cloned from various species [54]. In contrast to the single copy of SNAT present in animals [28], plant SNAT exists as a gene family containing at least 3 isogenes with very low amino acid sequence homology [25]. These SNAT genes include rice SNAT1 [29,55] and SNAT2 [38], Arabidopsis SNAT1 [56] and SNAT2 [11], tobacco SNAT1 and SNAT2 [57], apple SNAT3 [58], red algal SNAT [59], and cyanobacterial SNAT [30]. All plant SNAT1 and SNAT2 expression is localized in chloroplasts, except apple SNAT3, which is expressed in mitochondria [58]. A mitochondrial apple SNAT3 orthologue has been found in rice, but its function as an SNAT enzyme in rice remains unknown [25]. Collectively, rice likely contains up to four SNAT isogenes including an archaeal orthologue, which is expressed in the cytoplasm, in accordance with human Naa50, another archaeal SNAT orthologue [31]. The archaeal SNAT gene was recently cloned and characterized from archaeal GNAT family genes [3]. This archaeal SNAT from Thermoplasma volcanium was previously annotated as TvArd1 (arrest-defective-1), encoding a protein with N-terminal acetyltransferase activity that transfers an acetyl group from acetyl-coenzyme A to the N-termini of various proteins [60,61,62]. Ard1 is also called Naa10, and it is one of six NAT enzyme complexes containing Nat10 to Nat60 [63]. The closest orthologue of archaeal SNAT in humans is Naa50, N-alpha-acetyltransferase 50, which was recently revealed to possess SNAT enzyme activity [31]. These data suggest that archaeal SNAT proteins with N-acetyltransferase activity represent a novel class of SNAT family genes. In support of this possibility, E. coli RimI, an archaeal SNAT orthologue that exhibits protein N-acetyltransferase activity, also showed SNAT enzyme activity, indicating that E. coli have the genetic capacity to synthesize melatonin from its penultimate precursor, serotonin, as well as the direct substrate 5-methoxytryptamine (Figure 2). Phylogenetic analysis indicated that EcRimI belongs to the archaea SNAT family comprising TvSNAT [3] and human Naa50 [31] and is distantly related to the animal SNAT and plant SNAT family, suggesting that EcRimI is a functional E. coli orthologue of archaeal SNAT. In addition to the SNAT orthologue genes found in animals, plants, and archaea, enhanced intracellular survival (Eis) proteins containing two NAT domains and one sterol carrier domain also exhibit SNAT enzyme activity; however, Eis homolog proteins are present mainly in mycobacteria and certain Gram-positive bacteria, but are not found in animals and plants [64].

Table 1.
Enzyme kinetics of SNAT proteins from various organism.
The different activities of SNAT enzymes derived from plants and archaea have triggered fundamental questions about their biological functions, such as whether transgenic phenotypes showing gain or loss of SNAT gene function can be attributed to changes in melatonin or protein acetylation. Arabidopsis SNAT1 exhibits N-acetyltransferase activity toward chloroplast proteins [65], and its knockout mutant of SNAT1 (snat1) exhibited stunted growth depending on light intensity [12]. Whether the main reason for altered phenotypes in the snat1 mutant of Arabidopsis is decreased biosynthesis of melatonin, altered levels of chloroplast protein acetylation, or the combined effects of both changes remains unclear [12,20,65]. Analogously, Naa50 knockdown disrupted centromeric sister chromatid cohesion in Drosophila [66] and centromeric cohesion in HeLa cells [67], suggesting that protein N-acetyltransferase activity plays an important role in cell division. However, these findings do not eliminate the possible involvement of melatonin in cell division. This possibility remains to be elucidated in future research. Notably, the phenotypic abnormality observed in humans and Drosophila was not found in Naa50 knockdown yeast [68]. E. coli RimI is responsible for the N-terminal acetylation of the ribosomal protein S18 [69] and elongation factor Tu [36]. Consequently, an E. coli strain devoid of RimI showed slightly reduced growth on minimal medium, which was not observed in nutrient-rich medium, possibly driven by reduced efficiency of translation [36]. Assessing whether the growth retardation of the RimI-lacking strain is associated with melatonin biosynthesis remains an intriguing question.
All SNAT genes belong to the GCN5-related N-acetyltransferase (GNAT) superfamily of enzymes and share a common acetyl coenzyme A binding domain [70]. Although whether animal SNAT proteins such as sheep SNAT have protein N-acetyltransferase activity remains unclear, SNAT proteins derived from plants and archaea exhibit such activity [36,65]. The dual activity of SNAT proteins gives rise to the production of melatonin, N-acetylated proteins, or both simultaneously. Both products play diverse biological roles in organisms. Melatonin is a universal and pleiotropic molecule orchestrating the day–night waking cycle, seasonal reproduction, antitumor functions, and immune responses in animals [2,71] while acting as a master regulator of plant growth and development [72]. N-acetylation is a universal protein modification process, with 80–90% of soluble proteins being N-terminally acetylated [62], and regulates protein degradation, subcellular translocation, and protein complex formation [63].
As reported in animals and plants, melatonin may have certain biological functions, such as antioxidation, in E. coli [4,41,73,74]. The EcRimI mutant strain showed no growth inhibition in nutrient-rich LB medium [37]. In addition, the EcRimI overexpression strain exhibited far less N-acetylation activity toward E. coli proteins in vivo than other N-acetyltransferases such as YfiQ, Yjab, and YiaC, suggesting that the role of EcRimI as a protein N-acetyltransferase is minimal [37]. Although no direct evidence connects melatonin with E. coli growth, a beneficial effect of melatonin on E. coli growth cannot be ruled out, as the EcRimI overexpression E. coli strain showed enhanced growth in conjunction with increased melatonin production in this study. Melatonin has long been proposed to be synthesized in bacteria, especially in E. coli, but no biosynthesis pathway has been found. Due to the discovery SNAT gene in E. coli, we open a new window to studying the function of melatonin in bacteria by way of gene manipulation and to engineer melatonin biosynthesis for its overproduction in E. coli. This is the first report of successful cloning of SNAT from E. coli. Further studies will shed light on the physiological roles of melatonin in E. coli during growth and under oxidative stress.
5. Conclusions
With the help of the first successful cloning of archaeal SNAT, we revealed that E. coli RimI, encoding a protein N-acetyltransferase, exhibited SNAT enzyme activity in vitro and that EcRimI overexpression in vivo was associated with enhanced melatonin production in E. coli. The EcRimI overexpression E. coli strain exhibited enhanced growth compared to the control strain. The EcRimI overexpression E. coli strain showed strong tolerance against stationary-phase stress based on its growth curves. This enhanced growth effect of EcRimI overexpression compared to that of the control remained in the presence of cadmium stress. In summary, E. coli clearly has the capacity to synthesize melatonin through the enzymatic activity of EcRimI using 5-methoxytryptamine present in the diet, and EcRimI overexpression plays an important role in E. coli growth, possibly associated with melatonin synthesis, protein acetylation, or both.
Author Contributions
Conceptualization, K.B.; formal analysis, K.B. and K.L.; investigation, K.L.; writing—original draft preparation, K.B.; writing—review and editing, K.B.; funding acquisition, K.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported from grants by the Basic Science Research Program of the National Research Foundation of Korea (NRF-2021R1I1A2042237) funded by the Ministry of Education.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the finding of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hardeland, R. Melatonin in the evolution of plants and other phototrophs. Melatonin Res. 2019, 2, 10–36. [Google Scholar] [CrossRef]
- Zhao, D.; Yu, Y.; Shen, Y.; Liu, Q.; Zhao, Z.; Sharma, R.; Reiter, R.J. Melatonin synthesis and function: Evolutionary history in animals and plants. Front. Endocrinol. 2019, 10, 249. [Google Scholar] [CrossRef]
- Lee, K.; Choi, G.H.; Back, K. Functional characterization of serotonin N-acetyltransferase in archaeon Thermoplasma volcanium. Antioxidants 2022, 11, 596. [Google Scholar] [CrossRef] [PubMed]
- Rosen, J.; Than, N.N.; Koch, D.; Poeggeler, B.; Laatsch, L.H.; Hardeland, R. Interactions of melatonin and its metabolites with the ABTS cation radical: Extension of the radical scavenger cascade and formation of a novel class of oxidation products, C2-substituted 3-indolinones. J. Pineal Res. 2006, 41, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.X.; Manchester, L.C.; Terron, M.P.; Flores, L.J.; Reiter, R.J. One molecule, many derivatives: A never-ending interaction of melatonin with reactive oxygen and nitrogen species? J. Pineal Res. 2007, 42, 28–42. [Google Scholar] [CrossRef]
- Ahmad, S.B.; Ali, A.; Bilal, M.; Rashid, S.M.; Wani, A.B.; Bhat, R.R.; Rehman, M.U. Melatonin and health: Insights of melatonin action, biological functions, and associated disorders. Cell Mol. Neurobiol. 2023, in press. [CrossRef]
- Cosme, P.; Rodríguez, A.B.; Garrido, M.; Espino, J. Coping with oxidative stress in reproductive pathophysiology and assisted reproduction: Melatonin as an emerging therapeutical tool. Antioxidants 2023, 12, 86. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Liu, L.; Wang, H.; Li, D.; Bai, Z.; Zhang, Y.; Sun, H.; Zhang, K.; Li, C. Exogenous melatonin accelerates seed germination in cotton (Gossypium hirsutum L.). PLoS ONE 2019, 14, e0216575. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Back, K. 2-Hydroxymelatonin promotes seed germination by increasing reactive oxygen species production and gibberellin synthesis in Arabidopsis thaliana. Antioxidants 2022, 11, 737. [Google Scholar] [CrossRef]
- Hwang, O.J.; Back, K. Melatonin is involved in skotomorphogenesis by regulating brassinosteroid biosynthesis in plants. J. Pineal Res. 2018, 65, e12495. [Google Scholar] [CrossRef]
- Lee, H.Y.; Lee, K.; Back, K. Knockout of Arabidopsis serotonin N-acetyltransferase-2 reduces melatonin levels and delays flowering. Biomolecules 2019, 9, 712. [Google Scholar] [CrossRef]
- Lee, H.Y.; Back, K. Melatonin regulates chloroplast protein quality control via a mitogen-activated protein kinase signaling pathway. Antioxidants 2021, 10, 511. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-Q.; Zhang, Z.-W.; Chen, Y.-E.; Ding, C.-B.; Yuan, S.; Reiter, R.J.; Yuan, M. Melatonin: A potential agent in delaying leaf senescence. Crit. Rev. Plant Sci. 2021, 40, 1–22. [Google Scholar] [CrossRef]
- Jiang, Y.; Huang, S.; Ma, L.; Kong, L.; Pan, S.; Tang, X.; Tian, H.; Duan, M.; Mo, Z. Effects of exogenous melatonin application on the grain yield and antioxidant capacity in aromatic rice under combined lead-cadmium stress. Antioxidants 2022, 11, 776. [Google Scholar] [CrossRef]
- Muhammad, I.; Yang, L.; Ahmad, S.; Mosaad, I.S.M.; Al-Ghamdi, A.A.; Abbasi, A.M.; Zhou, X.-B. Melatonin application alleviates stress-induced photosynthetic inhibition and oxidative damage by regulating antioxidant defense system of maize: A meta-analysis. Antioxidants 2022, 11, 512. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin and its relationship to plant hormones. Ann. Bot. 2018, 121, 195–207. [Google Scholar] [CrossRef]
- Hwang, O.J.; Back, K. Molecular regulation of antioxidant melatonin biosynthesis by brassinosteroid acting as an endogenous elicitor of melatonin induction in rice seedling. Antioxidants 2022, 11, 918. [Google Scholar] [CrossRef]
- Altaf, M.A.; Shahid, R.; Ren, M.X.; Naz, S.; Altaf, M.M.; Khan, L.U.; Tiwari, R.K.; Lal, M.K.; Shahid, M.A.; Kumar, R.; et al. Melatonin improves drought tolerance of tomato by modulating plant growth, root architecture, photosynthesis, and antioxidant defense system. Antioxidants 2022, 11, 309. [Google Scholar] [CrossRef]
- Bhardwaj, R.; Pareek, S.; Domínguez-Avila, J.A.; Gonzalez-Aguilar, G.A.; Valero, D.; Serrano, M. An exogenous pre-storage melatonin alleviates chilling injury in some mango fruit cultivars, by acting on the enzymatic and non-enzymatic antioxidant system. Antioxidants 2022, 11, 384. [Google Scholar] [CrossRef]
- Lee, H.Y.; Hwang, O.J.; Back, K. Phytomelatonin as a signaling molecule for protein quality control via chaperone, autophagy, and ubiquitin–proteasome systems in plants. J. Exp. Bot. 2022, 73, 5863–5873. [Google Scholar] [CrossRef] [PubMed]
- Manchester, L.C.; Poeggeler, B.; Alvares, F.L.; Ogden, G.B.; Reiter, R.J. Melatonin immunoreactivity in the photosynthetic prokaryote Rhodospirillum rubrum: Implications for an ancient antioxidant system. Cell Mol. Biol. Res. 1995, 41, 391–395. [Google Scholar]
- Tilden, A.R.; Becker, M.A.; Amma, L.L.; Arciniega, J.; McGaw, A.K. Melatonin production in an aerobic photosynthetic bacterium: An evolutionarily early association with darkness. J. Pineal Res. 1997, 22, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Ma, Y.; Chen, S.; Liu, C.; Song, Y.; Qin, Y.; Yuan, C.; Liu, Y. Melatonin-producing endophytic bacteria from grapevine roots promote the abiotic stress-induced production of endogenous melatonin in their hosts. Front. Plant Sci. 2016, 7, 1387. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, C.; Laborda, P.; Zhao, Y.; Palmer, I.; Fu, Z.Q.; Qiu, J.; Liu, F. Melatonin treatment inhibits the growth of Xanthomonas oryzae pv. oryzae. Front. Microbiol. 2018, 9, 2280. [Google Scholar] [CrossRef] [PubMed]
- Back, K. Melatonin metabolism, signaling and possible roles in plants. Plant J. 2021, 105, 376–391. [Google Scholar] [CrossRef]
- Tan, D.X.; Hardeland, R.; Back, K.; Manchester, L.C.; Alatorre-Jimenez, M.A.; Reiter, R.J. On the significance of an alternate pathway of melatonin synthesis via 5-methoxytryptamine: Comparisons across species. J. Pineal Res. 2016, 61, 27–40. [Google Scholar] [CrossRef]
- Coon, S.L.; Roseboom, P.H.; Baler, R.; Weller, J.L.; Nambroodiri, M.A.A.; Koonin, E.V.; Klein, D.C. Pineal serotonin N-acetyltransferase: Expression cloning and molecular analysis. Science 1995, 270, 1681–1683. [Google Scholar] [CrossRef]
- Coon, S.L.; Klein, D.C. Evolution of arylalkylamine N-acetyltransferase: Emergence and divergence. Mol. Cell Endocrinol. 2006, 252, 2–10. [Google Scholar] [CrossRef]
- Kang, K.; Lee, K.; Park, S.; Byeon, Y.; Back, K. Molecular cloning of rice serotonin N-acetyltransferase, the penultimate gene in plant melatonin biosynthesis. J. Pineal Res. 2013, 55, 7–13. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, K.; Park, Y.I.; Park, S.; Back, K. Molecular cloning and functional analysis of serotonin N-acetyltransferase from the cyanobacterium Synechocystis sp. PCC 6803. J. Pineal Res. 2013, 55, 371–376. [Google Scholar]
- Lee, K.; Back, K. Human Naa50 harbors serotonin N-acetyltransferase activity and its overexpression enhances melatonin biosynthesis resulting in osmotic stress tolerance in rice. Antioxidants 2023, 12, 319. [Google Scholar] [CrossRef]
- Balzer, I.; Hocker, B.; Kappm, H.; Bartolomaeus, B. Occurrence and comparative physiology of melatonin in evolutionary diverse organisms. In The Redox State and Circadian Rhythms; Vanden Driessche, T., Guisset, J.-L., Petiau-de Vries, G.M., Eds.; Kluwer: Dordrecht, The Netherlands; Boston, MA, USA; London, UK, 2000; pp. 95–119. [Google Scholar]
- Dereeper, A.; Audic, S.; Claverie, J.M.; Blanc, G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 2010, 10, 8. [Google Scholar] [CrossRef]
- Byeon, Y.; Back, K. Melatonin production in Escherichia coli by dual expression of serotonin N-acetyltransferase and caffeic acid O-methyltransferase. Appl. Microbiol. Biotechnol. 2016, 100, 6683–6691. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, A.; Isono, S.; Sheback, A.; Isono, K. Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol. Gen. Genet. 1987, 209, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Pletnev, P.I.; Shulenina, O.; Evfratov, S.; Treshin, V.; Subach, M.F.; Serebryakova, M.V.; Osterman, I.A.; Paleskava, A.; Bogdanov, A.A.; Dontsova, O.A.; et al. Ribosomal protein S18 acetyltransferase RimI is responsible for the acetylation of elongation factor Tu. J. Biol. Chem. 2022, 298, 101914. [Google Scholar] [CrossRef] [PubMed]
- Christensen, D.G.; Meyer, J.G.; Baumagartner, J.T.; D’Souza, A.K.; Nelson, W.C.; Payne, S.H.P.; Kuhn, M.L.; Schilling, B.; Wolfe, A.J. Identification of novel protein lysine acetyltransferases in Escherichia coli. Mol. Biol. Physiol. 2018, 9, e01905-18. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.; Lee, H.Y.; Back, K. Cloning and characterization of the serotonin N-acetyltransferase-2 gene (SNAT2) in rice (Oryza sativa). J. Pineal Res. 2016, 61, 198–207. [Google Scholar] [CrossRef]
- Schein, C.H.; Noteborn, M.H.M. Formation of soluble recombinant proteins in Escherichia coli is favored by lower growth temperature. Nat. Biotech. 1988, 6, 291–294. [Google Scholar] [CrossRef]
- Choi, G.H.; Lee, H.Y.; Back, K. Chloroplast overexpression of rice caffeic acid O-methyltransferase increase melatonin production in chloroplasts via the 5-methoxytryptamine pathway in transgenic rice plants. J. Pineal Res. 2017, 63, e12412. [Google Scholar] [CrossRef]
- Tan, D.X.; Reiter, R.J.; Zimmerman, S.; Hardeland, R. Melatonin: Both a messenger of darkness and a participant in the cellular actions of non-visible solar radiation of near infrared light. Biology 2023, 12, 89. [Google Scholar] [CrossRef]
- Thapa, G.; Das, D.; Gunupuru, L.R. Expression of Echmr gene from Eichhornia offers multiple stress tolerance to Cd sensitive Escherichia coli Delta gsh mutants. Biochem. Biophys Res. Commun. 2016, 478, 101–109. [Google Scholar] [CrossRef]
- Qin, W.; Zhao, J.; Yu, X.; Liu, X.; Chu, X.; Tian, J.; Wu, N. Improving cadmium resistance in Escherichia coli through continuous genome evolution. Front. Microbiol. 2019, 10, 278. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.B.; Case, J.D.; Takahashi, Y. Isolation of melatonin, a pineal factor that lightness melanocytes. J. Am. Soc. 1958, 80, 2587. [Google Scholar] [CrossRef]
- Dubbels, R.; Reiter, R.J.; Klenke, E.; Goebel, A.; Schnakenberg, E.; Ehlers, C.; Schiwara, H.W.; Schloot, W. Melatonin in edible plants identified by radioimmunoassay and by high performance liquid chromatography-mass spectrometry. J. Pineal Res. 1995, 18, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Hattori, A.; Migitaka, H.; Iigo, M.; Itoh, M.; Yamamoto, K.; Ohtani-Kaneko, R.; Hara, M.; Suzuki, T.; Reiter, R.J. Identification of melatonin in plants and its effects on plasma melatonin levels and binding to melatonin receptors in vertebrates. Biochem. Mol. Biol. Int. 1995, 35, 627–634. [Google Scholar] [PubMed]
- Voisin, P.; Namboodiri, M.A.; Klein, D.C. Arylamine N-acetyltransferase and arylalkylamine N-acetyltransferase in the mammalian pineal gland. J. Biol. Chem. 1984, 259, 10913–10918. [Google Scholar] [CrossRef]
- Liu, G.; Hu, Q.; Zhang, X.; Jiang, J.; Zhang, Y.; Zhang, Z. Melatonin biosynthesis and signal transduction in plants in response to environmental conditions. J. Exp. Bot. 2022, 73, 5818–5827. [Google Scholar] [CrossRef]
- Ferry, G.; Loynel, A.; Kucharczyk, N.; Bertin, S.; Rodriguez, M.; Delagrange, P.; Galizzi, J.-P.; Jacoby, E.; Volland, J.-P.; Lesieur, D.; et al. Substrate specificity and inhibition studies of human serotonin N-acetyltransferase. J. Biol. Chem. 2000, 275, 8794–8805. [Google Scholar] [CrossRef]
- Ganguly, S.; Mummaneni, P.; Steinbach, P.J.; Klein, D.C.; Coon, S.L. Characterization of the Saccharomyces cerevisiae homolog of the melatonin rhythm enzyme arylalkylamine N-acetyltransferase (EC 2.3.1.87). J. Biol. Chem. 2001, 276, 47239–47247. [Google Scholar] [CrossRef]
- Hintermann, E.; Grieder, N.C.; Amherd, R.; Brodbeck, D.; Meyer, U.A. Cloning of an arylalkylamine N-acetyltransferase (aaNAT1) from Drosophila melanogaster expressed in the nervous system and the gut. Proc. Natl. Acad. Sci. USA 1996, 93, 12315–12320. [Google Scholar] [CrossRef]
- Hwang, O.J.; Back, K. Functional characterization of arylalkylamine N-acetyltransferase, a pivotal gene in antioxidant melatonin biosynthesis from Chlamydomonas reinhardtii. Antioxidants 2022, 11, 1531. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, Y.; Laborda, P.; Yang, Y.; Liu, F. Molecular cloning and characterization of a serotonin N-acetyltransferase gene, xoSNAT3, from Xanthomonas oryzae pv. oryzae. Int. J. Environ. Res. Public Health 2023, 20, 1865. [Google Scholar] [CrossRef]
- Yang, X.; Chen, J.; Ma, Y.; Huang, M.; Qiu, T.; Bian, H.; Han, N.; Wang, J. Function, mechanism, and application of plant melatonin: An update with a focus on the cereal crop, barley (Hordeum vulgare L.). Antioxidants 2022, 11, 634. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.; Lee, H.Y.; Lee, K.; Park, S.; Back, K. Cellular localization and kinetics of the rice melatonin biosynthetic enzymes SNAT and ASMT. J. Pineal Res. 2014, 56, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Byeon, Y.; Lee, K.; Lee, H.J.; Back, K. Cloning of Arabidopsis serotonin N-acetyltransferase and its role with caffeic acid O-methyltransferase in the biosynthesis of melatonin in vitro despite their different subcellular localization. J. Pineal Res. 2014, 57, 418–426. [Google Scholar] [CrossRef]
- Lee, H.Y.; Hwang, O.J.; Back, K. Functional characterization of tobacco (Nicotiana benthamiana) serotonin N-acetyltransferases (NbSNAT1 and NbSNAT2). Melatonin Res. 2021, 4, 507–521. [Google Scholar]
- Wang, L.; Feng, C.; Zheng, X.; Guo, Y.; Zhou, F.; Shan, D.; Liu, X.; Kong, J. Plant mitochondria synthesize melatonin and enhance the tolerance of plants to drought stress. J. Pineal Res. 2017, 63, e12429. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, H.Y.; Choi, D.W.; Back, K. Chloroplast encoded serotonin N-acetyltransferase in the red alga Pyropia yezoensis: Gene transition to the nucleus from chloroplasts. J. Exp. Bot. 2015, 66, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Arnesen, T.; Anderson, D.; Torsvik, J.; Halseth, H.B.; Varhaug, J.E.; Lillehaug, J.R. Cloning and characterization of hNAT5/hSAN: An evolutionarily conserved component of the NatA protein N-α-acetyltransferase complex. Gene 2006, 371, 291–295. [Google Scholar] [CrossRef]
- Ma, C.; Pathak, C.; Jang, S.; Lee, S.J.; Nam, M.; Kim, S.J.; Im, H.; Lee, B.J. Structure of Thermoplasma volcanium Ard1 belongs to N-acetyltransferase family member suggesting multiple ligand binding modes with acetyl coenzyme A and coenzyme A. Biochim. Biophys. Acta 2014, 1844, 1790–1797. [Google Scholar] [CrossRef]
- Linster, E.; Wirtz, M. N-terminal acetylation: An essential protein modification emerges as an important regulator of stress responses. J. Exp. Bot. 2018, 69, 4555–4568. [Google Scholar] [CrossRef]
- Starheim, K.K.; Gevaert, K.; Arnesen, T. Protein N-terminal acetyltransferases: When the start matters. Trends Biochem. Sci. 2012, 37, 152–161. [Google Scholar] [CrossRef]
- Pan, Q.; Zhao, F.-L.; Ye, B.-C. Eis, a novel family of arylalkylamine N-acetyltransferase (EC 2.3.1.87). Sci. Rep. 2018, 8, 2435. [Google Scholar] [CrossRef] [PubMed]
- Koskela, M.M.; Brünje, A.; Ivanauskaite, A.; Grabsztunowicz, M.; Lassowskat, I.; Neumann, U.; Dinh, T.V.; Sindlinger, J.; Schwarzer, D.; Wirtz, M.; et al. Chloroplast acetyltransferase NSI is required for state transition in Arabidopsis thaliana. Plant Cell 2018, 30, 1695–1709. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.C.; Garrett-Engele, C.M.; Li, Z.; Williams, E.V.; Rosenman, E.D.; Goldberg, M.L. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr. Biol. 2003, 13, 2025–2036. [Google Scholar] [CrossRef] [PubMed]
- Hou, F.; Chu, C.W.; Kong, X.; Yokomori, K.; Zou, H. The acetylase activity of San stabilizes the mitotic cohesin at the centromeres in a shugoshin-independent manner. J. Cell Biol. 2007, 177, 587–597. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, X.; Li, Y.; Kim, B.J.; Jia, J.; Huang, Z.; Yang, T.; Fu, X.; Jung, S.Y.; Wang, Y.; et al. Acetylation of Smc3 by EcoI is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell 2008, 31, 143–151. [Google Scholar] [CrossRef]
- Vetting, M.W.; Bareich, D.C.; Yu, M.; Blanchard, J.S. Crystal structure of RimI from Salmonella typhimurium LT2, the GNAT responsible for Nα-acetylation of ribosomal protein S18. Protein Sci. 2008, 17, 1781–1790. [Google Scholar] [CrossRef]
- Vetting, M.W.; Carvalho, L.P.; Yu, M.; Hegde, S.S.; Magnet, S.; Roderick, S.L.; Blanchard, J.S. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2005, 433, 212–226. [Google Scholar] [CrossRef]
- Florido, J.; Rodriguez-Santana, C.; Martinez-Ruiz, L.; López-Rodríguez, A.; Acuña-Castroviejo, D.; Rusanova, I.; Escames, G. Understanding the mechanism of action of melatonin, which induces ROS production in cancer cells. Antioxidants 2022, 11, 1621. [Google Scholar] [CrossRef]
- Sun, C.; Liu, L.; Wang, L.; Li, B.; Jin, C.; Lin, X. Melatonin: A master regulator of plant development and stress responses. J. Integr. Plant Biol. 2021, 63, 126–145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, Y.; Ma, X.; Ouyang, Z.; Deng, L.; Shen, S.; Dong, X.; Du, N.; Dong, H.; Guo, Z.; et al. Melatonin alleviates copper toxicity via improving ROS metabolism and antioxidant defense response in tomato seedlings. Antioxidants 2022, 11, 758. [Google Scholar] [CrossRef] [PubMed]
- Ramos, E.; Gil-Martín, E.; Ríos, C.D.L.; Egea, J.; López-Muñoz, F.; Pita, R.; Juberías, A.; Torrado, J.J.; Serrano, D.R.; Reiter, R.J.; et al. Melatonin as modulator for sulfur and nitrogen mustard-induced inflammation, oxidative stress and DNA damage: Molecular therapeutics. Antioxidants 2023, 12, 397. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).