MiRNA-Based Therapies for Lung Cancer: Opportunities and Challenges?
Abstract
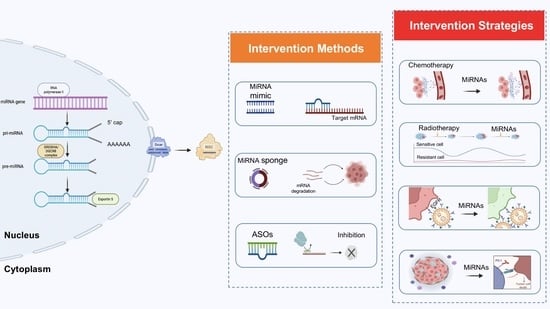
1. Introduction
2. MiRNA-Based Lung Cancer Therapies
3. MiRNAs as Complementary Therapies
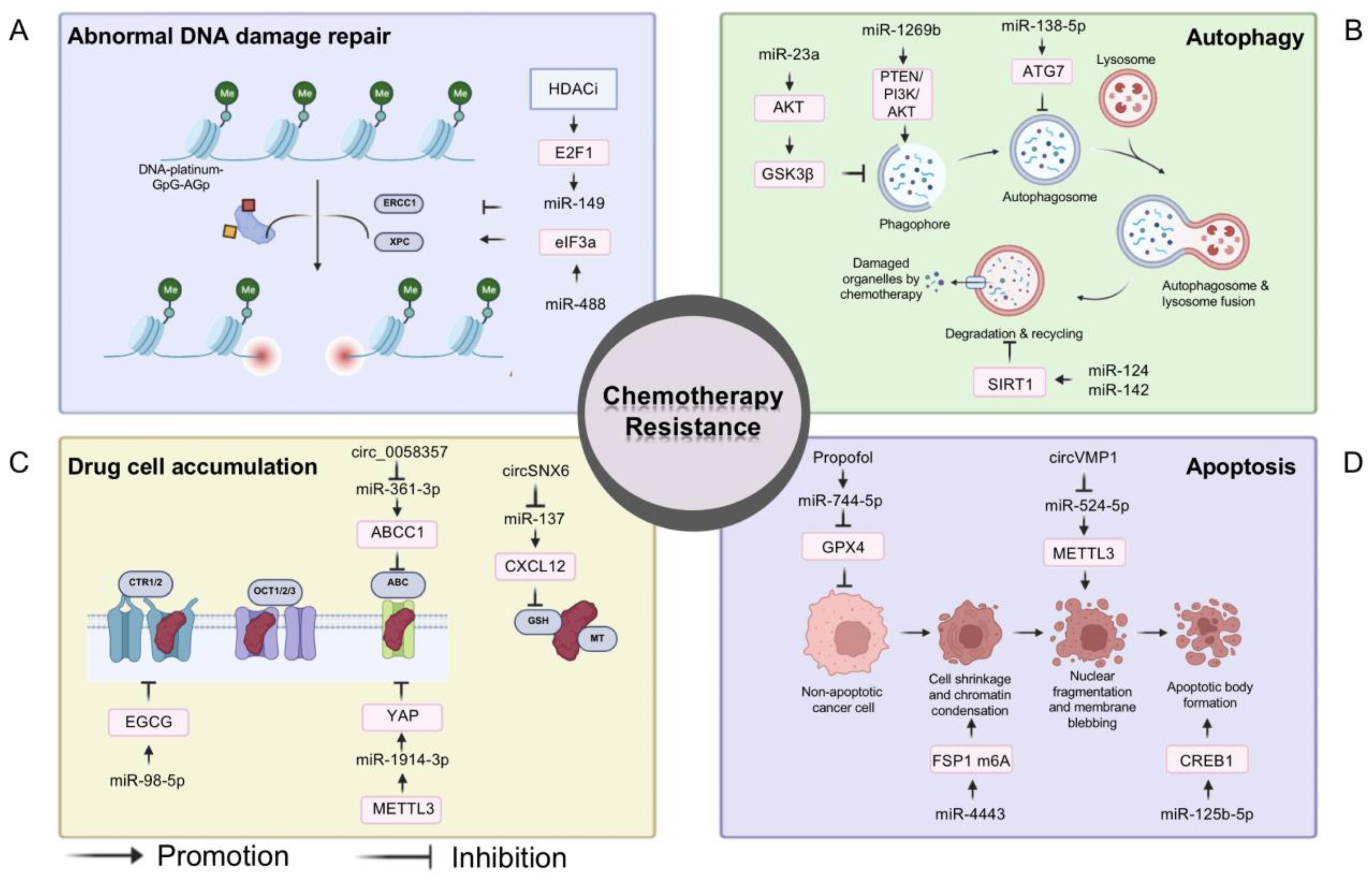
3.1. MiRNAs and Chemotherapy
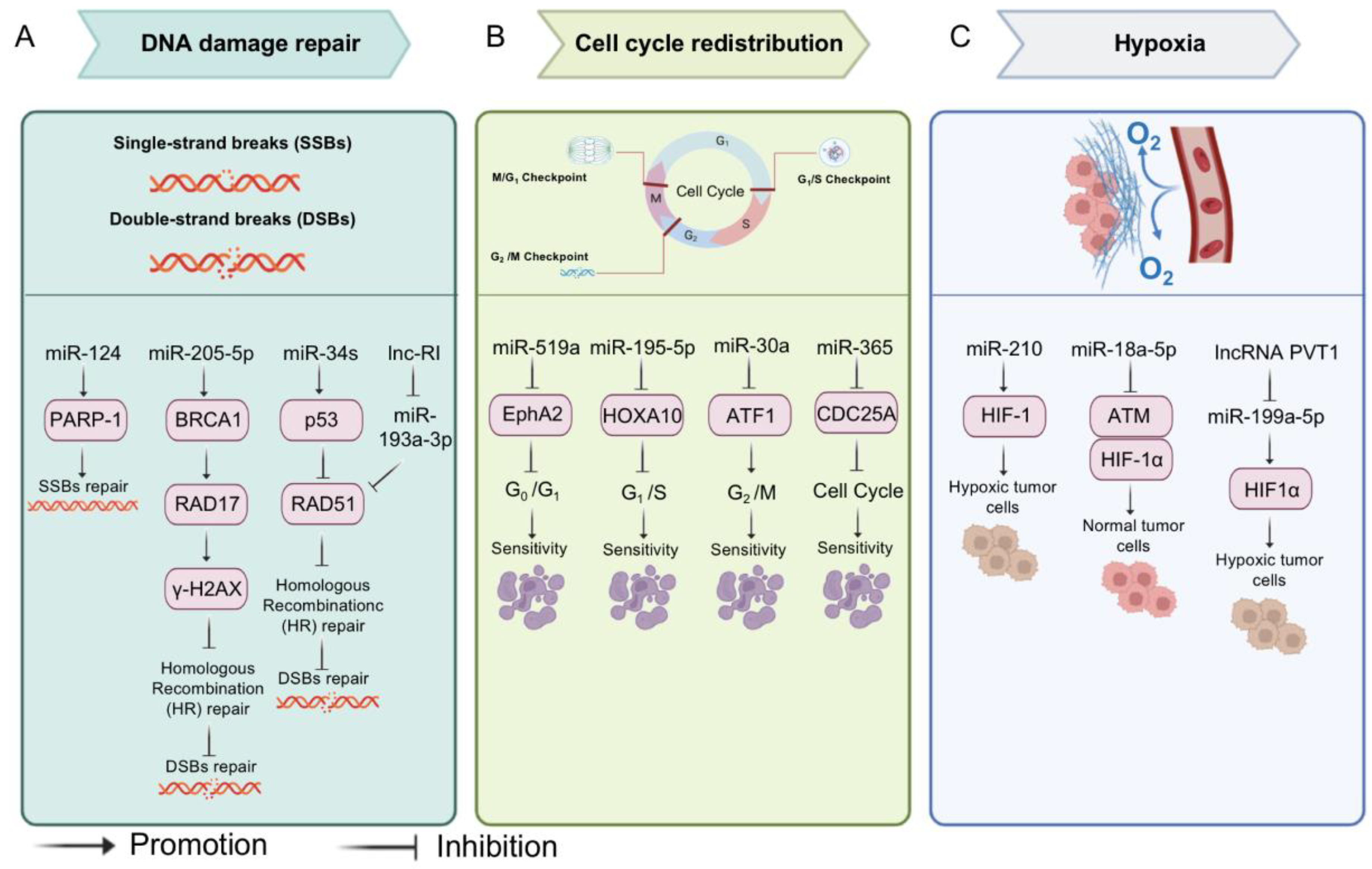
3.2. MiRNAs and Radiotherapy
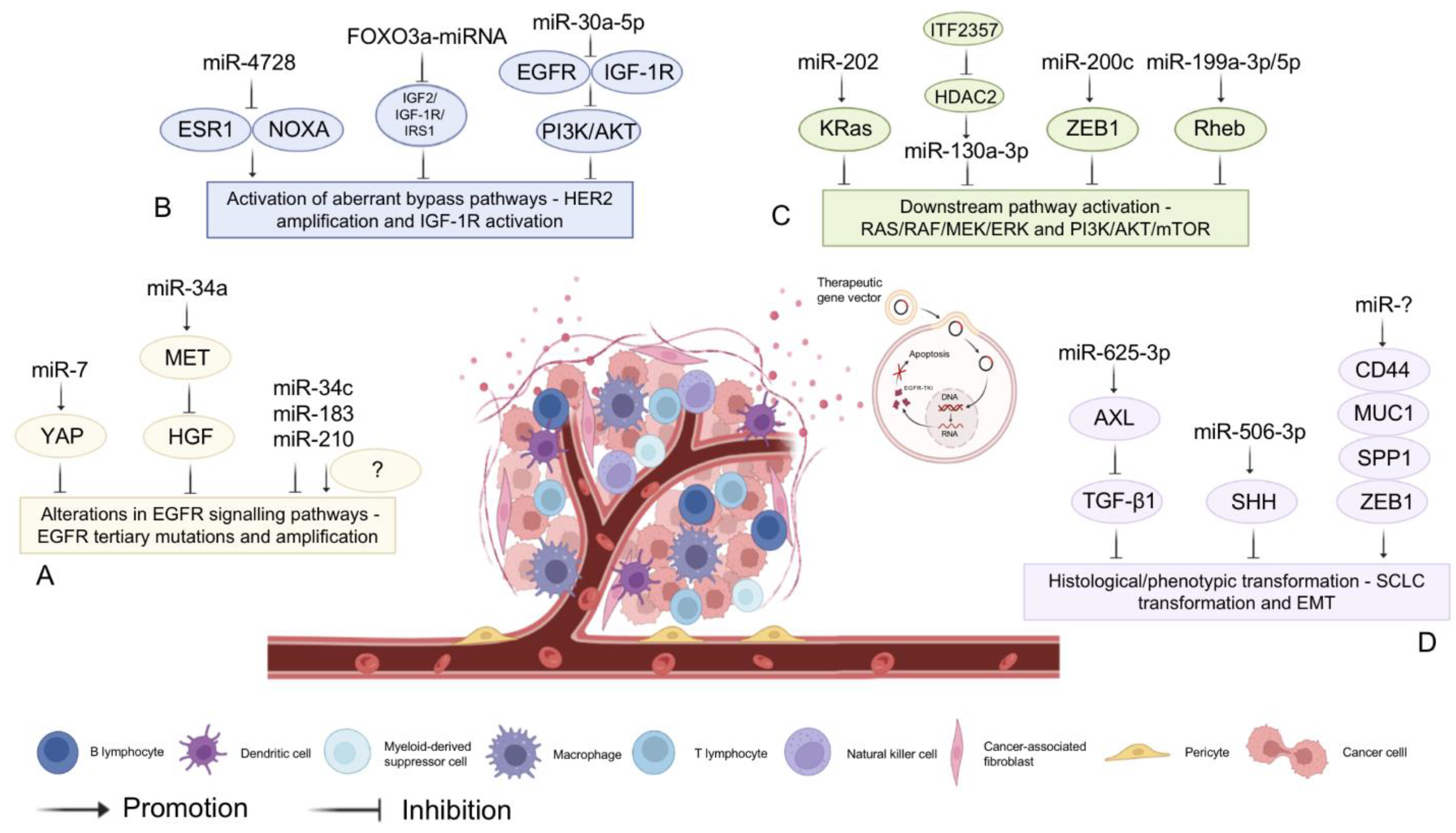
3.3. MiRNA-Assisted Targeted Therapy
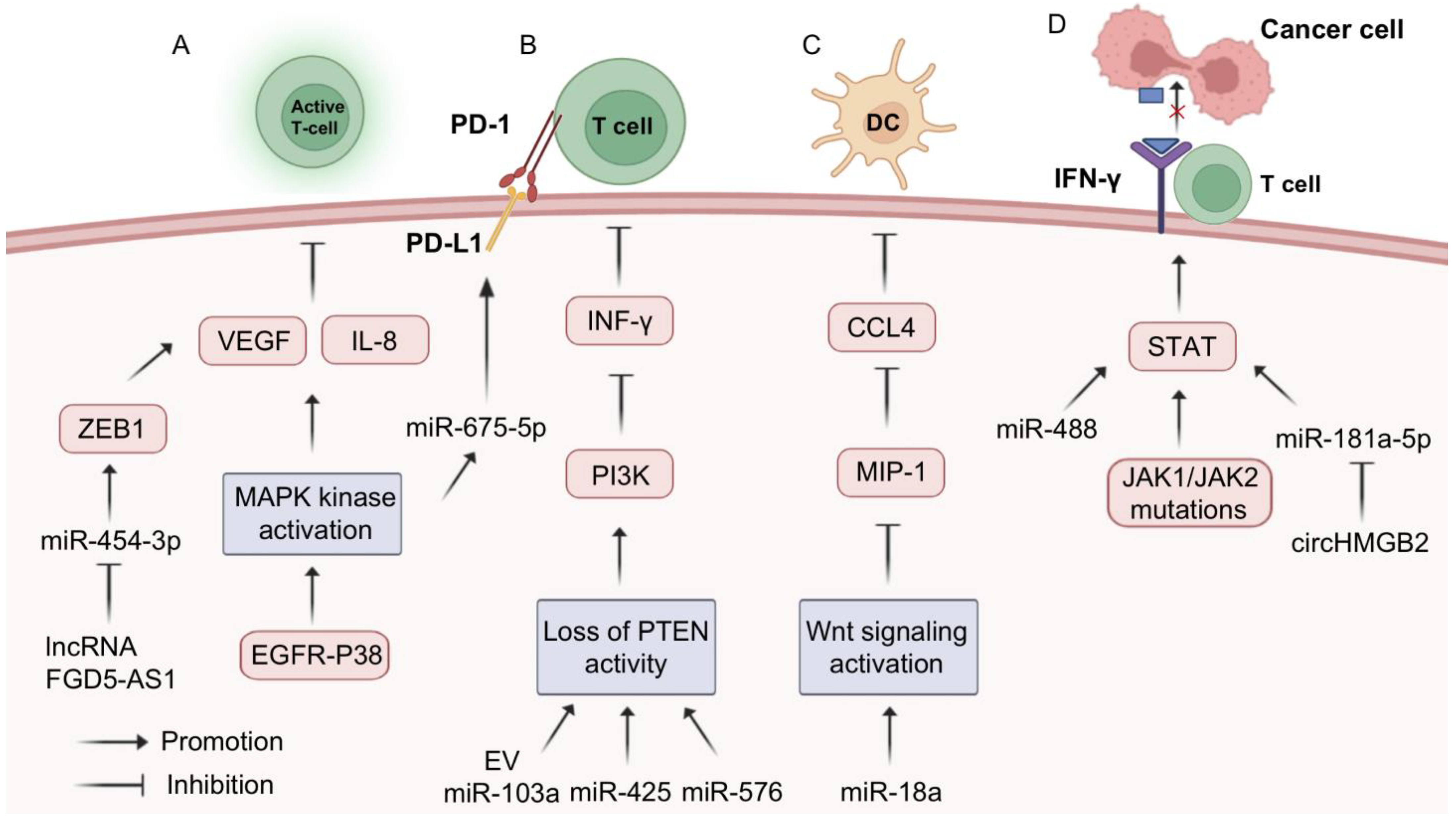
3.4. MiRNA-Assisted Immunotherapy
4. Clinical Trials Based on miRNAs
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Howlader, N.; Forjaz, G.; Mooradian, M.J.; Meza, R.; Kong, C.Y.; Cronin, K.A.; Mariotto, A.B.; Lowy, D.R.; Feuer, E.J. The Effect of Advances in Lung-Cancer Treatment on Population Mortality. N. Engl. J. Med. 2020, 383, 640–649. [Google Scholar] [CrossRef]
- Ganti, A.K.; Klein, A.B.; Cotarla, I.; Seal, B.; Chou, E. Update of Incidence, Prevalence, Survival, and Initial Treatment in Patients With Non-Small Cell Lung Cancer in the US. JAMA Oncol. 2021, 7, 1824–1832. [Google Scholar] [CrossRef]
- Bagcchi, S. Lung cancer survival only increases by a small amount despite recent treatment advances. Lancet Respir. Med. 2017, 5, 169. [Google Scholar] [CrossRef]
- Huang, Y.-A.; Hu, P.; Chan, K.C.C.; You, Z.-H. Graph convolution for predicting associations between miRNA and drug resistance. Bioinformatics 2020, 36, 851–858. [Google Scholar] [CrossRef]
- Boumahdi, S.; de Sauvage, F.J. The great escape: Tumour cell plasticity in resistance to targeted therapy. Nat. Reviews. Drug Discov. 2020, 19, 39–56. [Google Scholar] [CrossRef]
- Bagchi, S.; Yuan, R.; Engleman, E.G. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu. Rev. Pathol. Mech. Dis. 2021, 16, 223–249. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Takamizawa, J.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y.; et al. Reduced Expression of the let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Trang, P.; Wiggins, J.F.; Patrawala, L.; Cheng, A.; Ford, L.; Weidhaas, J.B.; Brown, D.; Bader, A.G.; Slack, F.J. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 2008, 7, 759–764. [Google Scholar] [CrossRef]
- Salim, H.; Akbar, N.S.; Zong, D.; Vaculova, A.H.; Lewensohn, R.; Moshfegh, A.; Viktorsson, K.; Zhivotovsky, B. miRNA-214 modulates radiotherapy response of non-small cell lung cancer cells through regulation of p38MAPK, apoptosis and senescence. Br. J. Cancer 2012, 107, 1361–1373. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Amri, J.; Molaee, N.; Karami, H.; Jamal, J. Up-Regulation of MiRNA-125a-5p Inhibits Cell Proliferation and Increases EGFR-TKI Induced Apoptosis in Lung Cancer Cells. Asian Pac. J. Cancer Prev. 2019, 20, 3361–3367. [Google Scholar] [CrossRef]
- Liu, W.; Liu, R.; Yuan, R.; Wang, X. MicroRNA-4458 Regulates PD-L1 Expression to Enhance Anti-tumor Immunity in NSCLC via Targeting STAT3. Mol. Biotechnol. 2021, 63, 1268–1279. [Google Scholar] [CrossRef]
- Iqbal, M.A.; Arora, S.; Prakasam, G.; Calin, G.A.; Syed, M.A. MicroRNA in lung cancer: Role, mechanisms, pathways and therapeutic relevance. Mol. Asp. Med. 2019, 70, 3–20. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Norouzi, M.; Yasamineh, S.; Montazeri, M.; Dadashpour, M.; Sheervalilou, R.; Abasi, M.; Pilehvar-Soltanahmadi, Y. Recent advances on nanomaterials-based fluorimetric approaches for microRNAs detection. Mater. Sci. Eng. C 2019, 104, 110007. [Google Scholar] [CrossRef]
- Liu, A.; Qu, H.; Gong, W.; Xiang, J.; Yang, M.; Zhang, W. LncRNA AWPPH and miRNA-21 regulates cancer cell proliferation and chemosensitivity in triple-negative breast cancer by interacting with each other. J. Cell Biochem. 2019, 120, 14860–14866. [Google Scholar] [CrossRef]
- Ji, Y.; Hocker, J.D.; Gattinoni, L. Enhancing adoptive T cell immunotherapy with microRNA therapeutics. Semin. Immunol. 2016, 28, 45–53. [Google Scholar] [CrossRef]
- Ricciuti, B.; Mecca, C.; Cenci, M.; Leonardi, G.C.; Perrone, L.; Mencaroni, C.; Crinò, L.; Grignani, F.; Baglivo, S.; Chiari, R.; et al. miRNAs and resistance to EGFR-TKIs in EGFR-mutant non-small cell lung cancer: Beyond ‘traditional mechanisms’ of resistance. Ecancermedicalscience 2015, 9, 569. [Google Scholar] [CrossRef]
- Mollaei, H.; Safaralizadeh, R.; Rostami, Z. MicroRNA replacement therapy in cancer. J. Cell Physiol. 2019, 234, 12369–12384. [Google Scholar] [CrossRef]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, J.; Chen, S.; Li, D.; Yang, J.; Zhao, X.; Qin, M.; Guo, M.; Chen, C.; He, Z.; et al. Let-7 as a Promising Target in Aging and Aging-Related Diseases: A Promise or a Pledge. Biomolecules 2022, 12, 1070. [Google Scholar] [CrossRef]
- He, X.; Duan, C.; Chen, J.; Ou-Yang, X.; Zhang, Z.; Li, C.; Peng, H. Let-7a elevates p21WAF1 levels by targeting of NIRF and suppresses the growth of A549 lung cancer cells. FEBS Lett. 2009, 583, 3501–3507. [Google Scholar] [CrossRef]
- He, X.-Y.; Chen, J.-X.; Zhang, Z.; Li, C.-L.; Peng, Q.; Peng, H.-M. The let-7a microRNA protects from growth of lung carcinoma by suppression of k-Ras and c-Myc in nude mice. J. Cancer Res. Clin. Oncol. 2010, 136, 1023–1028. [Google Scholar] [CrossRef]
- Li, Y.; Dong, R.; Lu, M.; Cheng, C.; Feng, Z.; Zhao, R.; Liang, J.; Han, J.; Jiang, J.; Xu-Welliver, M.; et al. Let-7b-3p inhibits tumor growth and metastasis by targeting the BRF2-mediated MAPK/ERK pathway in human lung adenocarcinoma. Transl. Lung Cancer Res. 2021, 10, 1841–1856. [Google Scholar] [CrossRef]
- Zhao, B.; Han, H.; Chen, J.; Zhang, Z.; Li, S.; Fang, F.; Zheng, Q.; Ma, Y.; Zhang, J.; Wu, N.; et al. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014, 342, 43–51. [Google Scholar] [CrossRef]
- Kasinski, A.L.; Kelnar, K.; Stahlhut, C.; Orellana, E.; Zhao, J.; Shimer, E.; Dysart, S.; Chen, X.; Bader, A.G.; Slack, F.J. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene 2015, 34, 3547–3555. [Google Scholar] [CrossRef]
- Trang, P.; Medina, P.P.; Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Homer, R.; Brown, D.; Bader, A.G.; Weidhaas, J.B.; et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene 2010, 29, 1580–1587. [Google Scholar] [CrossRef]
- Barh, D.; Malhotra, R.; Ravi, B.; Sindhurani, P. Microrna Let-7: An Emerging Next-Generation Cancer Therapeutic. Curr. Oncol. 2010, 17, 70–80. [Google Scholar] [CrossRef]
- Tang, L.; Chen, H.-Y.; Hao, N.-B.; Tang, B.; Guo, H.; Yong, X.; Dong, H.; Yang, S.-M. microRNA inhibitors: Natural and artificial sequestration of microRNA. Cancer Lett. 2017, 407, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Entezari, M.; Taheriazam, A.; Orouei, S.; Fallah, S.; Sanaei, A.; Hejazi, E.S.; Kakavand, A.; Rezaei, S.; Heidari, H.; Behroozaghdam, M.; et al. LncRNA-miRNA axis in tumor progression and therapy response: An emphasis on molecular interactions and therapeutic interventions. Biomed. Pharmacother.Biomed. Pharmacother. 2022, 154, 113609. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Zhang, D.-H.; Wu, N.; Xiao, J.-H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Mao, C.; Ouyang, L.; Liu, Y.; Lai, W.; Liu, N.; Shi, Y.; Chen, L.; Xiao, D.; Yu, F.; et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019, 26, 2329–2343. [Google Scholar] [CrossRef]
- Khan, P.; Siddiqui, J.A.; Lakshmanan, I.; Ganti, A.K.; Salgia, R.; Jain, M.; Batra, S.K.; Nasser, M.W. RNA-based therapies: A cog in the wheel of lung cancer defense. Mol. Cancer. 2021, 20, 54. [Google Scholar] [CrossRef]
- Dhuri, K.; Bechtold, C.; Quijano, E.; Pham, H.; Gupta, A.; Vikram, A.; Bahal, R. Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development. J. Clin. Med. 2020, 9, 2004. [Google Scholar] [CrossRef]
- Liang, H.; Jiao, Z.; Rong, W.; Qu, S.; Liao, Z.; Sun, X.; Wei, Y.; Zhao, Q.; Wang, J.; Liu, Y.; et al. 3′-Terminal 2′-O-methylation of lung cancer miR-21-5p enhances its stability and association with Argonaute 2. Nucleic Acids Res. 2020, 48, 7027–7040. [Google Scholar] [CrossRef]
- Scagliotti, G.V.; Parikh, P.; Von Pawel, J.; Biesma, B.; Vansteenkiste, J.; Serwatowski, C.M.; Serwatowski, P.; Gatzemeier, U.; Digumarti, R.; Zukin, M.; et al. Phase III Study Comparing Cisplatin Plus Gemcitabine With Cisplatin Plus Pemetrexed in Chemotherapy-Naive Patients With Advanced-Stage Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2008, 26, 3543–3551. [Google Scholar] [CrossRef]
- Pignon, J.P.; Tribodet, H.; Scagliotti, G.V.; Douillard, J.Y.; Shepherd, F.A.; Stephens, R.J.; Dunant, A.; Torri, V.; Rosell, R.; Seymour, L.; et al. Lung adjuvant cisplatin evaluation: A pooled analysis by the LACE Collaborative Group. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2008, 26, 3552–3559. [Google Scholar] [CrossRef]
- Wang, M.; Herbst, R.S.; Boshoff, C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat. Med. 2021, 27, 1345–1356. [Google Scholar] [CrossRef]
- Alexander, M.; Kim, S.Y.; Cheng, H. Update 2020: Management of Non-Small Cell Lung Cancer. Lung 2020, 198, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.; Camidge, D.; Gadgeel, S.; Rosell, R.; Dziadziuszko, R.; Kim, D.-W.; Pérol, M.; Ou, S.-H.; Ahn, J.; Shaw, A.; et al. Updated overall survival and final progression-free survival data for patients with treatment-naive advanced ALK-positive non-small-cell lung cancer in the ALEX study. Ann. Oncol. 2020, 31, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Landre, T.; Justeau, G.; Assié, J.-B.; Chouahnia, K.; Davoine, C.; Taleb, C.; Chouaïd, C.; Duchemann, B. Anti-PD-(L)1 for KRAS-mutant advanced non-small–cell lung cancers: A meta-analysis of randomized–controlled trials. Cancer Immunol. Immunother. 2022, 71, 719–726. [Google Scholar] [CrossRef]
- Pearl, L.H.; Schierz, A.C.; Ward, S.E.; Al-Lazikani, B.; Pearl, F.M. Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer 2015, 15, 166–180. [Google Scholar] [CrossRef]
- Kubisch, J.; Türei, D.; Földvári-Nagy, L.; Dunai, Z.A.; Zsákai, L.; Varga, M.; Vellai, T.; Csermely, P.; Korcsmáros, T. Complex regulation of autophagy in cancer–Integrated approaches to discover the networks that hold a double-edged sword. Semin. Cancer Biol. 2013, 23, 252–261. [Google Scholar] [CrossRef]
- Carneiro, B.A.; El-Deiry, W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, X.; Jin, S.; Chen, Y.; Guo, R. Ferroptosis in cancer therapy: A novel approach to reversing drug resistance. Mol. Cancer 2022, 21, 47. [Google Scholar] [CrossRef]
- He, Y.; Chen, D.; Yi, Y.; Zeng, S.; Liu, S.; Li, P.; Xie, H.; Yu, P.; Jiang, G.; Liu, H. Histone Deacetylase Inhibitor Sensitizes ERCC1-High Non-small-Cell Lung Cancer Cells to Cisplatin via Regulating miR-149. Mol. Ther. Oncolytics 2020, 17, 448–459. [Google Scholar] [CrossRef]
- Fang, C.; Chen, Y.-X.; Wu, N.-Y.; Yin, J.-Y.; Li, X.-P.; Huang, H.-S.; Zhang, W.; Zhou, H.-H.; Liu, Z.-Q. MiR-488 inhibits proliferation and cisplatin sensibility in non-small-cell lung cancer (NSCLC) cells by activating the eIF3a-mediated NER signaling pathway. Sci. Rep. 2017, 7, 40384. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J.; Hui, B.; Sun, W.; Li, B.; Shi, F.; Che, S.; Chai, L.; Song, L. Pristimerin enhances the effect of cisplatin by inhibiting the miR-23a/Akt/GSK3β signaling pathway and suppressing autophagy in lung cancer cells. Int. J. Mol. Med. 2019, 43, 1382–1394. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Chen, Y.; Shen, Y.; Tantai, J. Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis. 2019, 10, 429. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Kong, F.; Zong, Z.; Ren, M.; Meng, Q.; Li, Y.; Sun, Z. miR-124 and miR-142 enhance cisplatin sensitivity of non-small cell lung cancer cells through repressing autophagy via directly targeting SIRT1. RSC Adv. 2019, 9, 5234–5243. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Xiao, W.; Cai, Z.; Jin, S.; Li, T. miR-1269b Drives Cisplatin Resistance of Human Non-Small Cell Lung Cancer via Modulating the PTEN/PI3K/AKT Signaling Pathway. OncoTargets Ther. 2020, 13, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Wu, X.; Wang, X.; Huang, W.; Feng, Q. NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget 2016, 7, 43337–43351. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Li, P.; Li, Y.; Shi, J.; Huang, S.; Jiao, P. Identification of circ_0058357 as a regulator in non-small cell lung cancer cells resistant to cisplatin by miR -361-3p/ABCC1 axis. Thorac. Cancer 2021, 12, 2894–2906. [Google Scholar] [CrossRef]
- Jin, D.; Guo, J.; Wu, Y.; Du, J.; Yang, L.; Wang, X.; Di, W.; Hu, B.; An, J.; Kong, L.; et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J. Hematol. Oncol. 2021, 14, 135. [Google Scholar] [CrossRef]
- Zhu, K.; Zhu, J.; Geng, J.; Zhang, Y.; Qin, Y.; Wang, F.; Weng, Y. circSNX6 (hsa_circ_0031608) enhances drug resistance of non-small cell lung cancer (NSCLC) via miR-137. Biochem. Biophys. Res. Commun. 2021, 567, 79–85. [Google Scholar] [CrossRef]
- Xie, H.; Yao, J.; Wang, Y.; Ni, B. Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Deliv. 2022, 29, 1257–1271. [Google Scholar] [CrossRef]
- Tan, Q.; Ma, J.; Zhang, H.; Wu, X.; Li, Q.; Zuo, X.; Jiang, Y.; Liu, H.; Yan, L. miR-125b-5p upregulation by TRIM28 induces cisplatin resistance in non-small cell lung cancer through CREB1 inhibition. BMC Pulm. Med. 2022, 22, 469. [Google Scholar] [CrossRef]
- Han, B.; Liu, Y.; Zhang, Q.; Liang, L. Propofol decreases cisplatin resistance of non-small cell lung cancer by inducing GPX4-mediated ferroptosis through the miR-744-5p/miR-615-3p axis. J. Proteom. 2023, 274, 104777. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Jia, G.; Ma, P.; Cang, S. Exosomal miR-4443 promotes cisplatin resistance in non-small cell lung carcinoma by regulating FSP1 m6A modification-mediated ferroptosis. Life Sci. 2021, 276, 119399. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Deng, T.; Liu, R.; Ning, T.; Yang, H.; Liu, D.; Zhang, Q.; Lin, D.; Ge, S.; Bai, M.; et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol. Cancer 2020, 19, 43. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, M.; Cui, X.; O’connell, D.; Yang, Y. Long noncoding RNA NEAT1 promotes ferroptosis by modulating the miR-362-3p/MIOX axis as a ceRNA. Cell Death Differ. 2022, 29, 1850–1863. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Song, W.; Gao, Y.; Dong, X.; Ji, X. Increased PD-L1 Expression in Acquired Cisplatin-Resistant Lung Cancer Cells via Mir-181a. Tohoku J. Exp. Med. 2022, 257, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Ba, Z.; Zhou, Y.; Yang, Z.; Xu, J.; Zhang, X. miR-324-5p upregulation potentiates resistance to cisplatin by targeting FBXO11 signalling in non-small cell lung cancer cells. J. Biochem. 2019, 166, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Y.; Li, Q.; Xu, R.; Huang, N. MiR-200c-3p and miR-485-5p overexpression elevates cisplatin sensitivity and suppresses the malignant phenotypes of non-small cell lung cancer cells through targeting RRM2. Thorac. Cancer 2022, 13, 1974–1985. [Google Scholar] [CrossRef]
- Chen, K.-B.; Yang, W.; Xuan, Y.; Lin, A.-J. miR-526b-3p inhibits lung cancer cisplatin-resistance and metastasis by inhibiting STAT3-promoted PD-L1. Cell Death Dis. 2021, 12, 748. [Google Scholar] [CrossRef]
- Qiu, H.; Shen, X.; Chen, B.; Chen, T.; Feng, G.; Chen, S.; Feng, D.; Xu, Q. miR-30b-5p inhibits cancer progression and enhances cisplatin sensitivity in lung cancer through targeting LRP8. Apoptosis 2021, 26, 261–276. [Google Scholar] [CrossRef]
- Wu, H.; Mu, X.; Liu, L.; Wu, H.; Hu, X.; Chen, L.; Liu, J.; Mu, Y.; Yuan, F.; Liu, W.; et al. Bone marrow mesenchymal stem cells-derived exosomal microRNA-193a reduces cisplatin resistance of non-small cell lung cancer cells via targeting LRRC1. Cell Death Dis. 2020, 11, 801. [Google Scholar] [CrossRef]
- Vinod, S.K.; Hau, E. Radiotherapy treatment for lung cancer: Current status and future directions. Respirology 2020, 25, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Bruno, D.; Grubb, W.; Biswas, T. The changing landscape of stage III lung cancer: A literature review. Expert Rev. Anticancer. Ther. 2020, 20, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, J.N.; Redmond, K.M.; Schettino, G.; Prise, K.M. DNA Double Strand Break Repair: A Radiation Perspective. Antioxid. Redox Signal. 2013, 18, 2458–2472. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, T.M.; Keyomarsi, K. Role of cell cycle in mediating sensitivity to radiotherapy. Int. J. Radiat. Oncol. 2004, 59, 928–942. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, R.; Wang, Q.; Qi, Z.; Hu, Y.; Zhou, P.; Wang, Z. MiR-34s negatively regulate homologous recombination through targeting RAD51. Arch. Biochem. Biophys. 2019, 666, 73–82. [Google Scholar] [CrossRef]
- Chen, S.-M.; Chou, W.-C.; Hu, L.-Y.; Hsiung, C.-N.; Chu, H.-W.; Huang, Y.-L.; Hsu, H.-M.; Yu, J.-C.; Shen, C.-Y. The Effect of MicroRNA-124 Overexpression on Anti-Tumor Drug Sensitivity. PLoS ONE 2015, 10, e0128472. [Google Scholar] [CrossRef]
- Valenti, F.; Sacconi, A.; Ganci, F.; Grasso, G.; Strano, S.; Blandino, G.; Di Agostino, S. The miR-205-5p/BRCA1/RAD17 Axis Promotes Genomic Instability in Head and Neck Squamous Cell Carcinomas. Cancers 2019, 11, 1347. [Google Scholar] [CrossRef]
- Shen, L.; Wang, Q.; Liu, R.; Chen, Z.; Zhang, X.; Zhou, P.-K.; Wang, Z. LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res. 2018, 46, 717–729. [Google Scholar] [CrossRef]
- Gong, S.; Li, Y.; Lv, L.; Men, W. Restored microRNA-519a enhances the radiosensitivity of non-small cell lung cancer via suppressing EphA2. Gene Ther. 2021, 29, 588–600. [Google Scholar] [CrossRef]
- Yuan, C.; Bai, R.; Gao, Y.; Jiang, X.; Li, S.; Sun, W.; Li, Y.; Huang, Z.; Gong, Y.; Xie, C. Effects of MicroRNA-195-5p on Biological Behaviors and Radiosensitivity of Lung Adenocarcinoma Cells via Targeting HOXA10. Oxidative Med. Cell Longev. 2022, 2021, 4522210. [Google Scholar] [CrossRef]
- Guo, Y.; Sun, W.; Gong, T.; Chai, Y.; Wang, J.; Hui, B.; Li, Y.; Song, L.; Gao, Y. miR-30a radiosensitizes non-small cell lung cancer by targeting ATF1 that is involved in the phosphorylation of ATM. Oncol. Rep. 2017, 37, 1980–1988. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jiang, M.; Cui, M.; Feng, G.; Dong, J.; Li, Y.; Xiao, H.; Fan, S. MiR-365 enhances the radiosensitivity of non-small cell lung cancer cells through targeting CDC25A. Biochem. Biophys. Res. Commun. 2019, 512, 392–398. [Google Scholar] [CrossRef]
- Mudassar, F.; Shen, H.; O’neill, G.; Hau, E. Targeting tumor hypoxia and mitochondrial metabolism with anti-parasitic drugs to improve radiation response in high-grade gliomas. J. Exp. Clin. Cancer Res. 2020, 39, 208. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wu, L.; Li, D.; Xu, Y.; Zhang, L.; Niu, K.; Kong, R.; Gu, J.; Xu, Z.; Chen, Z.; et al. Radiosensitizing effects of miR-18a-5p on lung cancer stem-like cells via downregulating both ATM and HIF-1α. Cancer Med. 2018, 7, 3834–3847. [Google Scholar] [CrossRef] [PubMed]
- Grosso, S.; Doyen, J.; Parks, S.K.; Bertero, T.; Paye, A.; Cardinaud, B.; Gounon, P.; Lacas-Gervais, S.; Noël, A.; Pouysségur, J.; et al. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 2013, 4, e544. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Wang, H.; Zhu, Y.; Feng, H.; Wang, G.; Wang, S. miR-199a-5p is involved in doxorubicin resistance of non-small cell lung cancer (NSCLC) cells. Eur. J. Pharmacol. 2020, 878, 173105. [Google Scholar] [CrossRef]
- Wang, X.-C.; Du, L.-Q.; Tian, L.-L.; Wu, H.-L.; Jiang, X.-Y.; Zhang, H.; Li, D.-G.; Wang, Y.-Y.; Wu, H.-Y.; She, Y.; et al. Expression and function of miRNA in postoperative radiotherapy sensitive and resistant patients of non-small cell lung cancer. Lung Cancer 2011, 72, 92–99. [Google Scholar] [CrossRef]
- Zhang, F.; Sang, Y.; Chen, D.; Wu, X.; Wang, X.; Yang, W.; Chen, Y. M2 macrophage-derived exosomal long non-coding RNA AGAP2-AS1 enhances radiotherapy immunity in lung cancer by reducing microRNA-296 and elevating NOTCH2. Cell Death Dis. 2021, 12, 467. [Google Scholar] [CrossRef]
- Wei, T.; Cheng, S.; Na Fu, X.; Feng, L.J. miR-219a-5p enhances the radiosensitivity of non-small cell lung cancer cells through targeting CD164. Biosci. Rep. 2020, 40, BSR20192795. [Google Scholar] [CrossRef]
- Jiang, K.; Zou, H. microRNA-20b-5p overexpression combing Pembrolizumab potentiates cancer cells to radiation therapy via repressing programmed death-ligand 1. Bioengineered 2022, 13, 917–929. [Google Scholar] [CrossRef]
- Sun, Y.; Hawkins, P.G.; Bi, N.; Dess, R.T.; Tewari, M.; Hearn, J.W.; Hayman, J.A.; Kalemkerian, G.P.; Lawrence, T.S.; Haken, R.K.T.; et al. Serum MicroRNA Signature Predicts Response to High-Dose Radiation Therapy in Locally Advanced Non-Small Cell Lung Cancer. Int. J. Radiat. Oncol. 2017, 100, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Zwitter, M. Toxicity and quality of life in published clinical trials for advanced lung cancer. Support. Care Cancer Off. J. Multinatl. Assoc. Support. Care Cancer 2018, 26, 3453–3459. [Google Scholar] [CrossRef] [PubMed]
- Melosky, B.; Wheatley-Price, P.; Juergens, R.A.; Sacher, A.; Leighl, N.B.; Tsao, M.-S.; Cheema, P.; Snow, S.; Liu, G.; Card, P.B.; et al. The rapidly evolving landscape of novel targeted therapies in advanced non-small cell lung cancer. Lung Cancer 2021, 160, 136–151. [Google Scholar] [CrossRef]
- da Cunha Santos, G.; Shepherd, F.A.; Tsao, M.S. EGFR mutations and lung cancer. Annu. Rev. Pathol. 2011, 6, 49–69. [Google Scholar] [CrossRef] [PubMed]
- Golding, B.; Luu, A.; Jones, R.; Viloria-Petit, A.M. The function and therapeutic targeting of anaplastic lymphoma kinase (ALK) in non-small cell lung cancer (NSCLC). Mol. Cancer 2018, 17, 52. [Google Scholar] [CrossRef] [PubMed]
- Socinski, M.A.; Pennell, N.A.; Davies, K.D. MET Exon 14 Skipping Mutations in Non-Small-Cell Lung Cancer: An Overview of Biology, Clinical Outcomes, and Testing Considerations. JCO Precis. Oncol. 2021, 5, PO.20.00516. [Google Scholar]
- Reck, M.; Carbone, D.P.; Garassino, M.; Barlesi, F. Targeting KRAS in non-small-cell lung cancer: Recent progress and new approaches. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2021, 32, 1101–1110. [Google Scholar] [CrossRef]
- Nan, X.; Xie, C.; Yu, X.; Liu, J. EGFR TKI as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer. Oncotarget 2017, 8, 75712–75726. [Google Scholar] [CrossRef]
- Ercan, D.; Choi, H.G.; Yun, C.H.; Capelletti, M.; Xie, T.; Eck, M.J.; Gray, N.S.; Jänne, P.A. EGFR Mutations and Resistance to Irreversible Pyrimidine-Based EGFR Inhibitors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 3913–3923. [Google Scholar] [CrossRef]
- Le, X.; Puri, S.; Negrao, M.V.; Nilsson, M.B.; Robichaux, J.; Boyle, T.; Hicks, J.K.; Lovinger, K.L.; Roarty, E.; Rinsurongkawong, W.; et al. Landscape of EGFR-Dependent and -Independent Resistance Mechanisms to Osimertinib and Continuation Therapy Beyond Progression in EGFR-Mutant NSCLC. Clin. Cancer Res. 2018, 24, 6195–6203. [Google Scholar] [CrossRef]
- Chen, R.; Qian, Z.; Xu, X.; Zhang, C.; Niu, Y.; Wang, Z.; Sun, J.; Zhang, X.; Yu, Y. Exosomes-transmitted miR-7 reverses gefitinib resistance by targeting YAP in non-small-cell lung cancer. Pharmacol. Res. 2021, 165, 105442. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-Y.; Chen, X.; Zhao, J.; Bao, Z.; Chen, X.; Zhang, P.; Liu, Z.-F. MicroRNA-34a overcomes HGF-mediated gefitinib resistance in EGFR mutant lung cancer cells partly by targeting MET. Cancer Lett. 2014, 351, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Pak, M.G.; Lee, C.-H.; Lee, W.-J.; Shin, D.-H.; Roh, M.-S. Unique microRNAs in lung adenocarcinoma groups according to major TKI sensitive EGFR mutation status. Diagn. Pathol. 2015, 10, 99. [Google Scholar] [CrossRef]
- Chmielecki, J.; Mok, T.; Wu, Y.-L.; Han, J.-Y.; Ahn, M.-J.; Ramalingam, S.S.; John, T.; Okamoto, I.; Yang, J.C.-H.; Shepherd, F.A.; et al. Analysis of acquired resistance mechanisms to osimertinib in patients with EGFR-mutated advanced non-small cell lung cancer from the AURA3 trial. Nat. Commun. 2023, 14, 1071. [Google Scholar] [CrossRef]
- Manabe, T.; Yasuda, H.; Terai, H.; Kagiwada, H.; Hamamoto, J.; Ebisudani, T.; Kobayashi, K.; Masuzawa, K.; Ikemura, S.; Kawada, I.; et al. IGF2 Autocrine-Mediated IGF1R Activation Is a Clinically Relevant Mechanism of Osimertinib Resistance in Lung Cancer. Mol. Cancer Res. 2020, 18, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Floros, K.V.; Lochmann, T.L.; Hu, B.; Monterrubio, C.; Hughes, M.T.; Wells, J.D.; Morales, C.B.; Ghotra, M.S.; Costa, C.; Souers, A.J.; et al. Coamplification of miR-4728 protects HER2 -amplified breast cancers from targeted therapy. Proc. Natl. Acad. Sci. USA 2018, 115, E2594–E2603. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, Z.; Qiu, N.; Ling, L.; Jia, X.; Song, Y.; Li, H.; Li, J.; Lyu, H.; Liu, H.; et al. Disruption of FOXO3a-miRNA feedback inhibition of IGF2/IGF-1R/IRS1 signaling confers Herceptin resistance in HER2-positive breast cancer. Nat. Commun. 2021, 12, 2699. [Google Scholar] [CrossRef]
- Meng, F.; Wang, F.; Wang, L.; Wong, S.C.C.; Cho, W.C.S.; Chan, L.W.C. MiR-30a-5p Overexpression May Overcome EGFR-Inhibitor Resistance through Regulating PI3K/AKT Signaling Pathway in Non-small Cell Lung Cancer Cell Lines. Front. Genet. 2016, 7, 197. [Google Scholar] [CrossRef]
- Nakatani, K.; Yamaoka, T.; Ohba, M.; Fujita, K.-I.; Arata, S.; Kusumoto, S.; Taki-Takemoto, I.; Kamei, D.; Iwai, S.; Tsurutani, J.; et al. KRAS and EGFR Amplifications Mediate Resistance to Rociletinib and Osimertinib in Acquired Afatinib-Resistant NSCLC Harboring Exon 19 Deletion/T790M in EGFR. Mol. Cancer Ther. 2019, 18, 112–126. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Hu, Y.; Mileham, K.F.; Husain, H.; Costa, D.B.; Tracy, P.; Feeney, N.; Sholl, L.M.; Dahlberg, S.E.; Redig, A.J.; et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol. 2018, 4, 1527–1534. [Google Scholar] [CrossRef]
- Cui, J.; Xu, F.; Bai, W.; Zhao, T.; Hong, J.; Zuo, W. HDAC inhibitor ITF2357 reduces resistance of mutant-KRAS non-small cell lung cancer to pemetrexed through a HDAC2/miR-130a-3p-dependent mechanism. J. Transl. Med. 2023, 21, 125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, L.; Guo, H.; Zhang, Z.; Wang, C.; Chian, R.; Zhang, Z. MicroRNA-202 inhibits endometrial stromal cell migration and invasion by suppressing the K-Ras/Raf1/MEK/ERK signaling pathway. Int. J. Mol. Med. 2020, 46, 2078–2088. [Google Scholar] [CrossRef] [PubMed]
- Kharbanda, A.; Rajabi, H.; Jin, C.; Alam, M.; Wong, K.-K.; Kufe, D. MUC1-C confers EMT and KRAS independence in mutant KRAS lung cancer cells. Oncotarget 2014, 5, 8893–8905. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.; Chai, B.; Wu, Z.; Gu, Z.; Zou, H.; Zhang, H.; Li, Y.; Sun, Q.; Fang, W.; et al. miR-199a-3p/5p regulate tumorgenesis via targeting Rheb in non-small cell lung cancer. Int. J. Biol. Sci. 2022, 18, 4187–4202. [Google Scholar] [CrossRef]
- Marcoux, N.; Gettinger, S.N.; O’kane, G.; Arbour, K.C.; Neal, J.W.; Husain, H.; Evans, T.L.; Brahmer, J.R.; Muzikansky, A.; Bonomi, P.D.; et al. EGFR-Mutant Adenocarcinomas That Transform to Small-Cell Lung Cancer and Other Neuroendocrine Carcinomas: Clinical Outcomes. J. Clin. Oncol. 2019, 37, 278–285. [Google Scholar] [CrossRef]
- Nukaga, S.; Yasuda, H.; Tsuchihara, K.; Hamamoto, J.; Masuzawa, K.; Kawada, I.; Naoki, K.; Matsumoto, S.; Mimaki, S.; Ikemura, S.; et al. Amplification of EGFR Wild-Type Alleles in Non–Small Cell Lung Cancer Cells Confers Acquired Resistance to Mutation-Selective EGFR Tyrosine Kinase Inhibitors. Cancer Res 2017, 77, 2078–2089. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Sun, L.; Liu, T.; Zhu, J.; Zeng, Y.; Zhang, Y.; Wang, X.; Liu, Z.; Huang, J.A. The miR 625 3p/AXL axis induces non T790M acquired resistance to EGFR TKI via activation of the TGF β/Smad pathway and EMT in EGFR mutant non small cell lung cancer. Oncol. Rep. 2020, 44, 185–195. [Google Scholar] [CrossRef]
- Haque, I.; Kawsar, H.I.; Motes, H.; Sharma, M.; Banerjee, S.; Banerjee, S.K.; Godwin, A.K.; Huang, C.H. Downregulation of miR-506-3p Facilitates EGFR-TKI Resistance through Induction of Sonic Hedgehog Signaling in Non-Small-Cell Lung Cancer Cell Lines. Int. J. Mol. Sci. 2020, 21, 9307. [Google Scholar] [CrossRef]
- Zhang, W.C.; Skiados, N.; Aftab, F.; Moreno, C.; Silva, L.; Corbilla, P.J.A.; Asara, J.M.; Hata, A.N.; Slack, F.J. MicroRNA-21 guide and passenger strand regulation of adenylosuccinate lyase-mediated purine metabolism promotes transition to an EGFR-TKI-tolerant persister state. Cancer Gene Ther. 2022, 29, 1878–1894. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, B.; Wang, Y.; Zhao, Q.; Wu, W.; Zhang, P.; Miao, L.; Sun, S. NOTCH3 Overexpression and Posttranscriptional Regulation by miR-150 Were Associated With EGFR-TKI Resistance in Lung Adenocarcinoma. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2019, 27, 751–761. [Google Scholar] [CrossRef]
- Li, B.; Ren, S.; Li, X.; Wang, Y.; Garfield, D.; Zhou, S.; Chen, X.; Su, C.; Chen, M.; Kuang, P.; et al. MiR-21 overexpression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer 2014, 83, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Leonetti, A.; Capula, M.; Minari, R.; Mazzaschi, G.; Gregori, A.; El Hassouni, B.; Papini, F.; Bordi, P.; Verzè, M.; Avan, A.; et al. Dynamic Evaluation of Circulating miRNA Profile in EGFR-Mutated NSCLC Patients Treated with EGFR-TKIs. Cells 2021, 10, 1520. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Li, Y.; Zheng, Y.; Zhang, L.; Pan, Y.; Yu, J.; Yang, M. miR-608 and miR-4513 significantly contribute to the prognosis of lung adenocarcinoma treated with EGFR-TKIs. Lab. Investig. 2019, 99, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xue, C.; Zhu, X.; Zhu, X.; Xian, H.; Huang, Z. Suppression of microRNA-125a-5p upregulates the TAZ-EGFR signaling pathway and promotes retinoblastoma proliferation. Cell Signal. 2016, 28, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Shen, T.; Zhang, C.; Zhang, Q.-H.; Zhang, Z.-Q. MiR-125a-5p inhibits EMT of ovarian cancer cells by regulating TAZ/EGFR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8249–8256. [Google Scholar] [PubMed]
- Steven, A.; Fisher, S.A.; Robinson, B.W. Immunotherapy for lung cancer. Respirology 2016, 21, 821–833. [Google Scholar]
- Chikuma, S. CTLA-4, an Essential Immune-Checkpoint for T-Cell Activation. Curr. Top. Microbiol. Immunol. 2017, 410, 99–126. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, J.; Song, Q.; Sun, X.; Xue, M.; Yang, Z.; Shang, J. Comprehensive analysis of CTLA-4 in the tumor immune microenvironment of 33 cancer types. Int. Immunopharmacol. 2020, 85, 106633. [Google Scholar] [CrossRef]
- Jin, H.T.; Ahmed, R.; Okazaki, T. Role of PD-1 in regulating T-cell immunity. Curr. Top. Microbiol. Immunol. 2011, 350, 17–37. [Google Scholar]
- Kaushik, I.; Ramachandran, S.; Zabel, C.; Gaikwad, S.; Srivastava, S.K. The evolutionary legacy of immune checkpoint inhibitors. Semin. Cancer Biol. 86 Pt 2, 491–498. [CrossRef]
- Xiao, X.; Huang, S.; Chen, S.; Wang, Y.; Sun, Q.; Xu, X.; Li, Y. Mechanisms of cytokine release syndrome and neurotoxicity of CAR T-cell therapy and associated prevention and management strategies. J. Exp. Clin. Cancer Res. 2021, 40, 367. [Google Scholar] [CrossRef] [PubMed]
- Soularue, E.; Lepage, P.; Colombel, J.F.; Coutzac, C.; Faleck, D.; Marthey, L.; Collins, M.; Chaput, N.; Robert, C.; Carbonnel, F. Enterocolitis due to immune checkpoint inhibitors: A systematic review. Gut 2018, 67, 2056–2067. [Google Scholar] [CrossRef] [PubMed]
- Michot, J.; Lazarovici, J.; Tieu, A.; Champiat, S.; Voisin, A.; Ebbo, M.; Godeau, B.; Michel, M.; Ribrag, V.; Lambotte, O. Haematological immune-related adverse events with immune checkpoint inhibitors, how to manage? Eur. J. Cancer 2019, 122, 72–90. [Google Scholar] [CrossRef] [PubMed]
- Bancroft, C.C.; Chen, Z.; Dong, G.; Sunwoo, J.B.; Yeh, N.; Park, C.; Van Waes, C. Coexpression of proangiogenic factors IL-8 and VEGF by human head and neck squamous cell carcinoma involves coactivation by MEK-MAPK and IKK-NF-kappaB signal pathways. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2001, 7, 435–442. [Google Scholar]
- Błach, J.; Wojas-Krawczyk, K.; Nicoś, M.; Krawczyk, P. Failure of Immunotherapy—The Molecular and Immunological Origin of Immunotherapy Resistance in Lung Cancer. Int. J. Mol. Sci. 2021, 22, 9030. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Wang, W.C.; Shen, W.H.; Xu, K.; Hu, Y.Y.; Han, G.H.; Liu, Y.B. Thalidomide suppresses angiogenesis and immune evasion via lncRNA FGD5-AS1/miR-454-3p/ZEB1 axis-mediated VEGFA expression and PD-1/PD-L1 checkpoint in NSCLC. Chem. Biol. Interact. 2021, 349, 109652. [Google Scholar] [CrossRef]
- Liu, Z.; Ning, F.; Cai, Y.; Sheng, H.; Zheng, R.; Yin, X.; Lu, Z.; Su, L.; Chen, X.; Zeng, C.; et al. The EGFR-P38 MAPK axis up-regulates PD-L1 through miR-675-5p and down-regulates HLA-ABC via hexokinase-2 in hepatocellular carcinoma cells. Cancer Commun. 2021, 41, 62–78. [Google Scholar] [CrossRef]
- Parsa, A.T.; Waldron, J.S.; Panner, A.; Crane, C.A.; Parney, I.F.; Barry, J.J.; Cachola, K.E.; Murray, J.C.; Tihan, T.; Jensen, M.C.; et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat. Med. 2007, 13, 84–88. [Google Scholar] [CrossRef]
- Hsu, Y.-L.; Hung, J.-Y.; Chang, W.-A.; Jian, S.-F.; Lin, Y.-S.; Pan, Y.-C.; Wu, C.-Y.; Kuo, P.-L. Hypoxic Lung-Cancer-Derived Extracellular Vesicle MicroRNA-103a Increases the Oncogenic Effects of Macrophages by Targeting PTEN. Mol. Ther. 2018, 26, 568–581. [Google Scholar] [CrossRef]
- Hu, X.; Chen, Q.; Guo, H.; Li, K.; Fu, B.; Chen, Y.; Zhao, H.; Wei, M.; Li, Y.; Wu, H. Identification of Target PTEN-Based miR-425 and miR-576 as Potential Diagnostic and Immunotherapeutic Biomarkers of Colorectal Cancer With Liver Metastasis. Front. Oncol. 2021, 11, 657984. [Google Scholar] [CrossRef]
- Taube, J.M.; Anders, R.A.; Young, G.D.; Xu, H.; Sharma, R.; McMiller, T.L.; Chen, S.; Klein, A.P.; Pardoll, D.M.; Topalian, S.L.; et al. Colocalization of Inflammatory Response with B7-H1 Expression in Human Melanocytic Lesions Supports an Adaptive Resistance Mechanism of Immune Escape. Sci. Transl. Med. 2012, 4, 127ra37. [Google Scholar] [CrossRef] [PubMed]
- Nair, M.G.; VP, S.; Patil, S.; CE, A.; Mukherjee, G.; Kumar, R.V.; Prabhu, J.S.; TS, S. miR-18a Mediates Immune Evasion in ER-Positive Breast Cancer through Wnt Signaling. Cells 2022, 11, 1672. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Guo, R.; Hao, G.; Xiao, J.; Bao, Y.; Zhou, J.; Chen, Q.; Wei, X. The expression profiling and ontology analysis of noncoding RNAs in peritoneal fibrosis induced by peritoneal dialysis fluid. Gene 2015, 564, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-X.; Gao, J.; Long, X.; Zhang, P.-F.; Yang, X.; Zhu, S.-Q.; Pei, X.; Qiu, B.-Q.; Chen, S.-W.; Lu, F.; et al. The circular RNA circHMGB2 drives immunosuppression and anti-PD-1 resistance in lung adenocarcinomas and squamous cell carcinomas via the miR-181a-5p/CARM1 axis. Mol. Cancer 2022, 21, 110. [Google Scholar] [CrossRef]
- Miliotis, C.; Slack, F.J. miR-105-5p regulates PD-L1 expression and tumor immunogenicity in gastric cancer. Cancer Lett. 2021, 518, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, C.; He, Z.; Chen, S.; Li, L.; Sun, C. LncRNA PSMB8-AS1 contributes to pancreatic cancer progression via modulating miR-382-3p/STAT1/PD-L1 axis. J. Exp. Clin. Cancer Res. CR 2020, 39, 179. [Google Scholar] [CrossRef]
- Kolahi, S.; Farajzadeh, M.-J.; Alipour, S.; Abhari, A.; Farhadi, J.; Bahavarnia, N.; Mahdavi, A.M.; Khabbazi, A.; Sakhinia, E. Determination of mir-155 and mir-146a expression rates and its association with expression level of TNF-α and CTLA4 genes in patients with Behcet’s disease. Immunol. Lett. 2018, 204, 55–59. [Google Scholar] [CrossRef]
- Dezfuli, N.K.; Alipoor, S.D.; Roofchayee, N.D.; Seyfi, S.; Salimi, B.; Adcock, I.M.; Mortaz, E. Evaluation Expression of miR-146a and miR-155 in Non-Small-Cell Lung Cancer Patients. Front. Oncol. 2021, 11, 4356. [Google Scholar] [CrossRef]
- Boldrini, L.; Giordano, M.; Niccoli, C.; Melfi, F.; Lucchi, M.; Mussi, A.; Fontanini, G. Role of microRNA-33a in regulating the expression of PD-1 in lung adenocarcinoma. Cancer Cell Int. 2017, 17, 105. [Google Scholar] [CrossRef]
- Zhang, Q.; Pan, J.; Xiong, D.; Wang, Y.; Miller, M.S.; Sei, S.; Shoemaker, R.H.; Izzotti, A.; You, M. Pulmonary Aerosol Delivery of Let-7b microRNA Confers a Striking Inhibitory Effect on Lung Carcinogenesis through Targeting the Tumor Immune Microenvironment. Adv. Sci. 2021, 8, e2100629. [Google Scholar] [CrossRef]
- Sp, N.; Kang, D.Y.; Lee, J.M.; Jang, K.J. Mechanistic Insights of Anti-Immune Evasion by Nobiletin through Regulating miR-197/STAT3/PD-L1 Signaling in Non-Small Cell Lung Cancer (NSCLC) Cells. Int. J. Mol. Sci. 2021, 22, 9843. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Crawford, M.; Mao, Y.; Lee, R.J.; Davis, I.C.; Elton, T.S.; Lee, L.J.; Nana-Sinkam, S.P. Therapeutic Delivery of MicroRNA-29b by Cationic Lipoplexes for Lung Cancer. Mol. Ther. Nucleic Acids 2013, 2, e84. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Valdecanas, D.; Zhang, X.; Zhan, Y.; Bhardwaj, V.; Calin, G.A.; Komaki, R.; Giri, D.K.; Quini, C.C.; Wolfe, T.; et al. Therapeutic Delivery of miR-200c Enhances Radiosensitivity in Lung Cancer. Mol. Ther. J. Am. Soc. Gene Ther. 2014, 22, 1494–1503. [Google Scholar] [CrossRef]
- Nie, H.; Xie, X.; Zhang, D.; Zhou, Y.; Li, B.; Li, F.; Li, F.; Cheng, Y.; Mei, H.; Meng, H.; et al. Use of lung-specific exosomes for miRNA-126 delivery in non-small cell lung cancer. Nanoscale 2020, 12, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Dong, C.; Chen, M.; Yang, T.; Wang, X.; Gao, Y.; Wang, L.; Wen, Y.; Chen, G.; Wang, X.; et al. Extracellular vesicle-mediated delivery of miR-101 inhibits lung metastasis in osteosarcoma. Theranostics 2020, 10, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, H.; Pang, Y.; Wang, M.; Liu, S. UTMD-mediated delivery of miR-21-5p inhibitor suppresses the development of lung cancer. Tissue Cell 2022, 74, 101719. [Google Scholar] [CrossRef]
- Chen, T.; Xiao, Q.; Wang, X.; Wang, Z.; Hu, J.; Zhang, Z.; Gong, Z.; Chen, S. miR-16 regulates proliferation and invasion of lung cancer cells via the ERK/MAPK signaling pathway by targeted inhibition of MAPK kinase 1 (MEK1). J. Int. Med. Res. 2019, 47, 5194–5204. [Google Scholar] [CrossRef]
- Andriani, F.; Majorini, M.T.; Mano, M.; Landoni, E.; Miceli, R.; Facchinetti, F.; Mensah, M.; Fontanella, E.; Dugo, M.; Giacca, M.; et al. MiR-16 regulates the pro-tumorigenic potential of lung fibroblasts through the inhibition of HGF production in an FGFR-1- and MEK1-dependent manner. J. Hematol. Oncol. 2018, 11, 45. [Google Scholar] [CrossRef]
- Fanini, F.; Bandini, E.; Plousiou, M.; Carloni, S.; Wise, P.; Neviani, P.; Murtadha, M.; Foca, F.; Fabbri, F.; Vannini, I.; et al. MicroRNA-16 Restores Sensitivity to Tyrosine Kinase Inhibitors and Outperforms MEK Inhibitors in KRAS-Mutated Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2021, 22, 13357. [Google Scholar] [CrossRef]
- Van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet. Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Rokavec, M.; Li, H.; Jiang, L.; Hermeking, H. The p53/miR-34 axis in development and disease. J. Mol. Cell Biol. 2014, 6, 214–230. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, M.; Jeon, Y.J.; Nuovo, G.J.; Middleton, J.; Secchiero, P.; Joshi, P.; Alder, H.; Nazaryan, N.; Di Leva, G.; Romano, G.; et al. MiR-34a/c-Dependent PDGFR-α/β Downregulation Inhibits Tumorigenesis and Enhances TRAIL-Induced Apoptosis in Lung Cancer. PLoS ONE 2013, 8, e67581. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Ge, F.; Du, L.; Zhang, Z.; Liu, D. MiR-34b-3p represses cell proliferation, cell cycle progression and cell apoptosis in non-small-cell lung cancer (NSCLC) by targeting CDK4. J. Cell. Mol. Med. 2019, 23, 5282–5291. [Google Scholar] [CrossRef] [PubMed]
- Kasinski, A.L.; Slack, F.J. miRNA-34 Prevents Cancer Initiation and Progression in a Therapeutically Resistant K-ras and p53-Induced Mouse Model of Lung Adenocarcinoma. Cancer Res 2012, 72, 5576–5587. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Calin, G.A.; Lopez-Berestein, G.; Sood, A.K. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016, 6, 235–246. [Google Scholar] [CrossRef]
- Tili, E.; Michaille, J.-J.; Croce, C.M. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunol. Rev. 2013, 253, 167–184. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Wu, S.Y.; Pradeep, S.; Ivan, C.; Pecot, C.V.; Gharpure, K.M.; Nagaraja, A.S.; Armaiz-Pena, G.N.; McGuire, M.; Zand, B.; et al. Hypoxia-mediated downregulation of miRNA biogenesis promotes tumour progression. Nat. Commun. 2014, 5, 5202. [Google Scholar] [CrossRef]
- Lei, L.; Chen, C.; Zhao, J.; Wang, H.; Guo, M.; Zhou, Y.; Luo, J.; Zhang, J.; Xu, L. Targeted Expression of miR-7 Operated by TTF-1 Promoter Inhibited the Growth of Human Lung Cancer through the NDUFA4 Pathway. Mol. Ther. Nucleic Acids 2017, 6, 183–197. [Google Scholar] [CrossRef]
- Chen, S.; Guan, L.; Zhao, X.; Yang, J.; Chen, L.; Guo, M.; Zhao, J.; Chen, C.; Zhou, Y.; Han, Y.; et al. Optimized thyroid transcription factor-1 core promoter-driven microRNA-7 expression effectively inhibits the growth of human non-small-cell lung cancer cells. J. Zhejiang Univ. B 2022, 23, 915–930. [Google Scholar] [CrossRef]
- Abdou, P.; Wang, Z.; Chen, Q.; Chan, A.; Zhou, D.R.; Gunadhi, V.; Gu, Z. Advances in engineering local drug delivery systems for cancer immunotherapy. WIREs Nanomed. Nanobiotechnol. 2020, 12, e1632. [Google Scholar] [CrossRef]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef]
- Gomes, E.R.; Souza, F.R.; Cassali, G.D.; Sabino, A.P.; Barros, A.L.B.; Oliveira, M.C. Investigation of the Antitumor Activity and Toxicity of Tumor-Derived Exosomes Fused with Long-Circulating and pH-Sensitive Liposomes Containing Doxorubicin. Pharmaceutics 2022, 14, 2256. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Du, C.; Duan, X.; Yao, X.; Wan, J.; Jiang, Z.; Qin, Z.; Li, W.; Pan, L.; Gu, Z.; et al. Inhibiting collagen I production and tumor cell colonization in the lung via miR-29a-3p loading of exosome-/liposome-based nanovesicles. Acta Pharm. Sinica. B 2022, 12, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, Y.; Li, D.; Wang, H.; Zhao, X.; Yang, J.; Chen, L.; Guo, M.; Zhao, J.; Chen, C.; et al. Mechanisms Controlling MicroRNA Expression in Tumor. Cells 2022, 11, 2852. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Tao, Y.; Zhou, Y.; Qin, N.; Chen, C.; Tian, D.; Xu, L. MicroRNA-7: A promising new target in cancer therapy. Cancer Cell Int. 2015, 15, 103. [Google Scholar] [CrossRef]
- Zhao, J.; Chu, F.; Xu, H.; Guo, M.; Shan, S.; Zheng, W.; Tao, Y.; Zhou, Y.; Hu, Y.; Chen, C.; et al. C/EBPα/miR-7 Controls CD4+ T-Cell Activation and Function and Orchestrates Experimental Autoimmune Hepatitis in Mice. Hepatology 2021, 74, 379–396. [Google Scholar] [CrossRef]


| MiRNAs or lncRNAs | Expression | Cell Source | Target | Function | Ref. |
|---|---|---|---|---|---|
| MiRNA replacement therapies | |||||
| Let-7a | Down | A549 | NIRF | Inhibit the proliferation | [24] |
| Let-7a | Down | A549 | k-Ras c-Myc | Inhibit the growth | [25] |
| Let-7b-3p | Down | A549 PC9 H1299 etc. | BRF2 | Inhibit the proliferation and metastasis | [26] |
| Let-7c | Down | SKMES-1 H520 H157 etc. | ITGB3 MAP4K3 | Inhibit migration and invasion | [27] |
| Suppressive miRNA therapies | |||||
| LINC00336 | Up | A549 H358 | CBS | As a ceRNA of miR-6852 to inhibit the growth | [34] |
| MiR-21-5p | Up | A549 | PDCD4 | Promote apoptosis | [37] |
| MiRNAs | Target | Delivery Vehicle | Approach | Tumor Type | Results | Ref. |
|---|---|---|---|---|---|---|
| Preclinical Trials | ||||||
| Let-7b | Kras | Adenovirus | Intranasal | NSCLC | 66% reduction in orthotopic tumor burden; Reduced xenograft growth | [10] |
| MiR-29b | CDK6 | Cationic lipoplexes | Caudal | NSCLC | 60% xenograft growth inhibition | [152] |
| MiR-200c | PRDX2 GABP/Nrf2 SESN1 | Amphoteric liposome | Subcutaneous | NSCLC | MiR-200c plus radiotherapy delayed xenograft growth | [153] |
| MiR-34a and Let-7b | Kras p53 | Neutral lipid emulsion | Caudal | NSCLC | 40% increased survival with combination or miR-34a alone | [28] |
| MiR-126 | PTEN/PI3K/AKT | 231-Exosome | Intravenous | NSCLC | Inhibit the formulation of lung metastasis | [154] |
| MiR-101 | BCL6 | AD-MSC-EVs | Caudal | Osteosarcoma | Inhibit the formulation of lung metastasis | [155] |
| MiR-21-5p | BTG2 | UTMD | Caudal | NSCLC | Reduced the size and volume of xenograft growth | [156] |
| Clinical Trials | ||||||
| MiR-16 | EGFR | EDVs | Intravenous | NSCLC MPM | 5 × 109 TargomiRs once weekly was the maximum tolerated dose. | [160] |
| MiR-34 | Kras P53 PDGFR CDK4 etc. | Liposome | Intravenous | Solid tumors Hematologic malignancies | Trial was closed early due to serious immune-mediated AEs | [12] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Liu, Y.; Chen, L.; Zhao, J.; Guo, M.; Zhao, X.; Wen, Z.; He, Z.; Chen, C.; Xu, L. MiRNA-Based Therapies for Lung Cancer: Opportunities and Challenges? Biomolecules 2023, 13, 877. https://doi.org/10.3390/biom13060877
Yang H, Liu Y, Chen L, Zhao J, Guo M, Zhao X, Wen Z, He Z, Chen C, Xu L. MiRNA-Based Therapies for Lung Cancer: Opportunities and Challenges? Biomolecules. 2023; 13(6):877. https://doi.org/10.3390/biom13060877
Chicago/Turabian StyleYang, Han, Yufang Liu, Longqing Chen, Juanjuan Zhao, Mengmeng Guo, Xu Zhao, Zhenke Wen, Zhixu He, Chao Chen, and Lin Xu. 2023. "MiRNA-Based Therapies for Lung Cancer: Opportunities and Challenges?" Biomolecules 13, no. 6: 877. https://doi.org/10.3390/biom13060877
APA StyleYang, H., Liu, Y., Chen, L., Zhao, J., Guo, M., Zhao, X., Wen, Z., He, Z., Chen, C., & Xu, L. (2023). MiRNA-Based Therapies for Lung Cancer: Opportunities and Challenges? Biomolecules, 13(6), 877. https://doi.org/10.3390/biom13060877