Timing of Chromosome DNA Integration throughout the Yeast Cell Cycle
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Media
2.2. Molecular Biology Techniques and Microscopy
2.3. Transformation and Synchronization Procedure
2.4. Measurement of ROS
2.5. p-Value Calculation
Relative Efficiency (Er) Calculation
3. Results
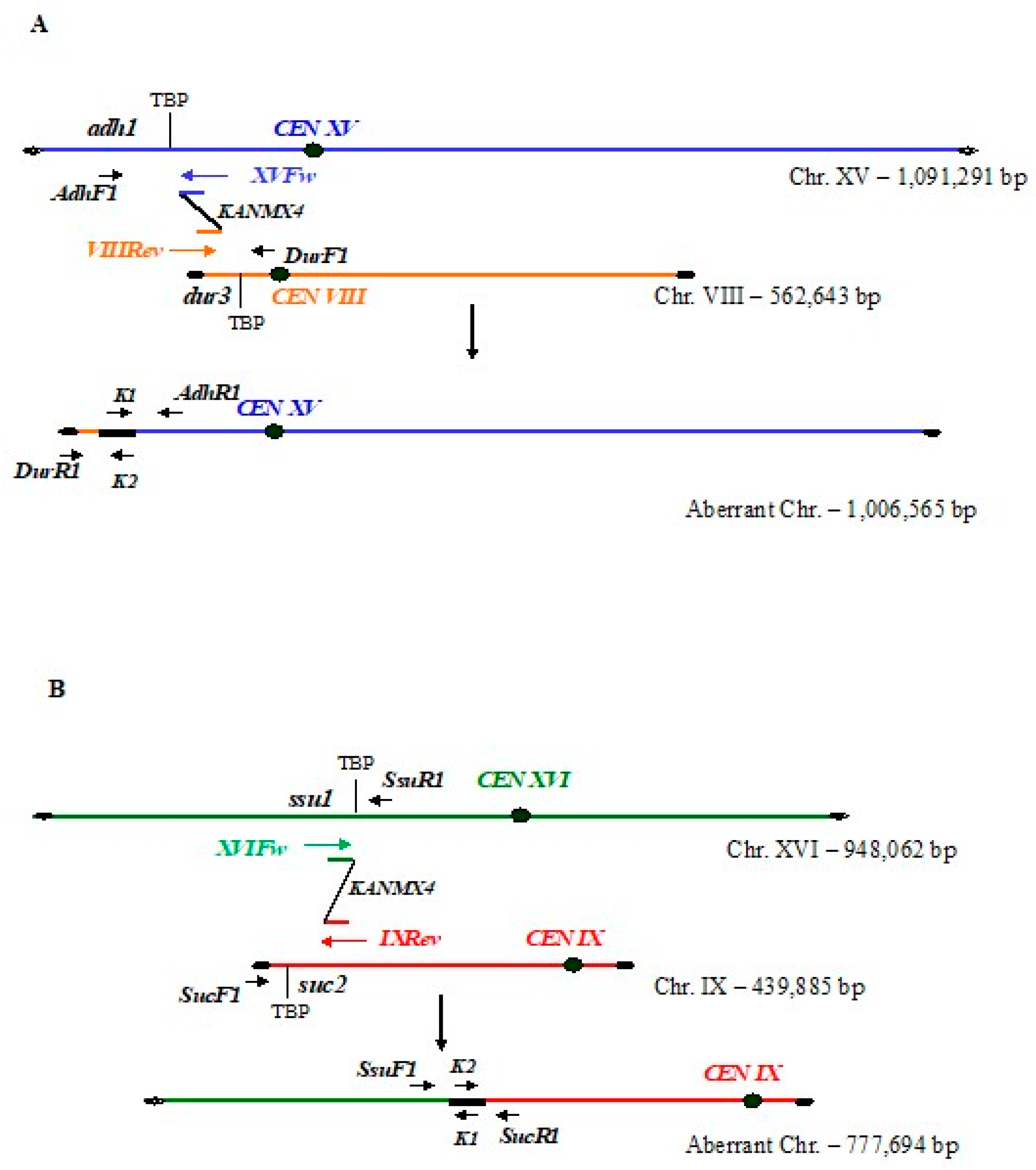
3.1. Integration of Bridge-Induced Translocation and Site-Specific DNA Cassettes
3.2. The Locus and the Cell Cycle Phase During Integration Strongly Affected Transformation Efficiency with Linear DNA Molecules
3.3. The S Phase May Have a Leading Role in Translocation Success
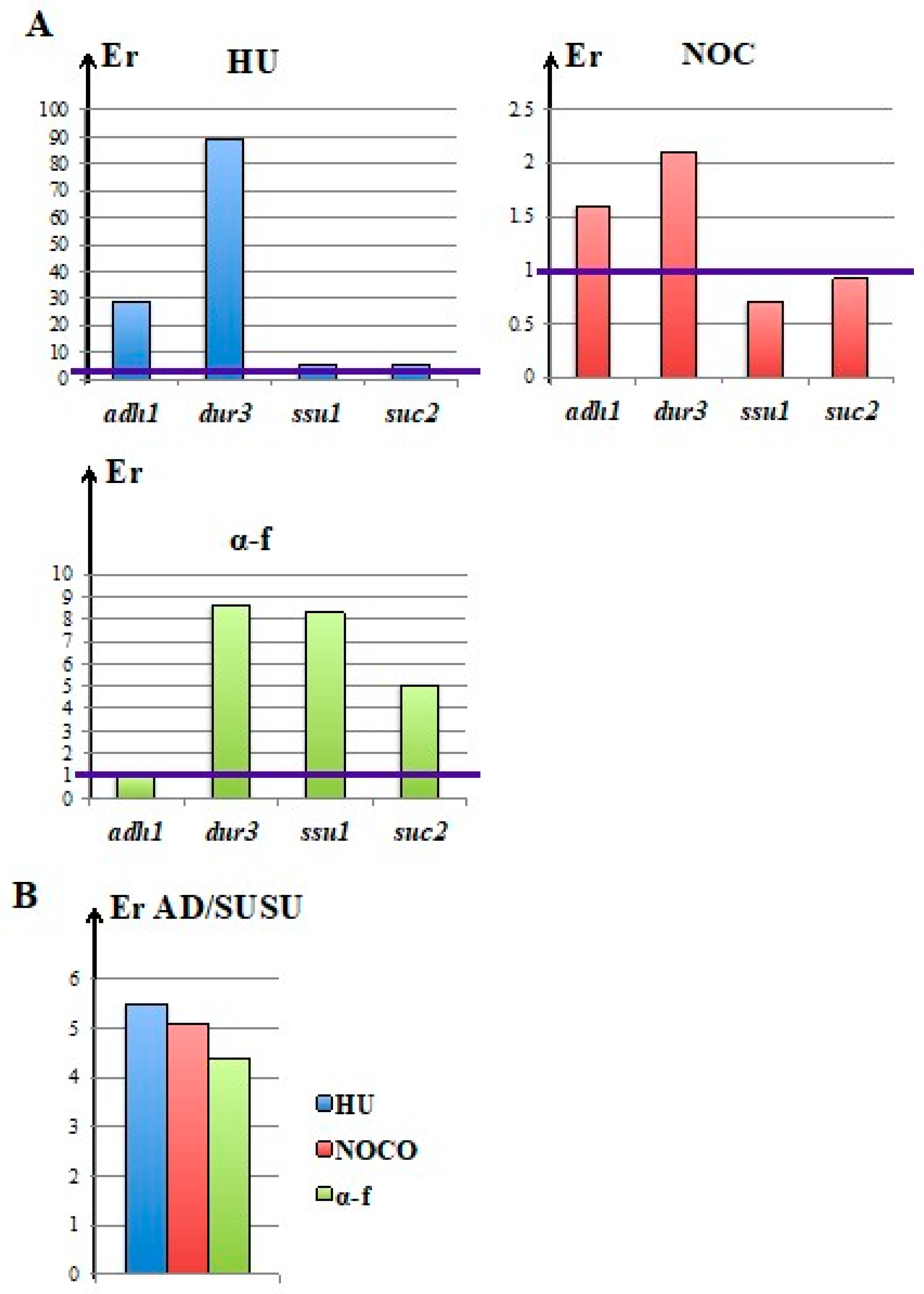
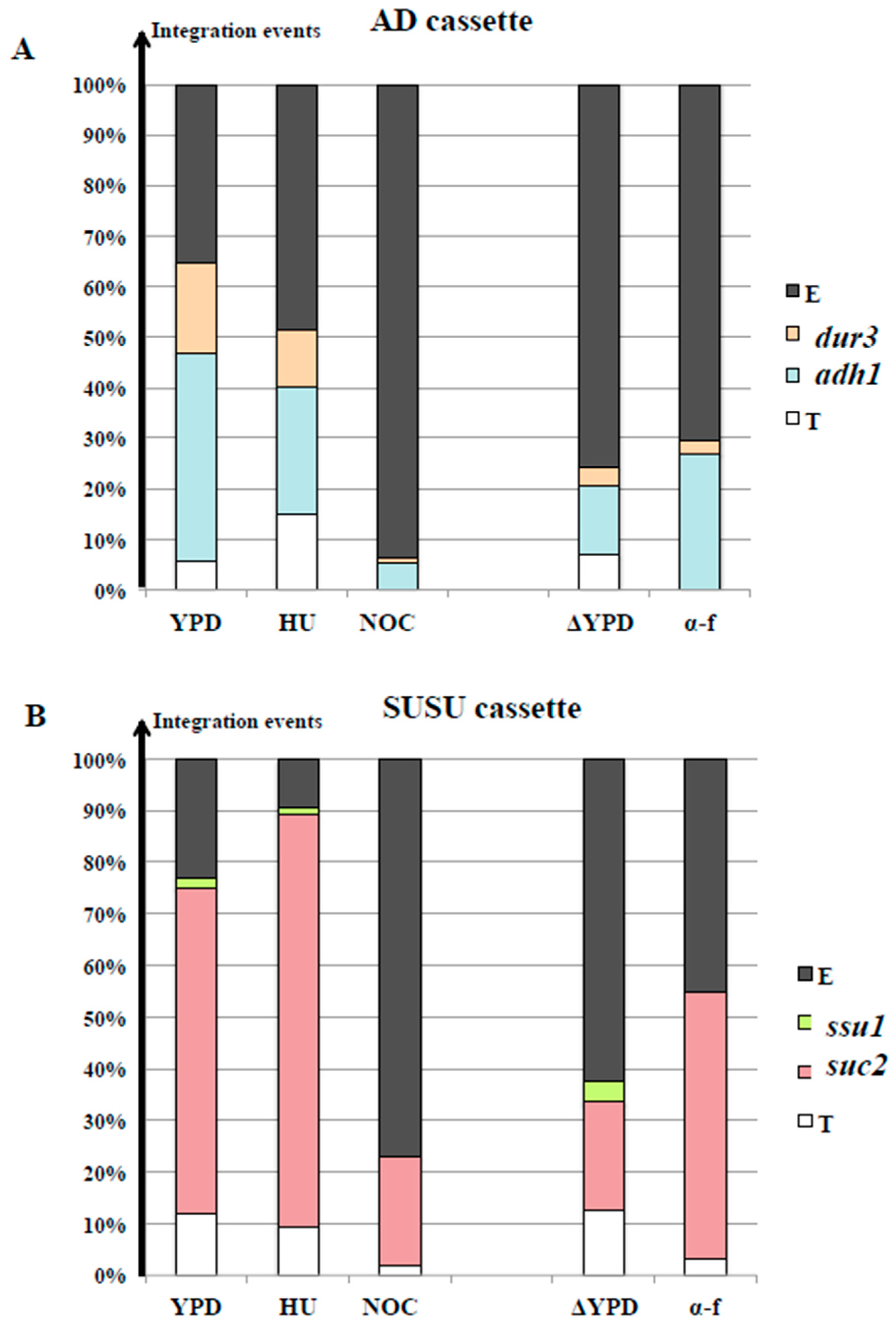
3.4. Nocodazole and α-Factor Strongly Decreased the Occurrence of Homologous Integration and Chromosomal Translocation
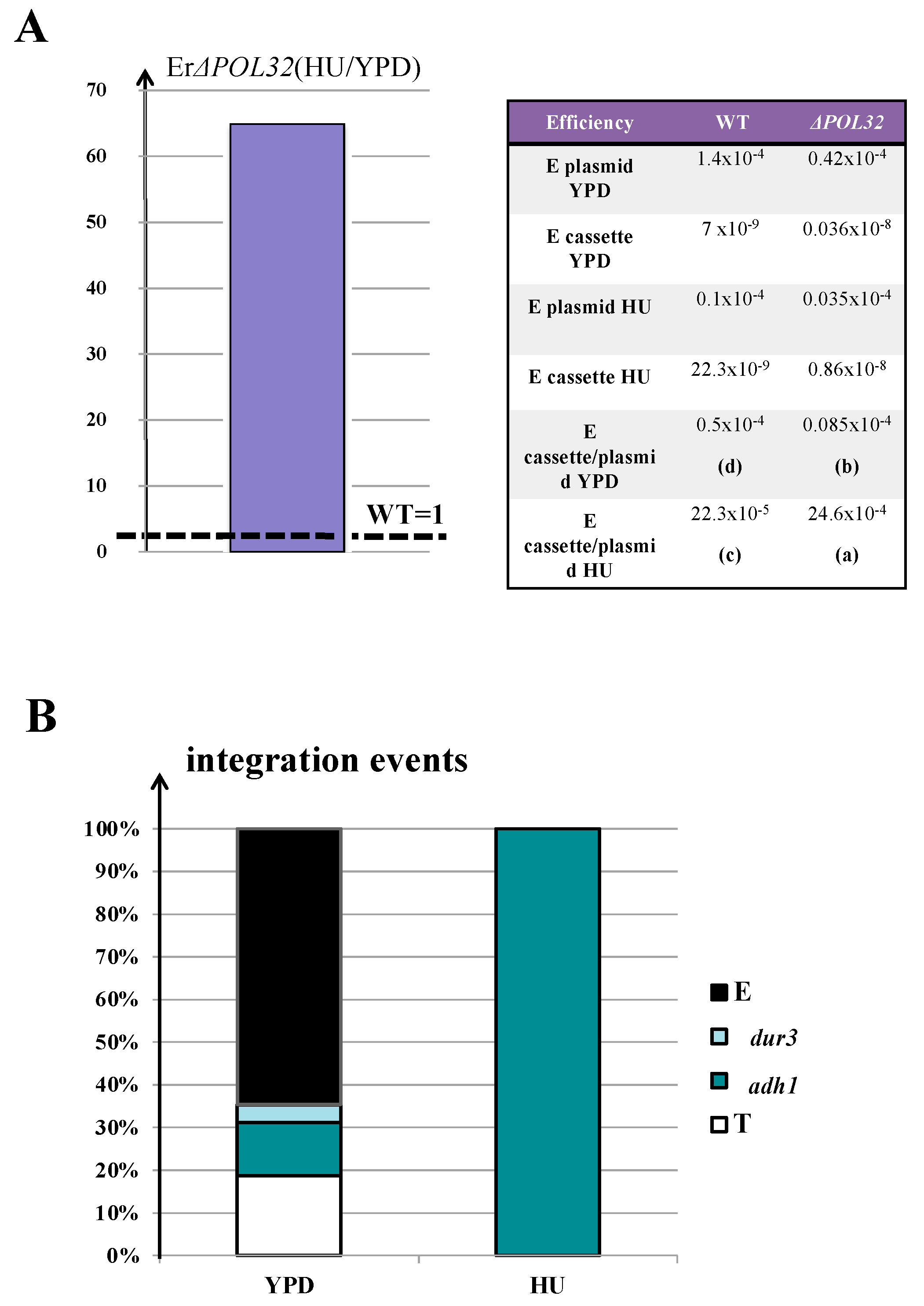
3.5. The Enhancement of AD-Translocation Frequency during Synthesis Is Pol32-Dependent
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tosato, V.; Waghmare, S.K.; Bruschi, C.V. Non-reciprocal chromosomal bridge-induced translocation (BIT) by targeted DNA integration in yeast. Chromosoma 2005, 114, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Sims, J.; Bruschi, C.V.; Bertin, C.; West, N.; Breitenbach, M.; Schroeder, S.; Eisenberg, T. High reactive oxygen species levels are detected at the end of the chronological life span of translocant yeast cells. Mol. Genet. Genom. 2016, 291, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Tosato, V.; Sims, J.; West, N.; Colombin, M.; Bruschi, C.V. Post-translocational adaptation drives evolution through genetic selection and transcriptional shift in S. cerevisiae. Curr. Genet. 2017, 63, 281–292. [Google Scholar] [CrossRef]
- Nikitin, D.; Tosato, V.; Zavec, A.B.; Bruschi, C.V. Cellular and molecular effects of nonreciprocal chromosome translocation in S. cerevisiae. Proc. Natl. Acad. Sci. USA 2008, 105, 9703–9708. [Google Scholar] [CrossRef] [PubMed]
- Bruschi, C.V.; Comer, A.R.; Howe, G.A. Specificity of DNA uptake during whole cell transformation of S. cerevisiae. Yeast 1987, 3, 131–137. [Google Scholar] [CrossRef]
- Chaustova, L.; Zimkus, A. Study of cell wall permeability properties of synchronous S. cerevisiae cells in different phases of cell cycle. Biologija 2005, 1, 38–40. [Google Scholar]
- Tsakraklides, V.; Brevnova, E.; Stephanopoulos, G.; Shaw, A.J. Improved gene targeting through cell cycle synchronization. PLoS ONE 2015, 10, e0133434. [Google Scholar] [CrossRef]
- Prado, F. Genetic instability is prevented by Mrc1-dependent spatio-temporal separation of replicative and repair activities of homologous recombination. BioEssays 2014, 36, 451–462. [Google Scholar] [CrossRef]
- Tosato, V.; Sidari, S.; Bruschi, C.V. Bridge-induced chromosome translocation in yeast relies upon a Rad/Rdh54 dependent, Pol32-independ pathway. PLoS ONE 2013, 8, e60926. [Google Scholar] [CrossRef]
- Lydeard, J.R.; Jain, S.; Yamaguchi, M.; Haber, J.E. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 2007, 448, 820–823. [Google Scholar] [CrossRef]
- Falbo, K.B.; Alabert, C.; Katou, Y.; Wu, S.; Han, J.; Wehr, T.; Xiao, J.; He, X.; Zhang, Z.; Shi, Y.; et al. Involvement of a chromatin remodeling complex in damage tolerance during DNA replication. Nat. Struct. Mol. Biol. 2009, 16, 1167–1172. [Google Scholar] [CrossRef]
- Bruschi, C.V.; Howe, G.A. High frequency FLP-independent homologous DNA recombination of 2μ plasmid in the yeast S. cerevisiae. Curr. Genet. 1988, 14, 191–199. [Google Scholar] [CrossRef]
- Lee, P.S.; Greenwell, P.W.; Dominska, M.; Gawel, M.; Hamilton, M.; Petes, T.D. A fine-structure map of spontaneous mitotic crossovers in the yeast S. cerevisiae. PLoS Genet. 2009, 5, e1000410. [Google Scholar] [CrossRef]
- Storici, F.; Coglievina, M.; Bruschi, C.V. A 2-micron DNA-based marker recycling system for multiple gene disruption in the yeast Saccharomyces cerevisiae. Yeast 1999, 15, 271–283. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Analysis of genomic DNA by Southern hybridization. In Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Wach, A.; Brachat, A.; Pöhlmann, R.; Philippsen, P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 1994, 10, 1793–1808. [Google Scholar] [CrossRef] [PubMed]
- Rossi, B.; Noel, P.; Bruschi, C.V. Different aneuploidies arise from the same bridge-induced chromosomal translocation event in Saccharomyces cerevisiae. Genetics 2010, 186, 775–790. [Google Scholar] [CrossRef][Green Version]
- Lieberman, H.B. Cell Cycle Checkpoint Control Protocols. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2004. [Google Scholar]
- Rose, M.D.; Novick, P.; Thomas, J.H.; Botstein, D.; Fink, G.R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 1987, 60, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Nyberg, K.A.; Michelson, R.J.; Putnam, C.W.; Weinert, T.A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002, 36, 617–656. [Google Scholar] [CrossRef]
- Branzei, D.; Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell. Biol. 2008, 9, 297–308. [Google Scholar] [CrossRef]
- Symington, L.S. Mechanism and regulation of DNA end resection in Eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 195–212. [Google Scholar] [CrossRef]
- Moore, J.K.; Haber, J.E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in S. cerevisiae. Mol. Cell. Biol. 1996, 16, 2164–2173. [Google Scholar] [CrossRef]
- Aylon, Y.; Liefshitz, B.; Kupiec, M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004, 23, 4868–4875. [Google Scholar] [CrossRef]
- Tosato, V.; Nicolini, C.; Bruschi, C.V. DNA bridging of yeast chromosomes VIII leads to near-reciprocal translocation and loss of heterozygosity with minor cellular defects. Chromosoma 2009, 118, 179–191. [Google Scholar] [CrossRef]
- Tosato, V.; Bruschi, C.V. Per aspera ad astra: When harmful chromosomal translocation become a plus value in genetic evolution. Lessons from S. cerevisiae. Microb. Cell. 2015, 2, 363–375. [Google Scholar] [CrossRef]
- Liu, H.; Liang, F.; Jin, F.; Wang, Y. The coordination of centromere replication, spindle formation and kinetochore microtubule interaction in budding yeast. PLoS Genet. 2008, 4, e1000262. [Google Scholar] [CrossRef]
- Klinner, U.; Schäfer, B. Genetic aspects of targeted insertion mutagenesis in yeast. FEMS Microbiol. Rev. 2004, 28, 201–223. [Google Scholar] [CrossRef][Green Version]
- Gerton, J.L.; DeRisi, J.; Shroff, R.; Lichten, M.; Brown, P.O.; Petes, T.D. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 11383–11390. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Beach, A.; Haber, J.E. Homology requirements and competition between Gene Conversion and Break-Induced Replication during Double-Strand Break Repair. Mol. Cell. 2017, 65, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Deem, A.; Keszthelyi, A.; Blackgrove, T.; Vayl, A.; Coffey, B.; Mathur, R.; Chabes, A.; Malkova, A. Break-induced replication is highly inaccurate. PLoS Biol. 2011, 9, e1000594. [Google Scholar] [CrossRef] [PubMed]
- Donnianni, R.A.; Symington, L.S. Break-induced replication occurs by conservative DNA synthesis. Proc. Natl. Acad. Sci. USA 2013, 110, 13475–13480. [Google Scholar] [CrossRef]
- Ruiz, J.F.; Gómez-González, B.; Aguilera, A. Chromosomal translocations caused by either pol32-dependent or pol32-independent triparental break-induced replication. Mol. Cell. Biol. 2009, 29, 5441–5454. [Google Scholar] [CrossRef] [PubMed]
- Malkova, A.; Naylor, M.L.; Yamaguchi, M.; Ira, G.; Haber, J.E. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol. Cell. Biol. 2005, 25, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Karras, G.I.; Jentsch, S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional behyond S phase. Cell 2010, 141, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Burgers, P.M.; Gerik, K.J. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 1998, 273, 19756–19762. [Google Scholar] [CrossRef]
- Tumini, E.; Barroso, S.; Perez-Calero, C.; Aguilera, A. Roles of human POLD1 and POLD3 in genome stability. Sci. Rep 2016, 6, 38873. [Google Scholar] [CrossRef]
- Dunlop, M.G.; Dobbins, S.E.; Farrington, S.M.; Jones, A.M.; Palles, C.; Whiffin, N.; Tenesa, A.; Spain, S.; Broderick, P.; Ooi, L.Y.; et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat. Genet. 2012, 44, 770–776. [Google Scholar] [CrossRef]
- Huang, M.E.; Le Douarin, B.; Henry, C.; Galibert, F. The Saccharomyces cerevisiae protein YJR043C (Pol32) interacts with the catalytic subunit of DNA polymerase alpha and is required for cell cycle progression in G2/M. Mol. Gen. Genet. 1999, 260, 541–550. [Google Scholar] [CrossRef]
- Jain, S.; Sugawara, N.; Lydeard, J.; Vaze, M.; Le Gac, N.T.; Haber, J.E. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 2009, 23, 291–303. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tosato, V.; Rossi, B.; Sims, J.; Bruschi, C.V. Timing of Chromosome DNA Integration throughout the Yeast Cell Cycle. Biomolecules 2023, 13, 614. https://doi.org/10.3390/biom13040614
Tosato V, Rossi B, Sims J, Bruschi CV. Timing of Chromosome DNA Integration throughout the Yeast Cell Cycle. Biomolecules. 2023; 13(4):614. https://doi.org/10.3390/biom13040614
Chicago/Turabian StyleTosato, Valentina, Beatrice Rossi, Jason Sims, and Carlo V. Bruschi. 2023. "Timing of Chromosome DNA Integration throughout the Yeast Cell Cycle" Biomolecules 13, no. 4: 614. https://doi.org/10.3390/biom13040614
APA StyleTosato, V., Rossi, B., Sims, J., & Bruschi, C. V. (2023). Timing of Chromosome DNA Integration throughout the Yeast Cell Cycle. Biomolecules, 13(4), 614. https://doi.org/10.3390/biom13040614




