NMR and Patch-Clamp Characterization of Yeast Mitochondrial Pyruvate Carrier Complexes
Abstract
1. Introduction
2. Materials and Methods
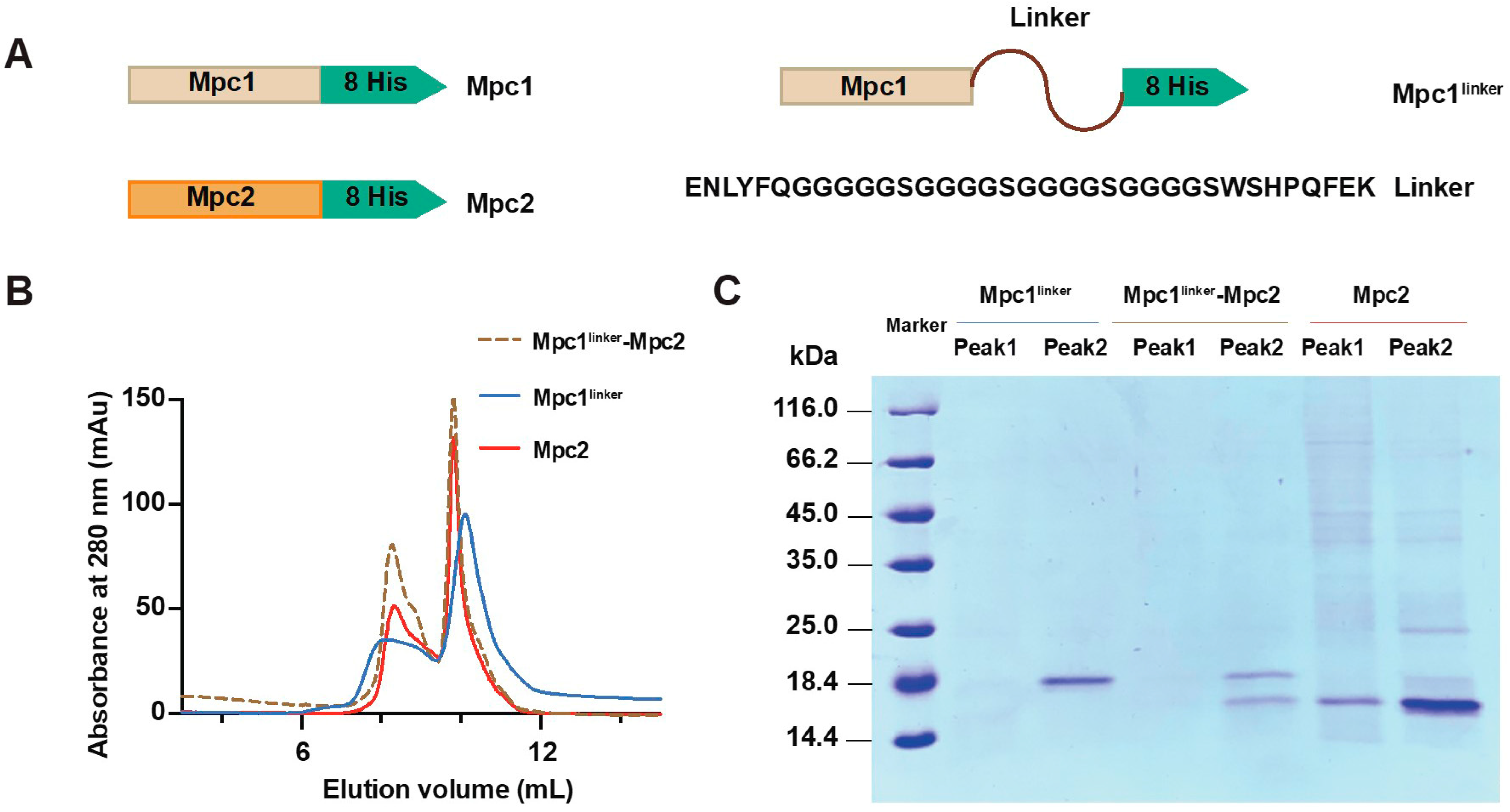
2.1. Plasmid Constructions
2.2. Protein Expression and Purifications
2.3. Size-Exclusion Chromatography with Multi-Angle Light Scattering (SEC-MALS)
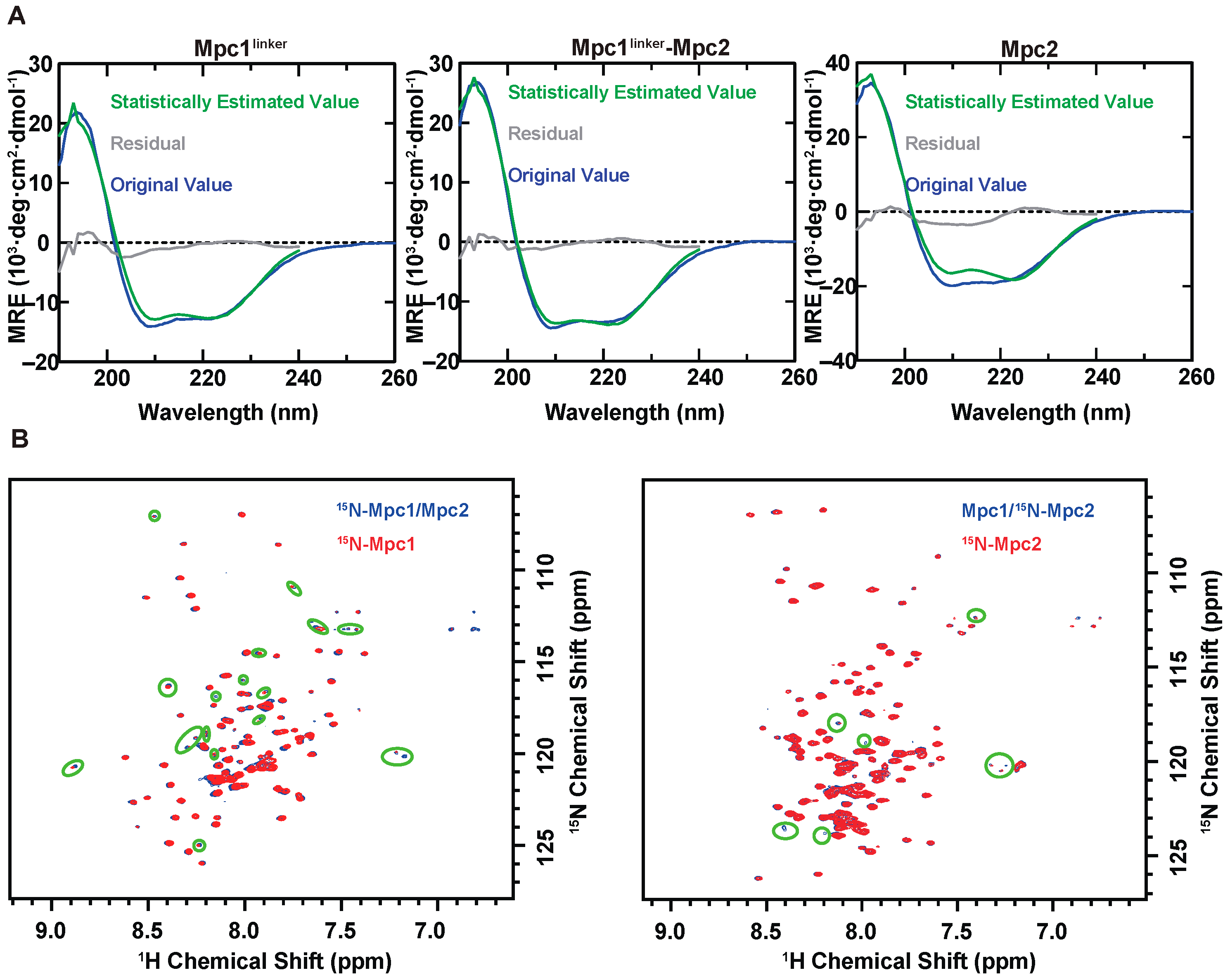
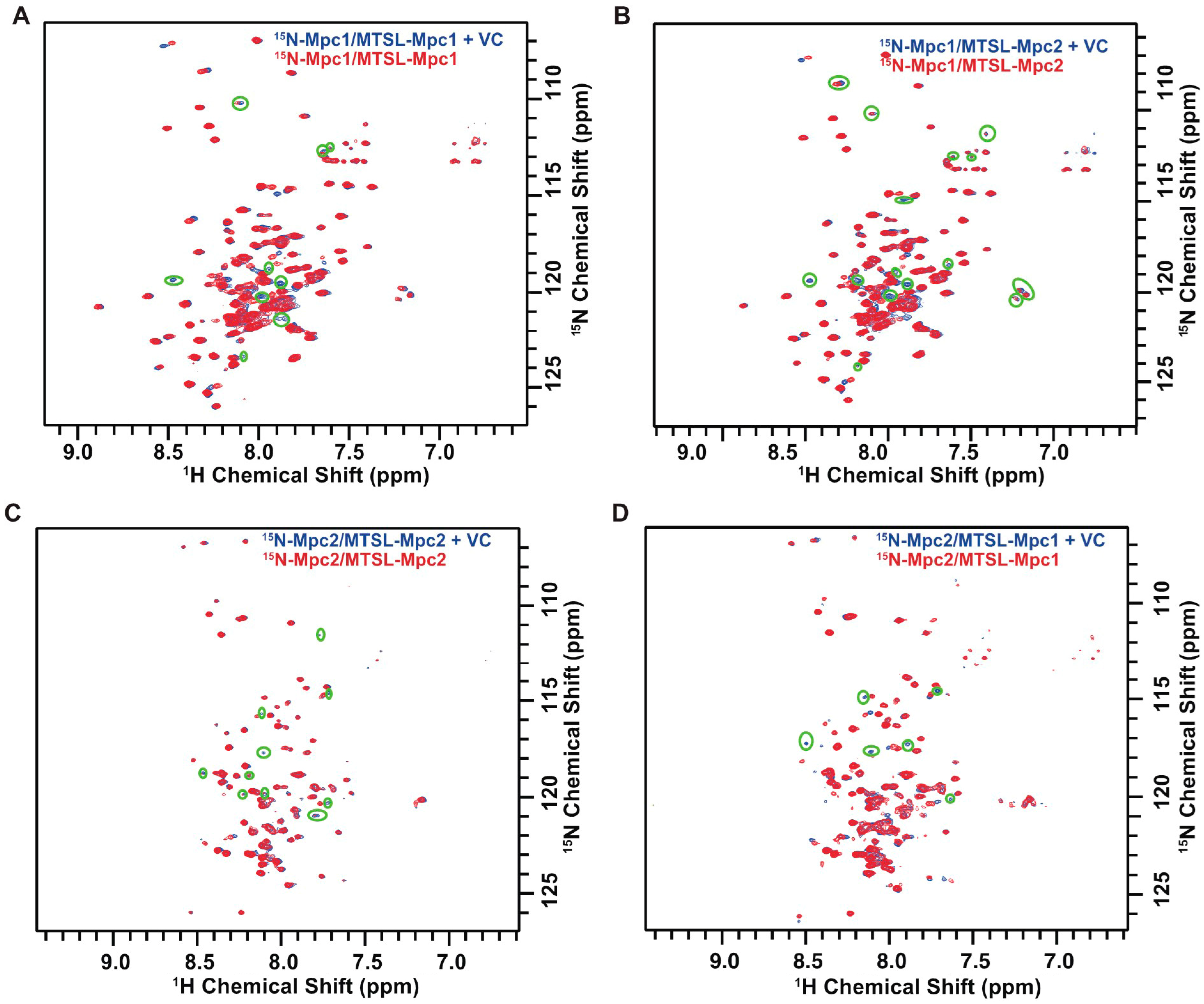
2.4. NMR Experiments
2.5. Circular Dichroism Spectroscopy
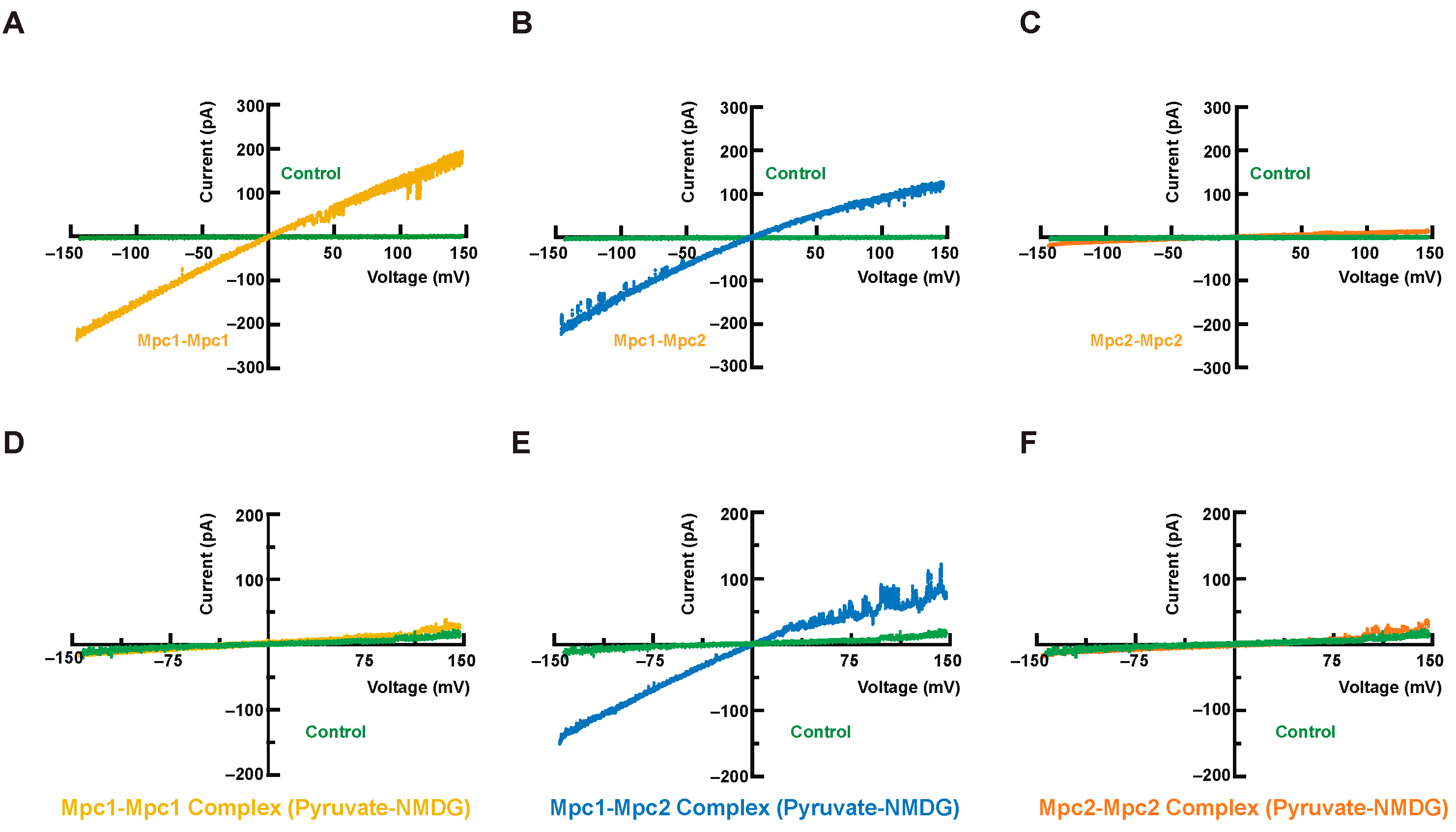
2.6. Patch-Clamp Assays
3. Results
3.1. Expression and Purification of Mpc Complexes
3.2. Characterization of Homo- and Hetero-Mpc Oligomer States
3.3. Analysis of Mpc Monomer Interactions Using PRE NMR
3.4. Thermostability of the Mpc Complexes
3.5. Transport Functional Verification of Mpc Complexes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bricker, D.K.; Taylor, E.B.; Schell, J.C.; Orsak, T.; Boutron, A.; Chen, Y.-C.; Cox, J.E.; Cardon, C.M.; Van Vranken, J.G.; Dephoure, N.; et al. A Mitochondrial Pyruvate Carrier Required for Pyruvate Uptake in Yeast, Drosophila, and Humans. Science 2012, 337, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Mccommis, K.S.; Finck, B.N. Mitochondrial pyruvate transport: A historical perspective and future research directions. Biochem. J. 2015, 466, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Yiew, N.K.H.; Finck, B.N. The mitochondrial pyruvate carrier at the crossroads of intermediary metabolism. Am. J. Physiol. -Endocrinol. Metab. 2022, 323, E33–E52. [Google Scholar] [CrossRef]
- Rogatzki, M.J.; Ferguson, B.S.; Goodwin, M.L.; Gladden, L.B. Lactate is always the end product of glycolysis. Front. Neurosci. 2015, 9. [Google Scholar] [CrossRef]
- Rauckhorst, A.J.; Taylor, E.B. Mitochondrial pyruvate carrier function and cancer metabolism. Curr. Opin. Genet. Dev. 2016, 38, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Papa, S.; Francavilla, A.; Paradies, G.; Meduri, B. The transport of pyruvate in rat liver mitochondria. FEBS Lett. 1971, 12, 285–288. [Google Scholar] [CrossRef]
- Halestrap, A.P.; Denton, R.M. Specific inhibition of pyruvate transport in rat liver mitochondria and human erythrocytes by alpha-cyano-4-hydroxycinnamate. Biochem. J. 1974, 138, 313–316. [Google Scholar] [CrossRef]
- Bender, T.; Pena, G.; Martinou, J.C. Regulation of mitochondrial pyruvate uptake by alternative pyruvate carrier complexes. EMBO J. 2015, 34, 911–924. [Google Scholar] [CrossRef]
- Compan, V.; Pierredon, S.; Vanderperre, B.; Krznar, P.; Marchiq, I.; Zamboni, N.; Pouyssegur, J.; Martinou, J.-C. Monitoring Mitochondrial Pyruvate Carrier Activity in Real Time Using a BRET-Based Biosensor: Investigation of the Warburg Effect. Mol. Cell 2015, 59, 491–501. [Google Scholar] [CrossRef]
- Herzig, S.; Raemy, E.; Montessuit, S.; Veuthey, J.-L.; Zamboni, N.; Westermann, B.; Kunji, E.R.S.; Martinou, J.-C. Identification and Functional Expression of the Mitochondrial Pyruvate Carrier. Science 2012, 337, 93–96. [Google Scholar] [CrossRef]
- Nagampalli, R.S.K.; Quesñay, J.E.N.; Adamoski, D.; Islam, Z.; Birch, J.; Sebinelli, H.G.; Girard, R.M.B.M.; Ascenção, C.F.R.; Fala, A.M.; Pauletti, B.A.; et al. Human mitochondrial pyruvate carrier 2 as an autonomous membrane transporter. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- McCommis, K.S.; Chen, Z.; Fu, X.; McDonald, W.G.; Colca, J.R.; Kletzien, R.F.; Burgess, S.C.; Finck, B.N. Loss of Mitochondrial Pyruvate Carrier 2 in the Liver Leads to Defects in Gluconeogenesis and Compensation via Pyruvate-Alanine Cycling. Cell Metab. 2015, 22, 682–694. [Google Scholar] [CrossRef] [PubMed]
- Takaoka, Y.; Konno, M.; Koseki, J.; Colvin, H.; Asai, A.; Tamari, K.; Satoh, T.; Mori, M.; Doki, Y.; Ogawa, K.; et al. Mitochondrial pyruvate carrier 1 expression controls cancer epithelial-mesenchymal transition and radioresistance. Cancer Sci. 2019, 110, 1331–1339. [Google Scholar] [CrossRef]
- Tang, X.-P.; Chen, Q.; Li, Y.; Wang, Y.; Zou, H.-B.; Fu, W.-J.; Niu, Q.; Pan, Q.-G.; Jiang, P.; Xu, X.-S.; et al. Mitochondrial pyruvate carrier 1 functions as a tumor suppressor and predicts the prognosis of human renal cell carcinoma. Lab. Investig. 2019, 99, 191–199. [Google Scholar] [CrossRef]
- Lee, J.; Jin, Z.; Lee, D.; Yun, J.-H.; Lee, W. Characteristic Analysis of Homo- and Heterodimeric Complexes of Human Mitochondrial Pyruvate Carrier Related to Metabolic Diseases. Int. J. Mol. Sci. 2020, 21, 3403. [Google Scholar] [CrossRef]
- Tavoulari, S.; Thangaratnarajah, C.; Mavridou, V.; Harbour, M.E.; Martinou, J.C.; Kunji, E.R. The yeast mitochondrial pyruvate carrier is a hetero-dimer in its functional state. EMBO J. 2019, 38, e100785. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Slotboom, D.J.; Duurkens, R.H.; Olieman, K.; Erkens, G.B. Static light scattering to characterize membrane proteins in detergent solution. Methods 2008, 46, 73–82. [Google Scholar] [CrossRef]
- Oxenoid, K.; Dong, Y.; Cao, C.; Cui, T.; Sancak, Y.; Markhard, A.L.; Grabarek, Z.; Kong, L.; Liu, Z.; Ouyang, B.; et al. Architecture of the mitochondrial calcium uniporter. Nature 2016, 533, 269–273. [Google Scholar] [CrossRef]
- Altenbach, C.; Oh, K.J.; Trabanino, R.J.; Hideg, K.; Hubbell, W.L. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: Experimental strategies and practical limitations. Biochemistry 2001, 40, 15471–15482. [Google Scholar] [CrossRef]
- Keller, B.U.; Hedrich, R.; Vaz, W.L.C.; Criado, M. Single channel recordings of reconstituted ion channel proteins: An improved technique. Pflug. Arch. -Eur. J. Physiol. 1988, 411, 94–100. [Google Scholar] [CrossRef]
- Zhou, S.; Ruan, M.; Li, Y.; Yang, J.; Bai, S.; Richter, C.; Schwalbe, H.; Xie, C.; Shen, B.; Wang, J. Solution structure of the voltage-gated Tim23 channel in complex with a mitochondrial presequence peptide. Cell Res. 2021, 31, 821–824. [Google Scholar] [CrossRef]
- Villarroel, A.; Burnashev, N.; Sakmann, B. Dimensions of the narrow portion of a recombinant nmda receptor-channel. Biophys. J. 1995, 68, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wong, N.C.; Cheng, Y.; Kehl, S.J.; Fedida, D. Control of voltage-gated K+ channel permeability to NMDG+ by a residue at the outer pore. J. Gen. Physiol. 2009, 133, 361–374. [Google Scholar] [CrossRef]
- Chen, H.; Wu, B.; Zhang, T.; Jia, J.; Lu, J.; Chen, Z.; Ni, Z.; Tan, T. Effect of Linker Length and Flexibility on the Clostridium thermocellum Esterase Displayed on Bacillus subtilis Spores. Appl. Biochem. Biotechnol. 2017, 182, 168–180. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Miyazaki, M. Refolding Techniques for Recovering Biologically Active Recombinant Proteins from Inclusion Bodies. Biomolecules 2014, 4, 235–251. [Google Scholar] [CrossRef]
- Li, L.; Wen, M.; Run, C.; Wu, B.; Ouyang, B. Experimental Investigations on the Structure of Yeast Mitochondrial Pyruvate Carriers. Membranes 2022, 12, 916. [Google Scholar] [CrossRef]
- Sjodt, M.; Clubb, R.T. Nitroxide Labeling of Proteins and the Determination of Paramagnetic Relaxation Derived Distance Restraints for NMR Studies. Bio-protocol 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Timmins, P.A.; Leonhard, M.; Weltzien, H.U.; Wacker, T.; Welte, W. A physical characterization of some detergents of potential use for membrane protein crystallization. Febs Lett. 1988, 238, 361–368. [Google Scholar] [CrossRef]
- Yu, D.; Volkov, A.N.; Tang, C. Characterizing Dynamic Protein-Protein Interactions Using Differentially Scaled Paramagnetic Relaxation Enhancement. J. Am. Chem. Soc. 2009, 131, 17291–17297. [Google Scholar] [CrossRef]


| Protein Complex | Original d-Value | Spline Interpolation Method | ||
|---|---|---|---|---|
| Linear | Cubic | Quadratic | ||
| Curve Change Area (deg m3·(3 × 109 mol)−1) | ||||
| Mpc1linker homo-dimer | 222.828 | 220.054 | 222.062 | 220.342 |
| Mpc1linker–Mpc2 hetero-dimer | 213.457 | 208.063 | 208.229 | 208.219 |
| Mpc2 homo-dimer | 267.816 | 259.925 | 260.230 | 260.297 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Ding, W.; Ruan, M.; Liu, Y.; Yang, J.; Zhang, H.; Shen, B.; Wang, J.; Li, Y. NMR and Patch-Clamp Characterization of Yeast Mitochondrial Pyruvate Carrier Complexes. Biomolecules 2023, 13, 719. https://doi.org/10.3390/biom13050719
Wang Z, Ding W, Ruan M, Liu Y, Yang J, Zhang H, Shen B, Wang J, Li Y. NMR and Patch-Clamp Characterization of Yeast Mitochondrial Pyruvate Carrier Complexes. Biomolecules. 2023; 13(5):719. https://doi.org/10.3390/biom13050719
Chicago/Turabian StyleWang, Zhen, Wen Ding, Maosen Ruan, Yong Liu, Jing Yang, Huiqin Zhang, Bing Shen, Junfeng Wang, and Yunyan Li. 2023. "NMR and Patch-Clamp Characterization of Yeast Mitochondrial Pyruvate Carrier Complexes" Biomolecules 13, no. 5: 719. https://doi.org/10.3390/biom13050719
APA StyleWang, Z., Ding, W., Ruan, M., Liu, Y., Yang, J., Zhang, H., Shen, B., Wang, J., & Li, Y. (2023). NMR and Patch-Clamp Characterization of Yeast Mitochondrial Pyruvate Carrier Complexes. Biomolecules, 13(5), 719. https://doi.org/10.3390/biom13050719







