Minimizing the Anticodon-Recognized Loop of Methanococcus jannaschii Tyrosyl-tRNA Synthetase to Improve the Efficiency of Incorporating Noncanonical Amino Acids
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bacterial Strains
2.3. Plasmids Construction
2.4. Expression of Orthogonal eGFP in E. coli and Fluorescence Quantification
2.5. Native PAGE Gel Electrophoresis
3. Results
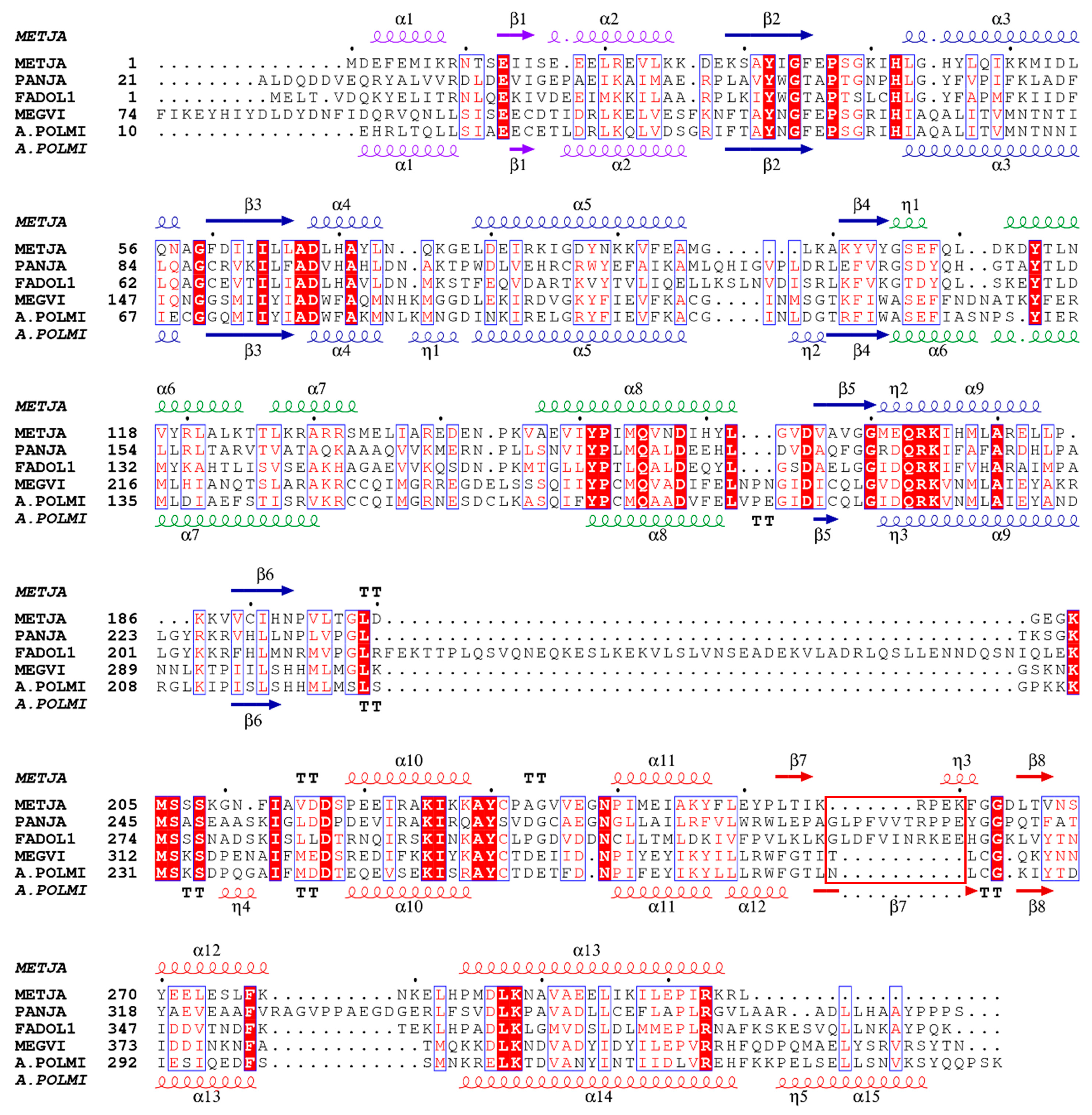
3.1. Gene mining of Giant Viruses
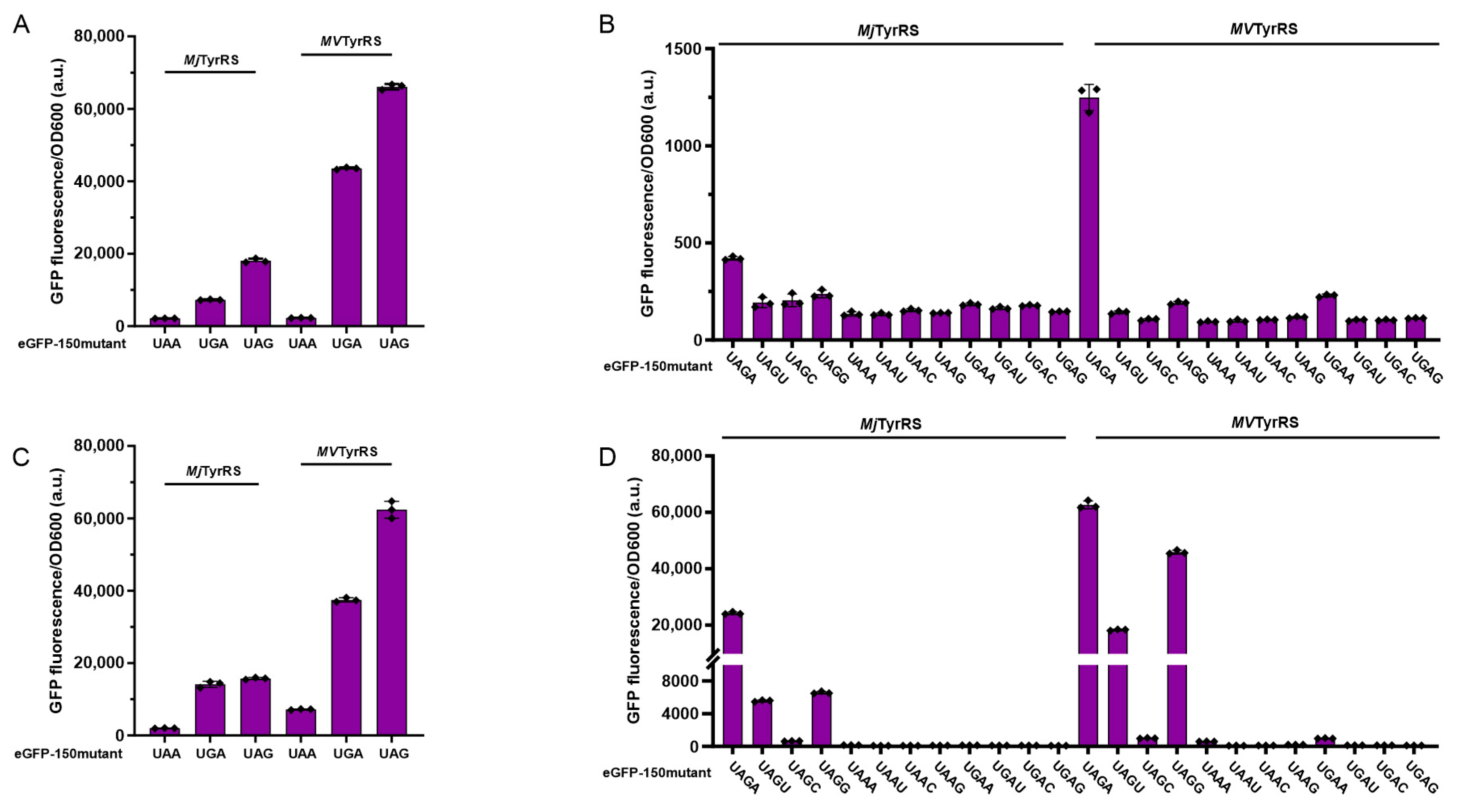
3.2. Activity Assay of Mimivirus TyrRS in Response to Triplet Codons
3.3. Activity Assay of Mimivirus TyrRS in Response to Quadruplet Codons
3.4. Crystal Structure Comparison of MVTyrRS
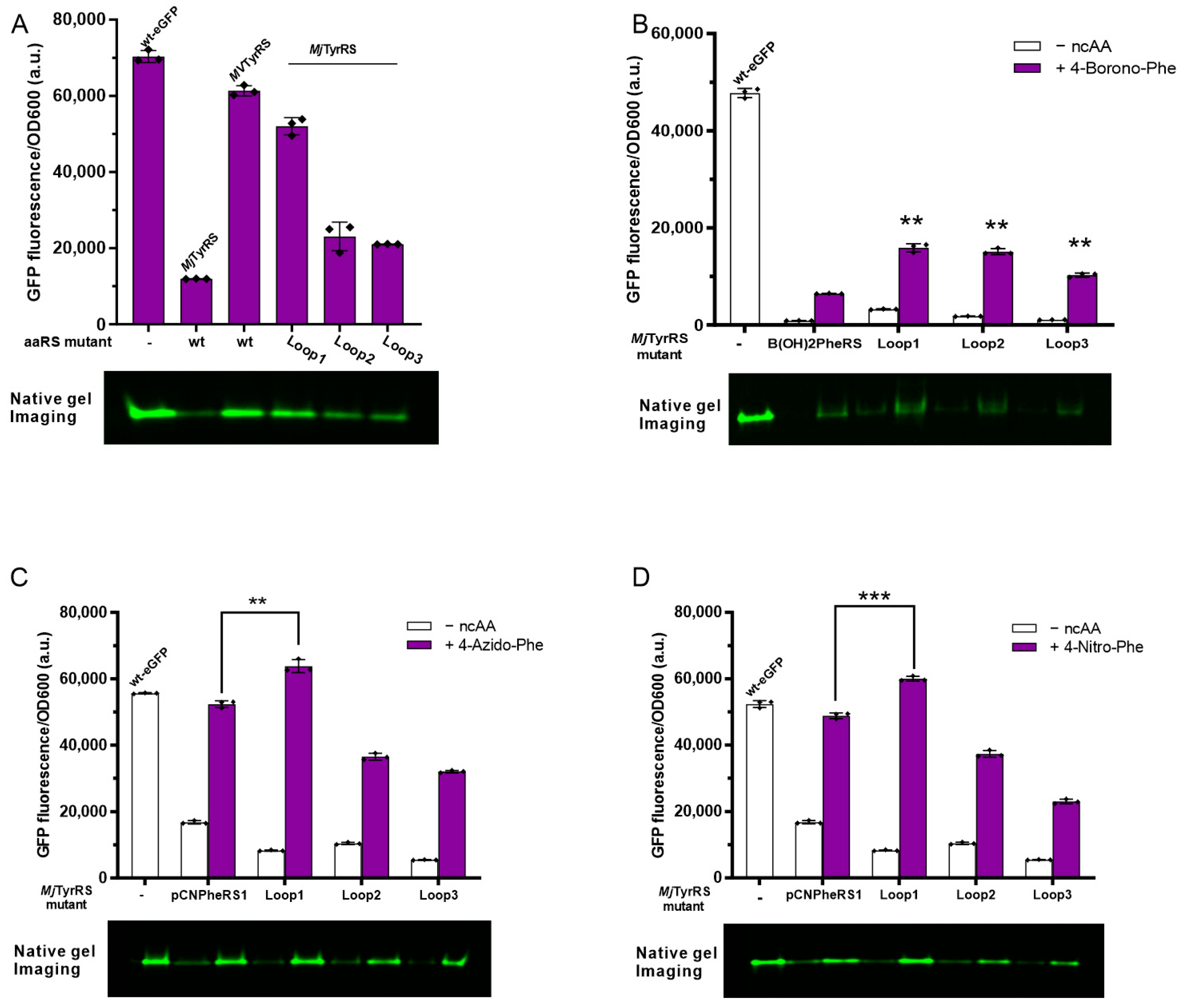
3.5. Activity Assay of Loop Minimized Wild-Type MjTyrRS
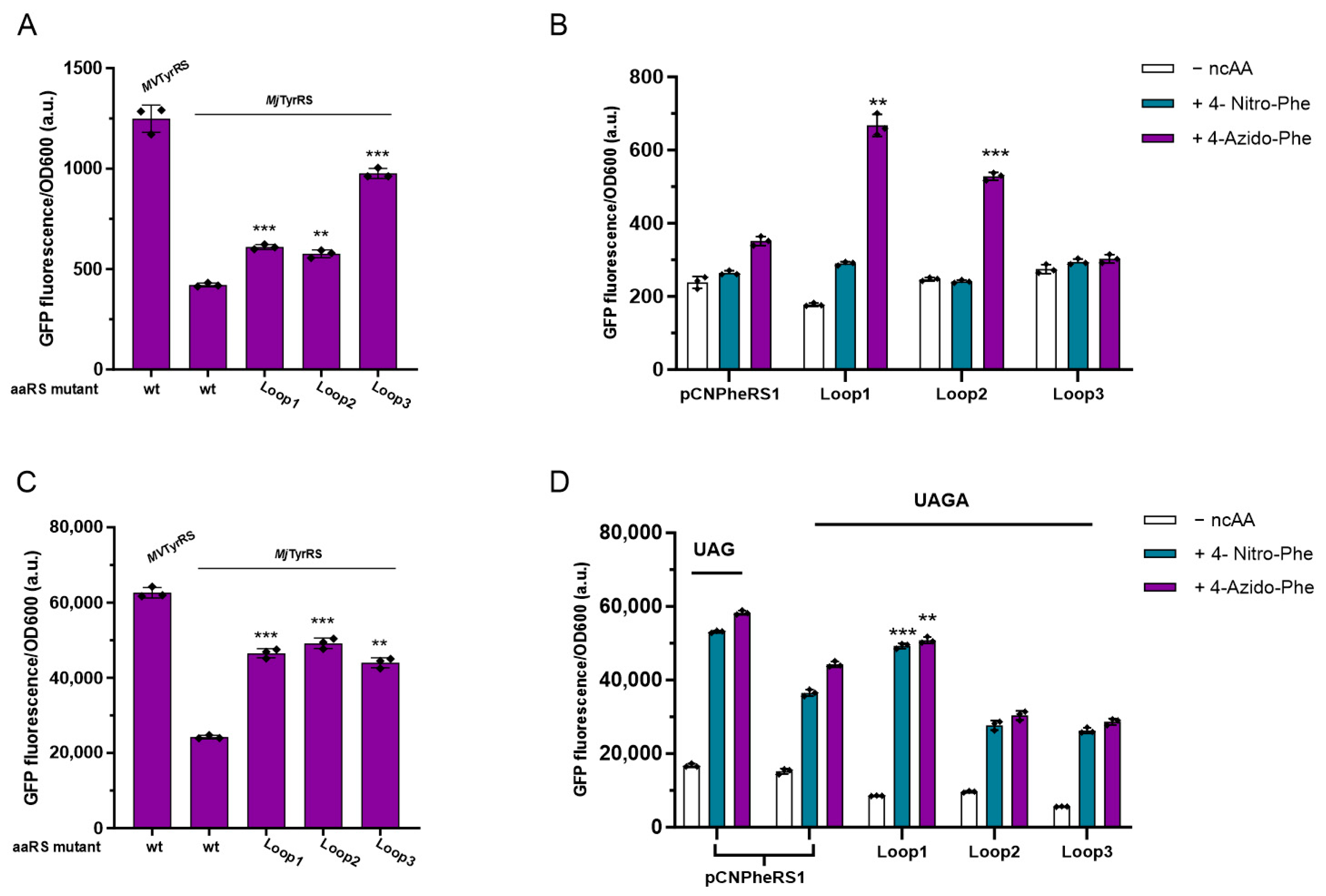
3.6. Activity Assay of Loop Minimized MjTyrRS Variants
3.7. Quadruplet Codons Suppression of Loop Minimization MjTyrRS Variants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hohsaka, T.; Sisido, M. Incorporation of non-natural amino acids into proteins. Curr. Opin. Chem. Biol. 2002, 6, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Schultz, P.G. Synthesis at the interface of chemistry and biology. J. Am. Chem Soc. 2009, 131, 12497–12515. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Jia, H.; Ramirez-Diaz, D.A.; Budisa, N.; Schwille, P. Fine-Tuning Protein Self-Organization by Orthogonal Chemo-Optogenetic Tools. Angew. Chem. Int. Ed. Engl. 2021, 60, 4501–4506. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liu, X.; Wang, J. Expansion of Redox Chemistry in Designer Metalloenzymes. Acc. Chem. Res. 2019, 52, 557–565. [Google Scholar] [CrossRef]
- Hauf, M.; Richter, F.; Schneider, T.; Faidt, T.; Martins, B.M.; Baumann, T.; Durkin, P.; Dobbek, H.; Jacobs, K.; Möglich, A.; et al. Photoactivatable Mussel-Based Underwater Adhesive Proteins by an Expanded Genetic Code. Chembiochem 2017, 18, 1819–1823. [Google Scholar] [CrossRef]
- Zhang, F.; Zhou, Q.; Yang, G.; An, L.; Li, F.; Wang, J. A genetically encoded (19)F NMR probe for lysine acetylation. Chem. Commun. 2018, 54, 3879–3882. [Google Scholar] [CrossRef]
- Völler, J.S.; Biava, H.; Hildebrandt, P.; Budisa, N. An expanded genetic code for probing the role of electrostatics in enzyme catalysis by vibrational Stark spectroscopy. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 3053–3059. [Google Scholar] [CrossRef]
- Wang, T.; Zhou, Q.; Li, F.; Yu, Y.; Yin, X.; Wang, J. Genetic Incorporation of N(ε)-Formyllysine, a New Histone Post-translational Modification. Chembiochem A Eur. J. Chem. Biol. 2015, 16, 1440–1442. [Google Scholar] [CrossRef]
- Ahmed, N.; De Graaf, J.F.; Ahmed, N.; Foss, D.V.; Delcorde, J.; Schultz, P.G.; Pezacki, J.P. Visualization of the Delivery and Release of Small RNAs Using Genetic Code Expansion and Unnatural RNA-Binding Proteins. Bioconjug. Chem. 2018, 29, 3982–3986. [Google Scholar] [CrossRef]
- Hallam, T.J.; Smider, V.V. Unnatural amino acids in novel antibody conjugates. Future Med. Chem. 2014, 6, 1309–1324. [Google Scholar] [CrossRef]
- Krebs, S.K.; Rakotoarinoro, N.; Stech, M.; Zemella, A.; Kubick, S. A CHO-Based Cell-Free Dual Fluorescence Reporter System for the Straightforward Assessment of Amber Suppression and scFv Functionality. Front. Bioeng. Biotechnol. 2022, 10, 873906. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yu, G.; Huang, Y.; Cheng, W.; Li, Y.; Sun, Y.; Ye, H.; Liu, T. Genetic-code-expanded cell-based therapy for treating diabetes in mice. Nat. Chem. Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Shi, N.; Yang, Q.; Zhang, H.; Lu, J.; Lin, H.; Yang, X.; Abulimiti, A.; Cheng, J.; Wang, Y.; Tong, L.; et al. Restoration of dystrophin expression in mice by suppressing a nonsense mutation through the incorporation of unnatural amino acids. Nat. Biomed. Eng. 2021. [Google Scholar] [CrossRef] [PubMed]
- Parrish, A.R.; She, X.; Xiang, Z.; Coin, I.; Shen, Z.; Briggs, S.P.; Dillin, A.; Wang, L. Expanding the genetic code of Caenorhabditis elegans using bacterial aminoacyl-tRNA synthetase/tRNA pairs. ACS Chem. Biol. 2012, 7, 1292–1302. [Google Scholar] [CrossRef]
- Elliott, T.S.; Townsley, F.M.; Bianco, A.; Ernst, R.J.; Sachdeva, A.; Elsasser, S.J.; Davis, L.; Lang, K.; Pisa, R.; Greiss, S.; et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat. Biotechnol. 2014, 32, 465–472. [Google Scholar] [CrossRef]
- Krogager, T.P.; Ernst, R.J.; Elliott, T.S.; Calo, L.; Beránek, V.; Ciabatti, E.; Spillantini, M.G.; Tripodi, M.; Hastings, M.H.; Chin, J.W. Labeling and identifying cell-specific proteomes in the mouse brain. Nat. Biotechnol. 2018, 36, 156–159. [Google Scholar] [CrossRef]
- Ding, W.; Zhao, H.; Chen, Y.; Zhang, B.; Yang, Y.; Zang, J.; Wu, J.; Lin, S. Chimeric design of pyrrolysyl-tRNA synthetase/tRNA pairs and canonical synthetase/tRNA pairs for genetic code expansion. Nat. Commun. 2020, 11, 3154. [Google Scholar] [CrossRef] [PubMed]
- DeBenedictis, E.A.; Carver, G.D.; Chung, C.Z.; Söll, D.; Badran, A.H. Multiplex suppression of four quadruplet codons via tRNA directed evolution. Nat. Commun. 2021, 12, 5706. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.; Schultz, P.G. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat. Methods 2006, 3, 263–265. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nureki, O.; Ishitani, R.; Yaremchuk, A.; Tukalo, M.; Cusack, S.; Sakamoto, K.; Yokoyama, S. Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nat. Struct. Biol. 2003, 10, 425–432. [Google Scholar] [CrossRef]
- Amiram, M.; Haimovich, A.D.; Fan, C.; Wang, Y.-S.; Aerni, H.-R.; Ntai, I.; Moonan, D.W.; Ma, N.J.; Rovner, A.J.; Hong, S.H.; et al. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat. Biotechnol. 2015, 33, 1272–1279. [Google Scholar] [CrossRef] [PubMed]
- Young, T.S.; Ahmad, I.; Yin, J.A.; Schultz, P.G. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol 2010, 395, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Xu, J.; Shen, Z.; Takimoto, J.K.; Schultz, M.D.; Schmitz, R.J.; Xiang, Z.; Ecker, J.R.; Briggs, S.P.; Wang, L. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat. Chem. Biol. 2011, 7, 779–786. [Google Scholar] [CrossRef]
- Johnson, D.B.; Wang, C.; Xu, J.; Schultz, M.D.; Schmitz, R.J.; Ecker, J.R.; Wang, L. Release factor one is nonessential in Escherichia coli. ACS Chem. Biol. 2012, 7, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Wu, I.L.; Patterson, M.A.; Carpenter Desai, H.E.; Mehl, R.A.; Giorgi, G.; Conticello, V.P. Multiple site-selective insertions of noncanonical amino acids into sequence-repetitive polypeptides. Chembiochem 2013, 14, 968–978. [Google Scholar] [CrossRef]
- Heinemann, I.U.; Rovner, A.J.; Aerni, H.R.; Rogulina, S.; Cheng, L.; Olds, W.; Fischer, J.T.; Söll, D.; Isaacs, F.J.; Rinehart, J. Enhanced phosphoserine insertion during Escherichia coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion. FEBS Lett. 2012, 586, 3716–3722. [Google Scholar] [CrossRef] [PubMed]
- Ohtake, K.; Sato, A.; Mukai, T.; Hino, N.; Yokoyama, S.; Sakamoto, K. Efficient decoding of the UAG triplet as a full-fledged sense codon enhances the growth of a prfA-deficient strain of Escherichia coli. J. Bacteriol. 2012, 194, 2606–2613. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, M.J.; Rovner, A.J.; Goodman, D.B.; Aerni, H.R.; Haimovich, A.D.; Kuznetsov, G.; Mercer, J.A.; Wang, H.H.; Carr, P.A.; Mosberg, J.A.; et al. Genomically recoded organisms expand biological functions. Science 2013, 342, 357–360. [Google Scholar] [CrossRef]
- Pósfai, G.; Plunkett, G., 3rd; Fehér, T.; Frisch, D.; Keil, G.M.; Umenhoffer, K.; Kolisnychenko, V.; Stahl, B.; Sharma, S.S.; de Arruda, M.; et al. Emergent properties of reduced-genome Escherichia coli. Science 2006, 312, 1044–1046. [Google Scholar] [CrossRef]
- Wang, K.; Neumann, H.; Peak-Chew, S.Y.; Chin, J.W. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 2007, 25, 770–777. [Google Scholar] [CrossRef]
- Al-Shayeb, B.; Sachdeva, R.; Chen, L.-X.; Ward, F.; Munk, P.; Devoto, A.; Castelle, C.J.; Olm, M.R.; Bouma-Gregson, K.; Amano, Y.; et al. Clades of huge phages from across Earth’s ecosystems. Nature 2020, 578, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Abergel, C.; Rudinger-Thirion, J.; Giegé, R.; Claverie, J.M. Virus-encoded aminoacyl-tRNA synthetases: Structural and functional characterization of mimivirus TyrRS and MetRS. J. Virol. 2007, 81, 12406–12417. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Iraha, F.; Ohtake, K.; Sakamoto, K. Pyrrolysyl-tRNA Synthetase with a Unique Architecture Enhances the Availability of Lysine Derivatives in Synthetic Genetic Codes. Molecules 2018, 23, 2460. [Google Scholar] [CrossRef] [PubMed]
- Koch, N.G.; Goettig, P.; Rappsilber, J.; Budisa, N. Engineering Pyrrolysyl-tRNA Synthetase for the Incorporation of Non-Canonical Amino Acids with Smaller Side Chains. Int. J. Mol. Sci. 2021, 22, 11194. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.T.; Wang, Y.S.; Nakamura, A.; Eiler, D.; Kavran, J.M.; Wong, M.; Kiessling, L.L.; Steitz, T.A.; O’Donoghue, P.; Soll, D. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc. Natl. Acad. Sci. USA 2014, 111, 16724–16729. [Google Scholar] [CrossRef]
- Liu, L.; Wang, B.; Li, S.; Xu, F.; He, Q.; Pan, C.; Gao, X.; Yao, W.; Song, X. Convenient Genetic Encoding of Phenylalanine Derivatives through Their alpha-Keto Acid Precursors. Biomolecules 2021, 11, 1358. [Google Scholar] [CrossRef]
- Brustad, E.; Bushey, M.L.; Lee, J.W.; Groff, D.; Liu, W.; Schultz, P.G. A genetically encoded boronate-containing amino acid. Angew. Chem. Int Ed. Engl. 2008, 47, 8220–8223. [Google Scholar] [CrossRef]
- Schultz, K.C.; Supekova, L.; Ryu, Y.; Xie, J.; Perera, R.; Schultz, P.G. A genetically encoded infrared probe. J. Am. Chem. Soc. 2006, 128, 13984–13985. [Google Scholar] [CrossRef]
- Nasir, A.; Kim, K.M.; Caetano-Anolles, G. Giant viruses coexisted with the cellular ancestors and represent a distinct supergroup along with superkingdoms Archaea, Bacteria and Eukarya. BMC Evol. Biol. 2012, 12, 156. [Google Scholar] [CrossRef]
- Yamada, T. Giant viruses in the environment: Their origins and evolution. Curr. Opin. Virol. 2011, 1, 58–62. [Google Scholar] [CrossRef]
- Yutin, N.; Wolf, Y.I.; Koonin, E.V. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology 2014, 466–467, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Lajoie, M.J.; Xiao, H.; Church, G.M.; Schultz, P.G. A bacterial strain with a unique quadruplet codon specifying non-native amino acids. Chembiochem 2014, 15, 1782–1786. [Google Scholar] [CrossRef] [PubMed]
- Steer, B.A.; Schimmel, P. Domain-domain communication in a miniature archaebacterial tRNA synthetase. Proc. Natl. Acad. Sci. USA 1999, 96, 13644–13649. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Ju, T.; Niu, W.; Guo, J. Fine-tuning interaction between aminoacyl-tRNA synthetase and tRNA for efficient synthesis of proteins containing unnatural amino acids. ACS Synth. Biol. 2015, 4, 207–212. [Google Scholar] [CrossRef]
- Wang, L.; Brock, A.; Herberich, B.; Schultz, P.G. Expanding the genetic code of Escherichia coli. Science 2001, 292, 498–500. [Google Scholar] [CrossRef]
- Chin, J.W.; Santoro, S.W.; Martin, A.B.; King, D.S.; Wang, L.; Schultz, P.G. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc. 2002, 124, 9026–9027. [Google Scholar] [CrossRef]
- Xie, J.; Wang, L.; Wu, N.; Brock, A.; Spraggon, G.; Schultz, P.G. The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination. Nat. Biotechnol. 2004, 22, 1297–1301. [Google Scholar] [CrossRef]
- Cooley, R.B.; Feldman, J.L.; Driggers, C.M.; Bundy, T.A.; Stokes, A.L.; Karplus, P.A.; Mehl, R.A. Structural basis of improved second-generation 3-nitro-tyrosine tRNA synthetases. Biochemistry 2014, 53, 1916–1924. [Google Scholar] [CrossRef]
- Fechter, P.; Rudinger, J.; Giege, R.; Theobald-Dietrich, A. Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: Production and activity. FEBS Lett. 1998, 436, 99–103. [Google Scholar] [CrossRef]
- Fechter, P.; Rudinger-Thirion, J.; Tukalo, M.; Giegé, R. Major tyrosine identity determinants in Methanococcus jannaschii and Saccharomyces cerevisiae tRNA(Tyr) are conserved but expressed differently. Eur. J. Biochem. 2001, 268, 761–767. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Z.; Liang, J.; Su, T.; Zhang, D.; Li, H.; Gao, X.; Yao, W.; Song, X. Minimizing the Anticodon-Recognized Loop of Methanococcus jannaschii Tyrosyl-tRNA Synthetase to Improve the Efficiency of Incorporating Noncanonical Amino Acids. Biomolecules 2023, 13, 610. https://doi.org/10.3390/biom13040610
Hu Z, Liang J, Su T, Zhang D, Li H, Gao X, Yao W, Song X. Minimizing the Anticodon-Recognized Loop of Methanococcus jannaschii Tyrosyl-tRNA Synthetase to Improve the Efficiency of Incorporating Noncanonical Amino Acids. Biomolecules. 2023; 13(4):610. https://doi.org/10.3390/biom13040610
Chicago/Turabian StyleHu, Zhiyang, Jinming Liang, Taogeng Su, Di Zhang, Hao Li, Xiangdong Gao, Wenbin Yao, and Xiaoda Song. 2023. "Minimizing the Anticodon-Recognized Loop of Methanococcus jannaschii Tyrosyl-tRNA Synthetase to Improve the Efficiency of Incorporating Noncanonical Amino Acids" Biomolecules 13, no. 4: 610. https://doi.org/10.3390/biom13040610
APA StyleHu, Z., Liang, J., Su, T., Zhang, D., Li, H., Gao, X., Yao, W., & Song, X. (2023). Minimizing the Anticodon-Recognized Loop of Methanococcus jannaschii Tyrosyl-tRNA Synthetase to Improve the Efficiency of Incorporating Noncanonical Amino Acids. Biomolecules, 13(4), 610. https://doi.org/10.3390/biom13040610




