Inhibition Analysis and High-Resolution Crystal Structure of Mus musculus Glutathione Transferase P1-1
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Cloning, Expression, and Purification of MmGSTP1-1
2.2.2. Assay of Protein and Enzyme Activity
2.2.3. Pesticides Screening
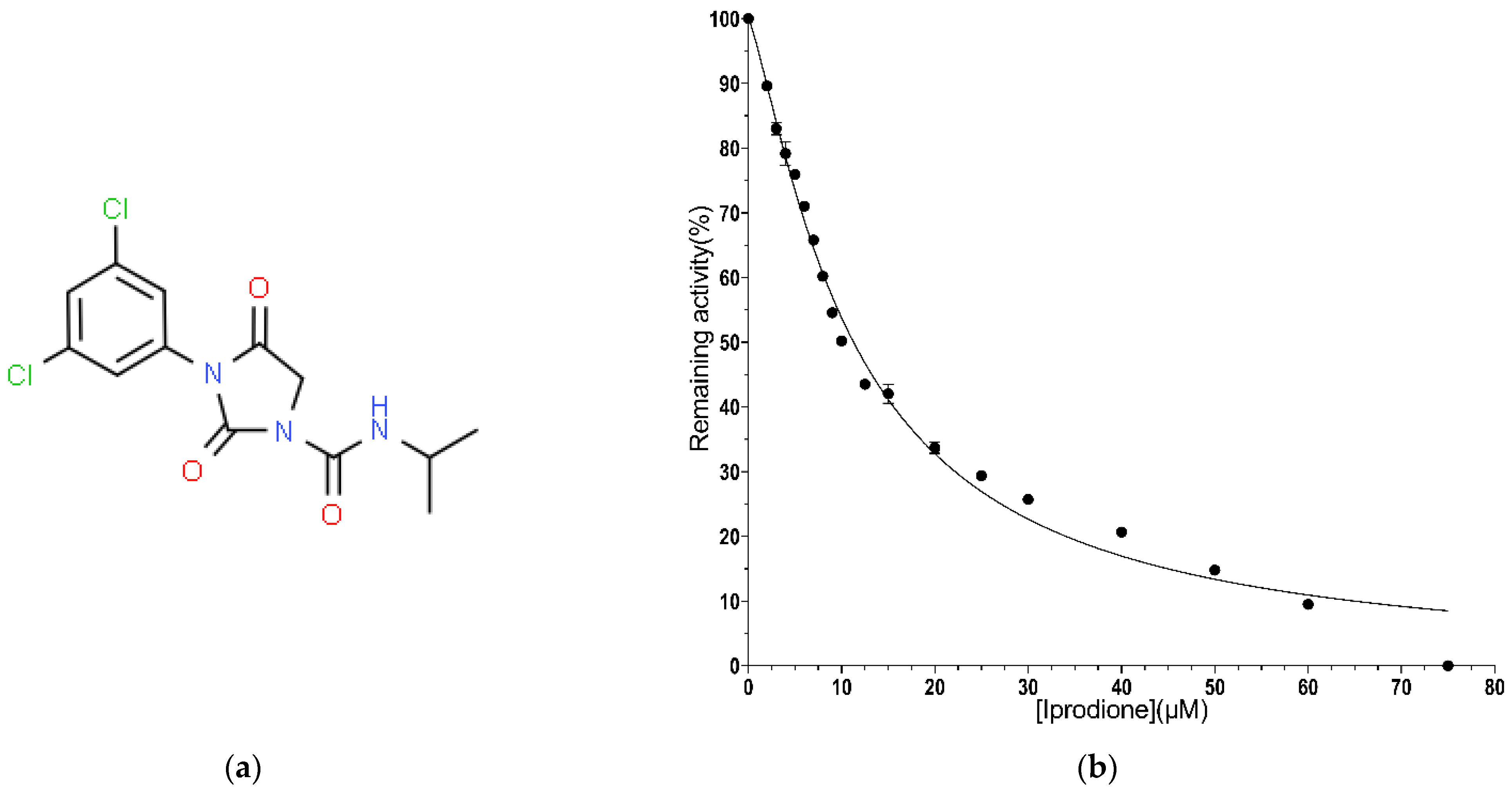
2.2.4. IC50 Value Determination for Iprodione
2.2.5. Kinetic Inhibition Study of MmGSTP1 in Presence of Iprodione
2.2.6. X-ray Crystallography
2.2.7. Molecular Docking of Iprodione
3. Results and Discussion
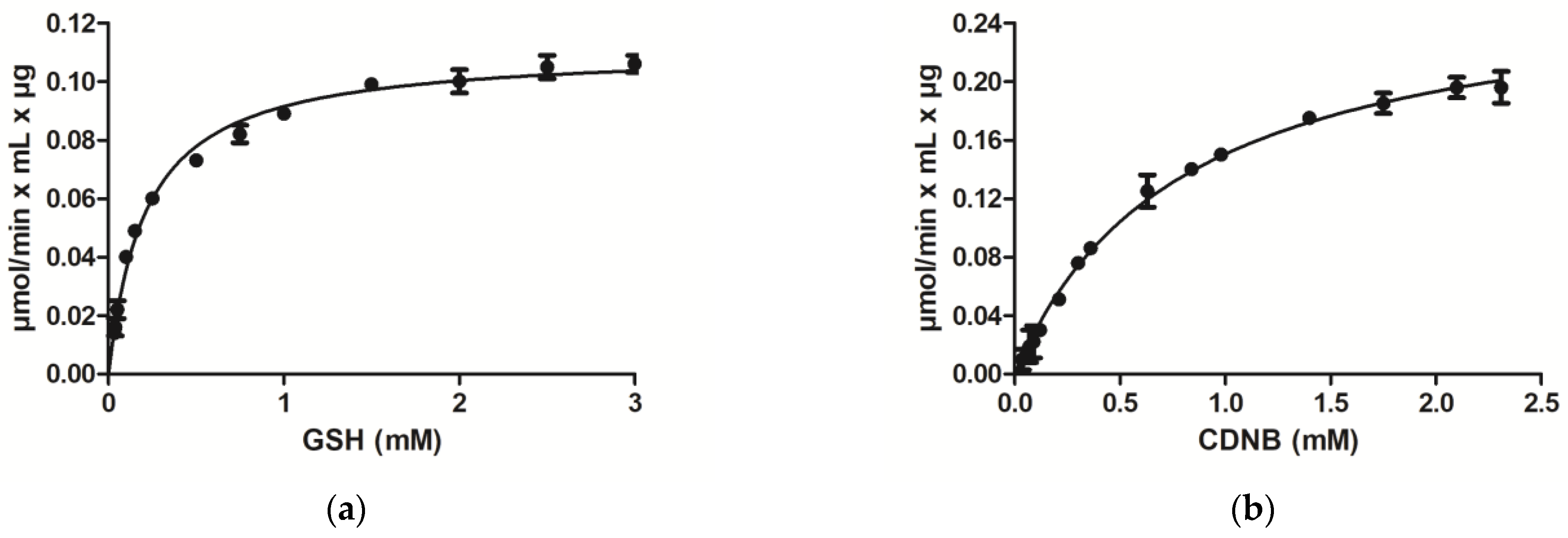
3.1. Purification and Kinetic Analysis
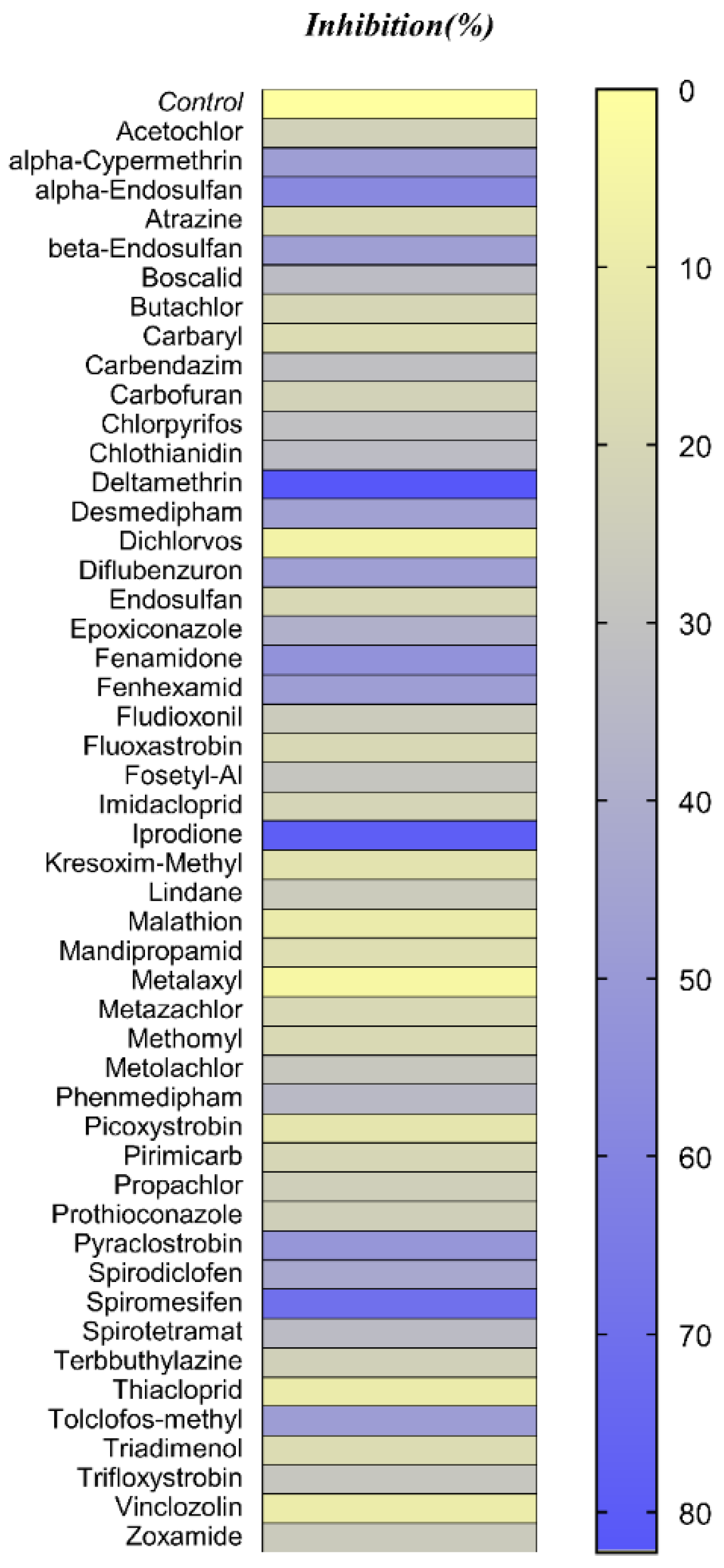
3.2. Pesticides Library Screening
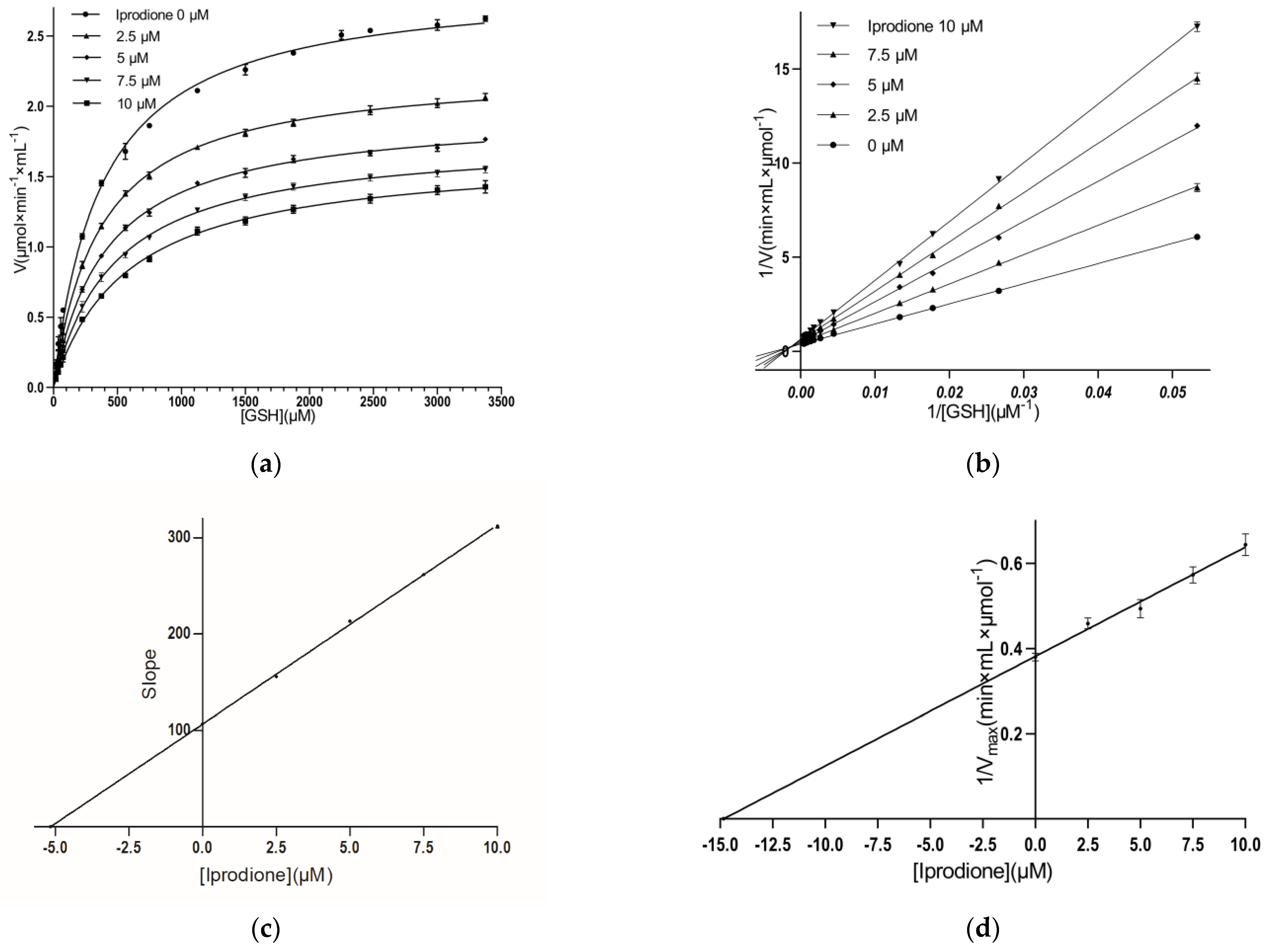
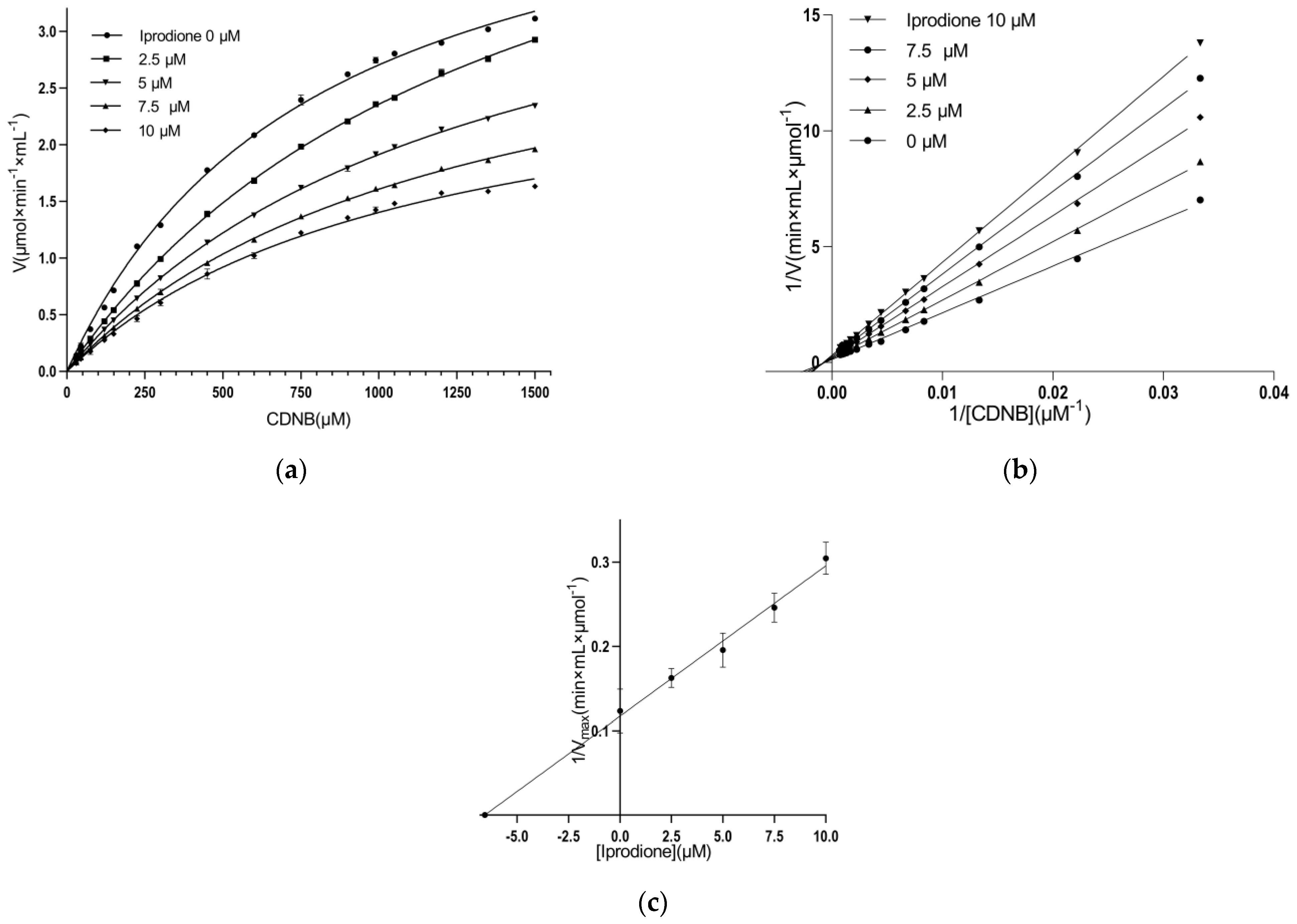
3.3. Kinetic Inhibition Study
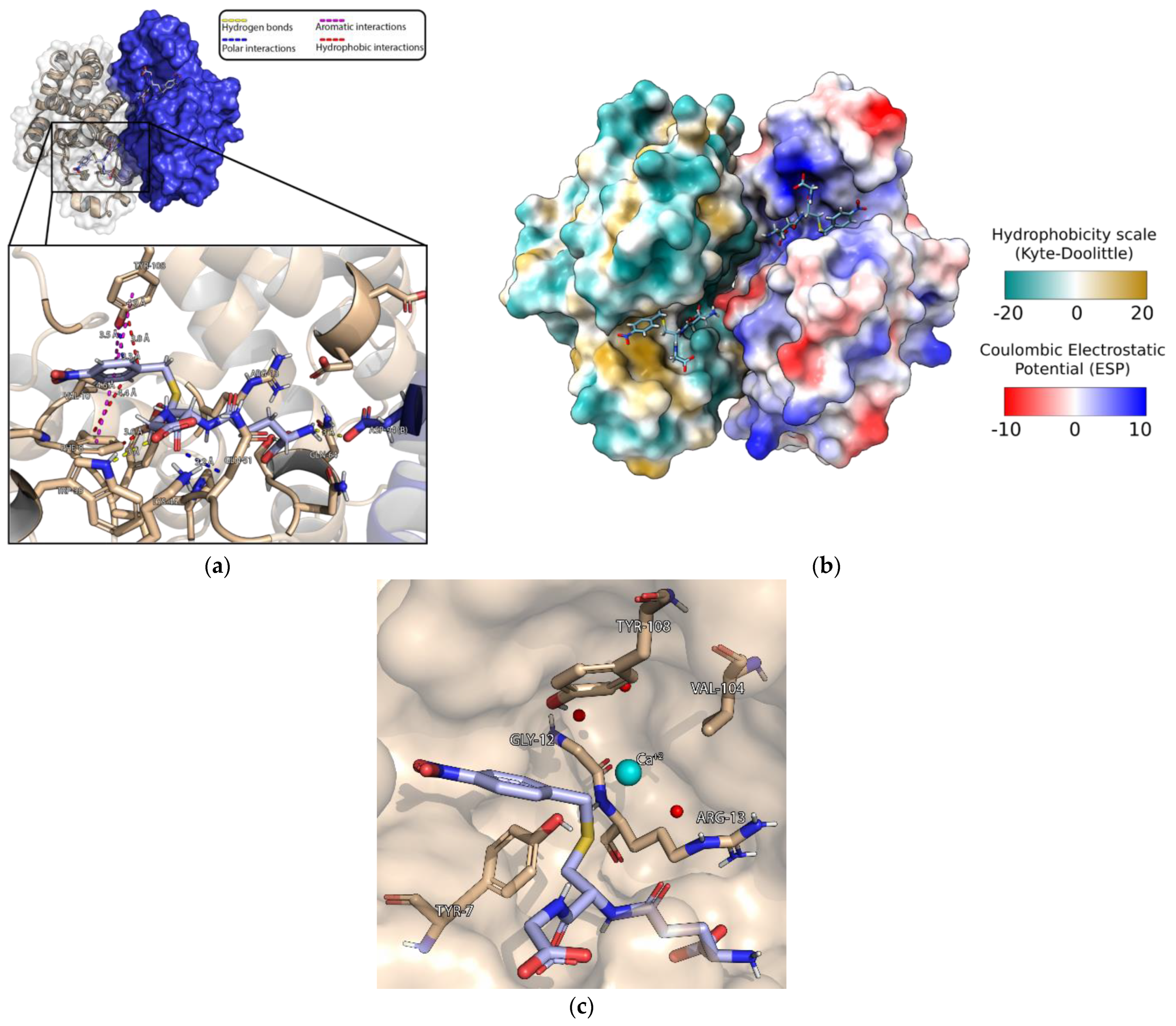
3.4. Crystallization of MmGSTP1-1 and Structural Analysis
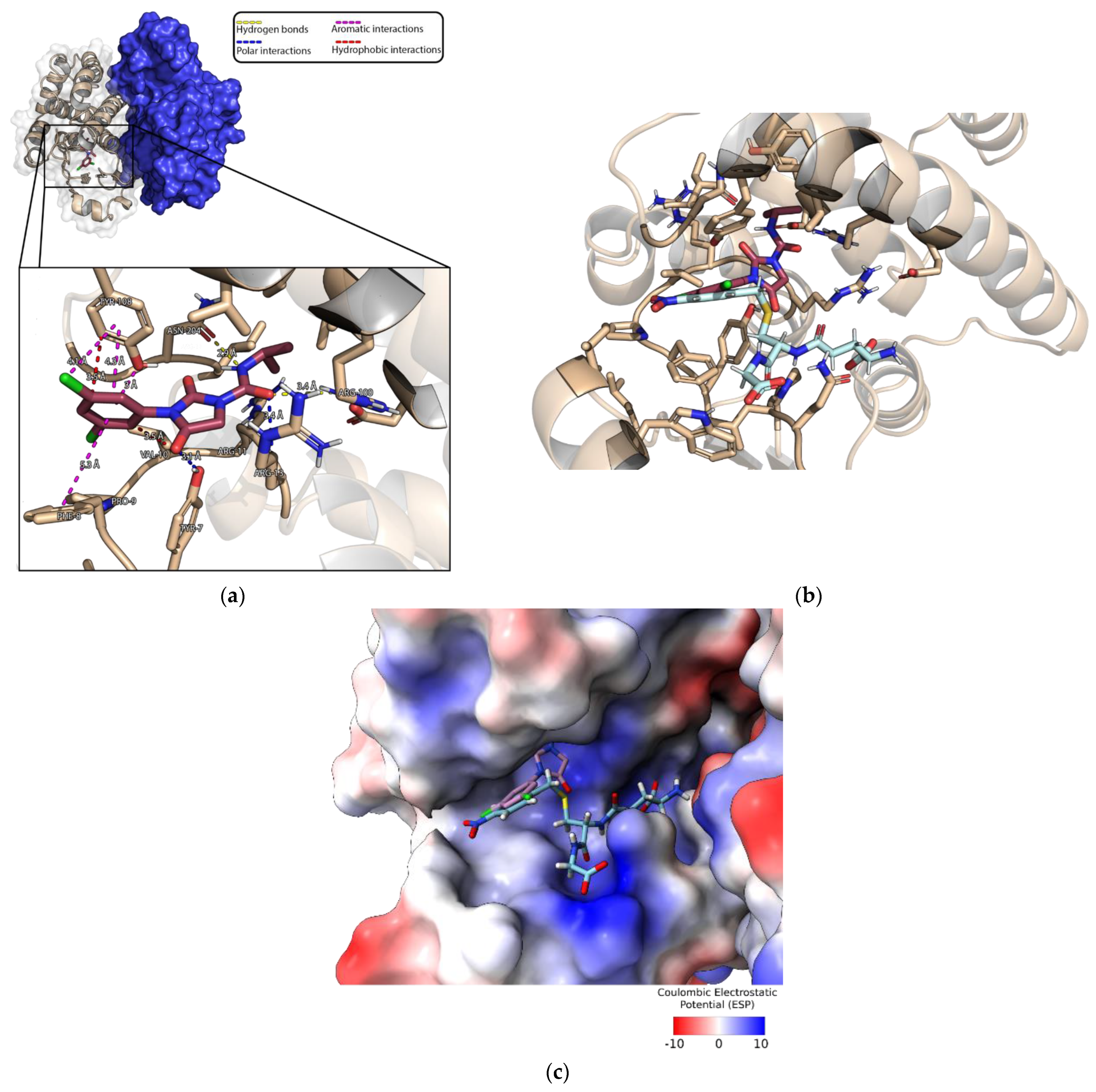
3.5. Characterization of the Iprodione Binding Site Using Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviation
References
- Mannervik, B. Evolution of Glutathione Transferases and Related Enzymes for the Protection of Cells against Electrophiles. Biochem. Soc. Trans. 1996, 24, 878–880. [Google Scholar] [CrossRef] [PubMed]
- Mannervik, B. The Isoenzymes of Glutathione Transferase. In Advances in Enzymology and Related Areas of Molecular Biology; Meister, A., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006; pp. 357–417. ISBN 978-0-470-12303-4. [Google Scholar]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. GLUTATHIONE TRANSFERASES. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef]
- Jakoby, W.B.; Ziegler, D.M. The Enzymes of Detoxication. J. Biol. Chem. 1990, 265, 20715–20718. [Google Scholar] [CrossRef]
- Perperopoulou, F.; Pouliou, F.; Labrou, N.E. Recent Advances in Protein Engineering and Biotechnological Applications of Glutathione Transferases. Crit. Rev. Biotechnol. 2018, 38, 511–528. [Google Scholar] [CrossRef] [PubMed]
- Vašková, J.; Kočan, L.; Vaško, L.; Perjési, P. Glutathione-Related Enzymes and Proteins: A Review. Molecules 2023, 28, 1447. [Google Scholar] [CrossRef] [PubMed]
- Board, P.G.; Menon, D. Glutathione Transferases, Regulators of Cellular Metabolism and Physiology. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3267–3288. [Google Scholar] [CrossRef]
- Oakley, A. Glutathione Transferases: A Structural Perspective. Drug Metab. Rev. 2011, 43, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Mannervik, B.; Berhane, K.; Castro, V.M.; Olin, B.; Ridderström, M.; Vignani, R.; Kozarich, J.W.; Ringborg, U. Glutathione-Linked Enzymes in Normal and Tumor Cells and Their Role in Resistance against Genotoxic Agents. Princess Takamatsu Symp. 1990, 21, 253–262. [Google Scholar]
- Mannervik, B.; Cameron, A.D.; Fernandez, E.; Gustafsson, A.; Hansson, L.O.; Jemth, P.; Jiang, F.; Alwyn Jones, T.; Larsson, A.-K.; Nilsson, L.O.; et al. An Evolutionary Approach to the Design of Glutathione-Linked Enzymes. Chem. Biol. Interact. 1998, 111–112, 15–21. [Google Scholar] [CrossRef]
- Kaur, G.; Gupta, S.K.; Singh, P.; Ali, V.; Kumar, V.; Verma, M. Drug-Metabolizing Enzymes: Role in Drug Resistance in Cancer. Clin. Transl. Oncol. 2020, 22, 1667–1680. [Google Scholar] [CrossRef]
- Pljesa-Ercegovac, M.; Savic-Radojevic, A.; Matic, M.; Coric, V.; Djukic, T.; Radic, T.; Simic, T. Glutathione Transferases: Potential Targets to Overcome Chemoresistance in Solid Tumors. Int. J. Mol. Sci. 2018, 19, 3785. [Google Scholar] [CrossRef] [PubMed]
- Bocedi, A.; Noce, A.; Marrone, G.; Noce, G.; Cattani, G.; Gambardella, G.; Di Lauro, M.; Di Daniele, N.; Ricci, G. Glutathione Transferase P1-1 an Enzyme Useful in Biomedicine and as Biomarker in Clinical Practice and in Environmental Pollution. Nutrients 2019, 11, 1741. [Google Scholar] [CrossRef] [PubMed]
- Laborde, E. Glutathione Transferases as Mediators of Signaling Pathways Involved in Cell Proliferation and Cell Death. Cell Death Differ. 2010, 17, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, G.; Yin, J.; Li, L.; Tan, Y.; Wei, H.; Liu, B.; Deng, L.; Tang, J.; Chen, Y.; et al. GSTP1 and Cancer: Expression, Methylation, Polymorphisms and Signaling (Review). Int. J. Oncol. 2020, 56, 867–878. [Google Scholar] [CrossRef]
- Sawers, L.; Ferguson, M.J.; Ihrig, B.R.; Young, H.C.; Chakravarty, P.; Wolf, C.R.; Smith, G. Glutathione S-Transferase P1 (GSTP1) Directly Influences Platinum Drug Chemosensitivity in Ovarian Tumour Cell Lines. Br. J. Cancer 2014, 111, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Potęga, A. Glutathione-Mediated Conjugation of Anticancer Drugs: An Overview of Reaction Mechanisms and Biological Significance for Drug Detoxification and Bioactivation. Molecules 2022, 27, 5252. [Google Scholar] [CrossRef] [PubMed]
- Dulhunty, A.; Gage, P.; Curtis, S.; Chelvanayagam, G.; Board, P. The Glutathione Transferase Structural Family Includes a Nuclear Chloride Channel and a Ryanodine Receptor Calcium Release Channel Modulator. J. Biol. Chem. 2001, 276, 3319–3323. [Google Scholar] [CrossRef]
- Wang, T.; Arifoglu, P.; Ronai, Z.; Tew, K.D. Glutathione S-Transferase P1–1 (GSTP1–1) Inhibits c-Jun N-Terminal Kinase (JNK1) Signaling through Interaction with the C Terminus. J. Biol. Chem. 2001, 276, 20999–21003. [Google Scholar] [CrossRef] [PubMed]
- Russell, T.M.; Richardson, D.R. The Good Samaritan Glutathione-S-Transferase P1: An Evolving Relationship in Nitric Oxide Metabolism Mediated by the Direct Interactions between Multiple Effector Molecules. Redox Biol. 2023, 59, 102568. [Google Scholar] [CrossRef]
- Kampranis, S.C.; Damianova, R.; Atallah, M.; Toby, G.; Kondi, G.; Tsichlis, P.N.; Makris, A.M. A Novel Plant Glutathione S-Transferase/Peroxidase Suppresses Bax Lethality in Yeast. J. Biol. Chem. 2000, 275, 29207–29216. [Google Scholar] [CrossRef]
- Wu, Y.; Fan, Y.; Xue, B.; Luo, L.; Shen, J.; Zhang, S.; Jiang, Y.; Yin, Z. Human Glutathione S-Transferase P1-1 Interacts with TRAF2 and Regulates TRAF2–ASK1 Signals. Oncogene 2006, 25, 5787–5800. [Google Scholar] [CrossRef]
- Duvoix, A.; Schnekenburger, M.; Delhalle, S.; Blasius, R.; Borde-Chiché, P.; Morceau, F.; Dicato, M.; Diederich, M. Expression of Glutathione S-Transferase P1-1 in Leukemic Cells Is Regulated by Inducible AP-1 Binding. Cancer Lett. 2004, 216, 207–219. [Google Scholar] [CrossRef]
- Georgakis, N.D.; Karagiannopoulos, D.A.; Thireou, T.N.; Eliopoulos, E.E.; Labrou, N.E.; Tsoungas, P.G.; Koutsilieris, M.N.; Clonis, Y.D. Concluding the Trilogy: The Interaction of 2,2′-Dihydroxy-Benzophenones and Their Carbonyl N-Analogues with Human Glutathione Transferase M1-1 Face to Face with the P1-1 and A1-1 Isoenzymes Involved in MDR. Chem. Biol. Drug Des. 2017, 90, 900–908. [Google Scholar] [CrossRef]
- Ertan-Bolelli, T.; Bolelli, K.; Musdal, Y.; Yildiz, I.; Aki-Yalcin, E.; Mannervik, B.; Yalcin, I. Design and Synthesis of 2-Substituted-5-(4-Trifluoromethylphenyl-Sulphonamido)Benzoxazole Derivatives as Human GST P1-1 Inhibitors. Artif. Cells Nanomed. Biotechnol. 2018, 46, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Bartolini, D.; Torquato, P.; Piroddi, M.; Galli, F. Targeting Glutathione S-Transferase P and Its Interactome with Selenium Compounds in Cancer Therapy. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Krasowska, D.; Iraci, N.; Santi, C.; Drabowicz, J.; Cieslak, M.; Kaźmierczak-Barańska, J.; Palomba, M.; Królewska-Golińska, K.; Magiera, J.; Sancineto, L. Diselenides and Benzisoselenazolones as Antiproliferative Agents and Glutathione-S-Transferase Inhibitors. Molecules 2019, 24, 2914. [Google Scholar] [CrossRef]
- Sha, H.; Wang, Z.; Dong, S.; Hu, T.; Liu, S.; Zhang, J.; Wu, Y.; Ma, R.; Wu, J.; Chen, D.; et al. 6-(7-Nitro-2,1,3-Benzoxadiazol-4-Ylthio) Hexanol: A Promising New Anticancer Compound. Biosci. Rep. 2018, 38, BSR20171440. [Google Scholar] [CrossRef] [PubMed]
- Fulci, C.; Rotili, D.; De Luca, A.; Stella, L.; della Rocca, B.M.; Forgione, M.; Di Paolo, V.; Mai, A.; Falconi, M.; Quintieri, L.; et al. A New Nitrobenzoxadiazole-Based GSTP1-1 Inhibitor with a Previously Unheard of Mechanism of Action and High Stability. J. Enzyme Inhib. Med. Chem. 2017, 32, 240–247. [Google Scholar] [CrossRef]
- De Luca, A.; Hartinger, C.G.; Dyson, P.J.; Lo Bello, M.; Casini, A. A New Target for Gold(I) Compounds: Glutathione-S-Transferase Inhibition by Auranofin. J. Inorg. Biochem. 2013, 119, 38–42. [Google Scholar] [CrossRef]
- Harshbarger, W.; Gondi, S.; Ficarro, S.B.; Hunter, J.; Udayakumar, D.; Gurbani, D.; Singer, W.D.; Liu, Y.; Li, L.; Marto, J.A.; et al. Structural and Biochemical Analyses Reveal the Mechanism of Glutathione S-Transferase Pi 1 Inhibition by the Anti-Cancer Compound Piperlongumine. J. Biol. Chem. 2017, 292, 112–120. [Google Scholar] [CrossRef]
- Larasati, Y.A.; Yoneda-Kato, N.; Nakamae, I.; Yokoyama, T.; Meiyanto, E.; Kato, J. Curcumin Targets Multiple Enzymes Involved in the ROS Metabolic Pathway to Suppress Tumor Cell Growth. Sci. Rep. 2018, 8, 2039. [Google Scholar] [CrossRef] [PubMed]
- Pantiora, P.; Furlan, V.; Matiadis, D.; Mavroidi, B.; Perperopoulou, F.; Papageorgiou, A.C.; Sagnou, M.; Bren, U.; Pelecanou, M.; Labrou, N.E. Monocarbonyl Curcumin Analogues as Potent Inhibitors against Human Glutathione Transferase P1-1. Antioxidants 2022, 12, 63. [Google Scholar] [CrossRef] [PubMed]
- Musdal, Y.; Hegazy, U.M.; Aksoy, Y.; Mannervik, B. FDA-Approved Drugs and Other Compounds Tested as Inhibitors of Human Glutathione Transferase P1-1. Chem. Biol. Interact. 2013, 205, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Labrou, N.E.; Mello, L.V.; Clonis, Y.D. Functional and Structural Roles of the Glutathione-Binding Residues in Maize (Zea Mays) Glutathione S-Transferase I. Biochem. J. 2001, 358, 101–110. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser Crystallographic Software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.-W.; Jain, S.; McCoy, A.J.; et al. Macromolecular Structure Determination Using X-Rays, Neutrons and Electrons: Recent Developments in Phenix. Acta Crystallogr. Sect. Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, Version 1.8; Schrödinger, LLC: New York, NY, USA, 2015.
- Zimniak, P.; Nanduri, B.; Pikula, S.; Bandorowicz-Pikula, J.; Singhal, S.S.; Srivastava, S.K.; Awasthi, S.; Awasthi, Y.C. Naturally Occurring Human Glutathione S-Transferase GSTP1-1 Isoforms with Isoleucine and Valine in Position 104 Differ in Enzymic Properties. Eur. J. Biochem. 1994, 224, 893–899. [Google Scholar] [CrossRef]
- Johansson, A.-S.; Stenberg, G.; Widersten, M.; Mannervik, B. Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governed by the H-Site Residue 105. J. Mol. Biol. 1998, 278, 687–698. [Google Scholar] [CrossRef]
- Hegazy, U.M.; Mannervik, B.; Stenberg, G. Functional Role of the Lock and Key Motif at the Subunit Interface of Glutathione Transferase P1-1. J. Biol. Chem. 2004, 279, 9586–9596. [Google Scholar] [CrossRef]
- Labrou, N.E.; Papageorgiou, A.C.; Pavli, O.; Flemetakis, E. Plant GSTome: Structure and Functional Role in Xenome Network and Plant Stress Response. Curr. Opin. Biotechnol. 2015, 32, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Kolm, R.H.; Danielson, U.H.; Zhang, Y.; Talalay, P.; Mannervik, B. Isothiocyanates as Substrates for Human Glutathione Transferases: Structure-Activity Studies. Biochem. J. 1995, 311, 453–459. [Google Scholar] [CrossRef] [PubMed]
- García-Sáez, I.; Párraga, A.; Phillips, M.F.; Mantle, T.J.; Coll, M. Molecular Structure at 1.8 A of Mouse Liver Class Pi Glutathione S-Transferase Complexed with S-(p-Nitrobenzyl)Glutathione and Other Inhibitors. J. Mol. Biol. 1994, 237, 298–314. [Google Scholar] [CrossRef]
- Jubb, H.C.; Higueruelo, A.P.; Ochoa-Montaño, B.; Pitt, W.R.; Ascher, D.B.; Blundell, T.L. Arpeggio: A Web Server for Calculating and Visualising Interatomic Interactions in Protein Structures. J. Mol. Biol. 2017, 429, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera: A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Yang, Z.; Lasker, K.; Schneidman-Duhovny, D.; Webb, B.; Huang, C.C.; Pettersen, E.F.; Goddard, T.D.; Meng, E.C.; Sali, A.; Ferrin, T.E. UCSF Chimera, MODELLER, and IMP: An Integrated Modeling System. J. Struct. Biol. 2012, 179, 269–278. [Google Scholar] [CrossRef]
- Axarli, I.; Labrou, N.; Petrou, C.; Rassias, N.; Cordopatis, P.; Clonis, Y. Sulphonamide-based bombesin prodrug analogues for glutathione transferase, useful in targeted cancer chemotherapy. Eur. J. Med. Chem. 2009, 44, 2009–2016. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
| Electrophile Substrates | U/mg (%) |
|---|---|
| 1-Chloro-2,4-dinitrobenzene | 100 |
| 1-Bromo-2,4-dinitrobenzene | 188.8 |
| 1-Iodo-2,4-dinitrobenzene | 14.4 |
| 4-Chloro-7-nitrobenzofurazan | - |
| p-Nitrobenzyl chloride | 1.7 |
| Bromosulfalein | 8.7 |
| Cumene hydroperoxide | 2.7 |
| tert-Butyl hydroperoxide | - |
| Dehydroascorbate | - |
| Sulphanilamide | 0.2 |
| 2,3-Dichloro-4-[2-methylene-butyryl]phenoxy)acetic acid (Ethacrynic acid) | 19.1 |
| trans-4-Phenyl-3-buten-2-one | - |
| Allyl isothiocyanate | - |
| Phenethyl isothiocyanate | 41.1 |
| Data Collection | MmGSTP1-1 |
|---|---|
| Beamline | P13 (EMBL, Hamburg) |
| Wavelength (Å) | 1.033 |
| Resolution (Å) | 56.62–1.28 (1.30–1.28) |
| Space group | P212121 |
| Unit cell (Å) a, b, c | 56.62, 77.37, 101.44 |
| No. of unique reflections | 114,851 (5532) |
| Completeness (%) | 99.7 (98.3) |
| Multiplicity | 6.4 (6.3) |
| Mosaicity (°) | 0.11 |
| Rmeas | 0.059 (2.485) |
| CC1/2 | 0.99 (0.35) |
| Mean (I/σ(I)) | 12.7 (0.9) |
| Wilson B-factor (Å2) | 18.3 |
| Refinement | |
| No. of reflections used | 114,744 |
| Rwork/Rfree | 0.175/0.199 |
| No. of non-H atoms (protein/ligand/solvent) | 3338/143/602 |
| Protein residues | 418 |
| RMSD in bonds (Å) | 0.006 |
| RMSD in angles (°) | 0.93 |
| Average B-factor (all/protein/ligands/solvent) (Å2) | 28.2/26.6/28.8/36.8 |
| Ramachandran favored/outliers (%) | 97.8/0.0 |
| Rotamer outliers (%) | 0.0 |
| Clashscore | 3.5 |
| PDB ID | 8C5D |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kupreienko, O.; Pouliou, F.; Konstandinidis, K.; Axarli, I.; Douni, E.; Papageorgiou, A.C.; Labrou, N.E. Inhibition Analysis and High-Resolution Crystal Structure of Mus musculus Glutathione Transferase P1-1. Biomolecules 2023, 13, 613. https://doi.org/10.3390/biom13040613
Kupreienko O, Pouliou F, Konstandinidis K, Axarli I, Douni E, Papageorgiou AC, Labrou NE. Inhibition Analysis and High-Resolution Crystal Structure of Mus musculus Glutathione Transferase P1-1. Biomolecules. 2023; 13(4):613. https://doi.org/10.3390/biom13040613
Chicago/Turabian StyleKupreienko, Oleksii, Fotini Pouliou, Konstantinos Konstandinidis, Irene Axarli, Eleni Douni, Anastassios C. Papageorgiou, and Nikolaos E. Labrou. 2023. "Inhibition Analysis and High-Resolution Crystal Structure of Mus musculus Glutathione Transferase P1-1" Biomolecules 13, no. 4: 613. https://doi.org/10.3390/biom13040613
APA StyleKupreienko, O., Pouliou, F., Konstandinidis, K., Axarli, I., Douni, E., Papageorgiou, A. C., & Labrou, N. E. (2023). Inhibition Analysis and High-Resolution Crystal Structure of Mus musculus Glutathione Transferase P1-1. Biomolecules, 13(4), 613. https://doi.org/10.3390/biom13040613









